Abstract
Pseudomonas syringae is an important phyllosphere colonist that utilizes flagellum-mediated motility both as a means to explore leaf surfaces, as well as to invade into leaf interiors, where it survives as a pathogen. We found that multiple forms of flagellum-mediated motility are thermo-suppressed, including swarming and swimming motility. Suppression of swarming motility occurs between 28° and 30°C, which coincides with the optimal growth temperature of P. syringae. Both fliC (encoding flagellin) and syfA (encoding a non-ribosomal peptide synthetase involved in syringafactin biosynthesis) were suppressed with increasing temperature. RNA-seq revealed 1440 genes of the P. syringae genome are temperature sensitive in expression. Genes involved in polysaccharide synthesis and regulation, phage and IS elements, type VI secretion, chemosensing and chemotaxis, translation, flagellar synthesis and motility, and phytotoxin synthesis and transport were generally repressed at 30°C, while genes involved in transcriptional regulation, quaternary ammonium compound metabolism and transport, chaperone/heat shock proteins, and hypothetical genes were generally induced at 30°C. Deletion of flgM, a key regulator in the transition from class III to class IV gene expression, led to elevated and constitutive expression of fliC regardless of temperature, but did not affect thermo-regulation of syfA. This work highlights the importance of temperature in the biology of P. syringae, as many genes encoding traits important for plant-microbe interactions were thermo-regulated.
Introduction
The plant pathogen Pseudomonas syringae has been studied extensively both as a saprophyte, living epiphytically on plant surfaces as well as a pathogen residing within the leaf apoplast. The epiphytic phase is an important part of the disease cycle for these pathogens since it provides inoculum for subsequent infection. Invasion of plants is apparently a relatively rare event, necessitating an abundant epiphytic population on leaves for disease to be likely to occur [1]. Thus most cells occur as epiphytes prior to invasion into the leaf interior. While an understanding of the biology of these pathogenic bacteria on leaf surfaces is of intrinsic scientific interest, it is also of practical significance since the traits that enable them to thrive as epiphytes contribute to diseases of important crop plants.
Leaf surfaces can be relatively hospitable under certain circumstances, but often provide a stressful habitat to bacteria due to conditions of periodic desiccation, UV irradiation, and nutrient limitation [2], [3]. Furthermore, the conditions on leaves are highly temporally variable, quickly switching between hospitable and inhospitable [2], [3]. Under stressful conditions (high irradiance and low relative humidity), the leaf surface, on balance, is inhospitable. However, some anatomical sites, such as the base of glandular trichomes or epidermal cell junctions, appear to offer protection against environmental extremes and are preferentially colonized by epiphytic bacteria [4]. These sites, often termed “preferred sites”, are likely sources of both water and nutrients [5].
Flagellar-mediated motility is an important trait for both the epiphytic and pathogenic lifestyles of P. syringae [6], [7], [8], [9]. Non-motile P. syringae mutants are more sensitive to desiccation and UV exposure than their motile counterparts presumably because they cannot access sites in which they can escape environmental stresses [6]. Additionally, non-motile mutants are severely reduced in their ability to invade the leaf interior or cause disease after topical application to plants [8], [9], [10]. Although motility is apparently beneficial for phyllosphere bacteria, such benefits may be conditional since motility would be dependent on the presence of at least some free water on plants to enable flagellar function [11]. The flagellum is a particularly costly organelle to synthesize, estimated to consume 2% of the cell’s biosynthetic energy expenditure in E. coli [12], so its expression is most likely highly suppressed under conditions that do not permit motility. Additionally, in many bacteria, including Pseudomonads, flagellum synthesis is suppressed when other traits such as the type III secretion system (T3SS) [9], [13], exopolysaccharide (EPS) production [9], [14], or biofilm formation [15] are expressed; all of these traits are critical for plant-pathogen interactions [16], [17].
Given the temporally and spatially variable nature of the leaf habitat, the conditional benefit of flagellar-mediated motility, and the inverse pattern of regulation of flagellum expression and other plant colonization traits, motility is likely regulated in a way that will maximize the fitness of P. syringae on leaves. By understanding the environmental parameters that modulate motility in P. syringae, we may gain insight into the conditions that promote invasion by this organism, as well as a sense of how different fitness factors are coordinately regulated under various environmental circumstances. In temperate regions, temperature may be a critical parameter influencing motility because it is a correlate with wetness of leaves. Leaves are unlikely to remain wet for lengthy periods under warm temperatures since this typically occurs during the day. In contrast, leaf wetness from either dew or rain is most likely to persist under cool temperatures that occur at night. Cells might therefore use temperature as a surrogate for leaf wetness and perhaps exhibit anticipatory behaviors that would maximize their fitness. Temperature is known to regulate certain virulence factors such as phaseolotoxin production in P. syringae pv. phaseolicola or coronatine in P. syringae pv. glycinea, reviewed in [18]. However, the more general role of temperature as a stimulus for changes in gene expression in P. syringae has not been well examined. Insights gained from an understanding of the temperature stimulon should illuminate the interactions of this pathogen with host plants.
In this report we investigate the effect of incubation temperature on global gene expression in P. syringae. Transcriptome analysis revealed that many genes known to be involved in plant colonization are temperature regulated. Notable among the genes affected by temperature were those involved in motility, including synthesis of the flagellum and syringafactin, a major surfactant produced by P. syringae.
Results
Swimming and Swarming Motility are Inhibited at Elevated Incubation Temperatures in P. syringae
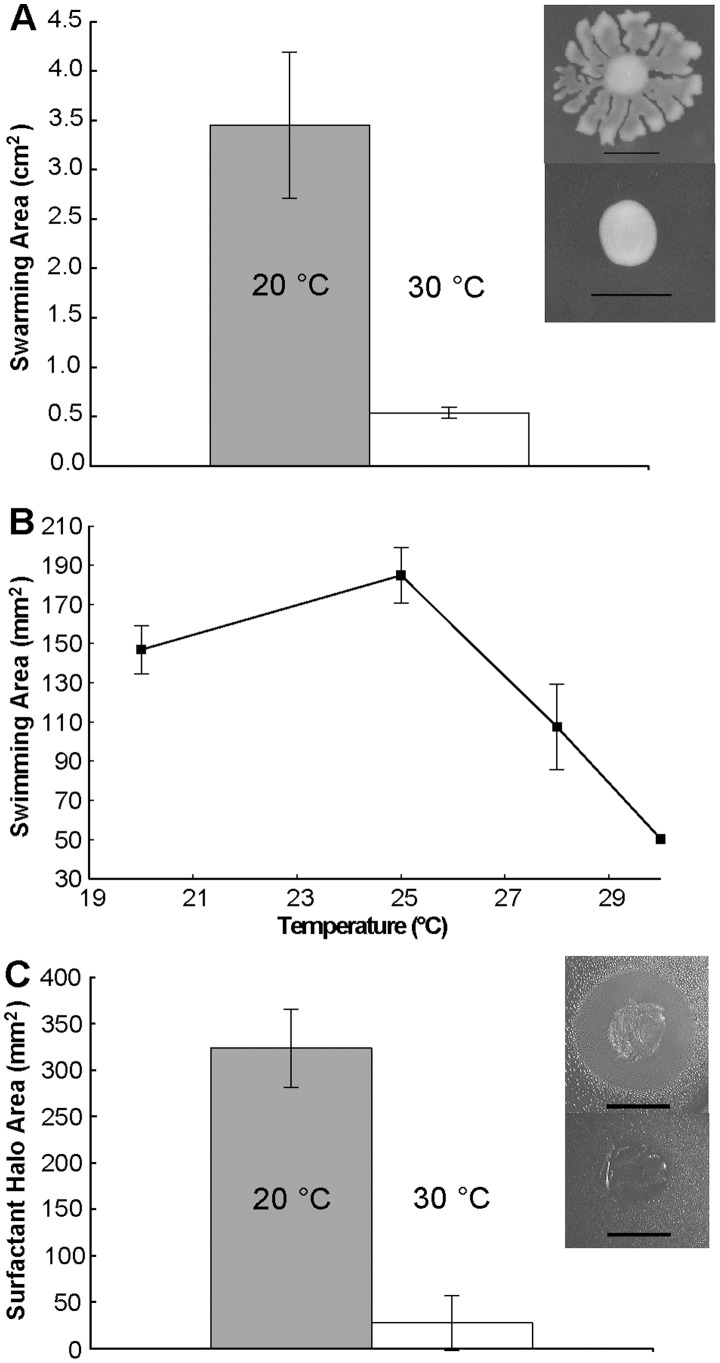
Swarming motility of P. syringae on the surface of low-agar plates was much greater in cells incubated at 20°C than at temperatures of 28–30°C (Fig. 1A). As swarming motility in most bacteria involves some form of surfactant production and requires a functional flagellum [19], we assayed swimming motility of P. syringae through soft agar (which is flagellum-dependent) as well as the abundance of surfactants produced by P. syringae at 20°C and 30°C using the atomized oil assay [20]. Both the rate of swimming and the amount of surfactant produced by P. syringae were significantly reduced at 30°C compared to 20°C (Fig. 1B and C), suggesting that biosurfactant production as well as flagellum synthesis or function was suppressed at the warmer temperature.
Figure 1. Thermo-regulation of motility in P. syringae.
Swarming of Pseudomonas syringae B728a at 20°C (gray bar, top photo of inset) and 30°C (white bar, bottom photo of inset) after 24 hours (A). Area of swimming colonies of Pseudomonas syringae B728a after 24 hours growth at various temperatures (B). Biosurfactant area on plates following 24 hours of incubation at 20°C (gray bar) or 30°C (white bar) (C). The vertical bars represent the standard deviation of the mean.
Syringafactin and Flagellin Expression are Repressed at Elevated Temperatures
Since swarming motility was strongly temperature-dependent, and temperature appeared to affect both flagellum and surfactant production, we investigated the influence of temperature on expression of genes associated with each trait. We measured the temperature-dependent expression of fliC, which encodes flagellin, and syfA, which encodes a non-ribosomal polypeptide synthase (NRPS) required for syringafactin (the major biosurfactant produced by P. syringae B728a) synthesis [20], [21], using both GFP reporter gene fusions and measurement of RNA abundance using qRT-PCR. The expression of syfA and fliC decreased greatly in the temperature range of 25 to 28°C (Fig. 2). While syfA is nearly fully repressed at 28°C the expression of fliC is only repressed by about 50% compared to its maximum expression at 19–20°C, but becomes nearly fully repressed at 30°C. The transcript abundances of both syfA and fliC as measured by qRT-PCR were also significantly lower in cells grown at 30°C compared to 20°C (Fig. 3).
Figure 2. Thermo-regulation of fliC and syfA expression.
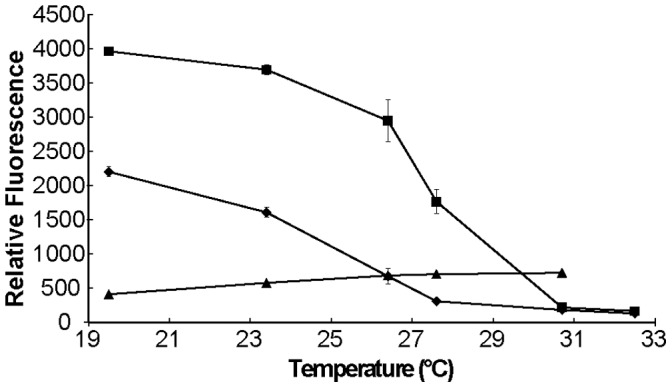
Cell normalized GFP fluorescence of cells grown for 48 hours at various temperatures that harbored a fusion with promoters of fliC (square), syfA (diamond), or nptII (triangle). The vertical bars represent the standard deviation of the mean.
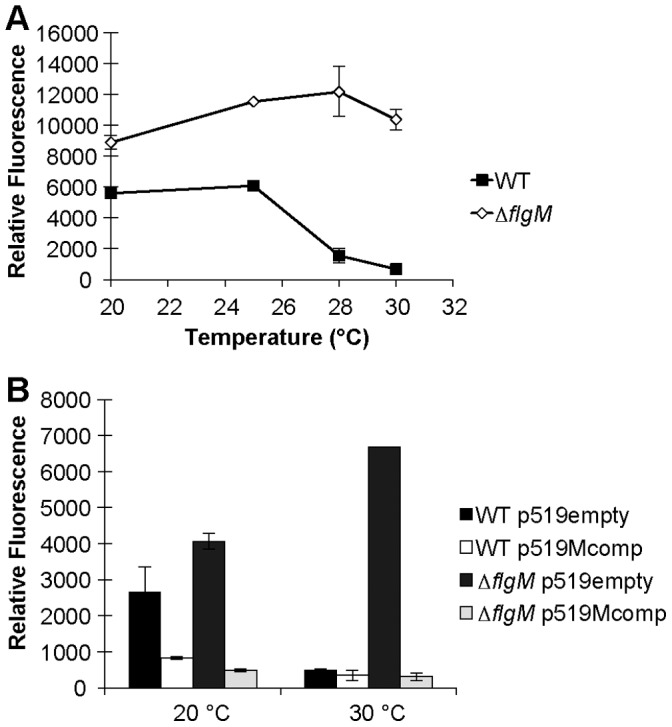
Figure 3. Lack of thermo-regulation of fliC in a ΔflgM mutant.
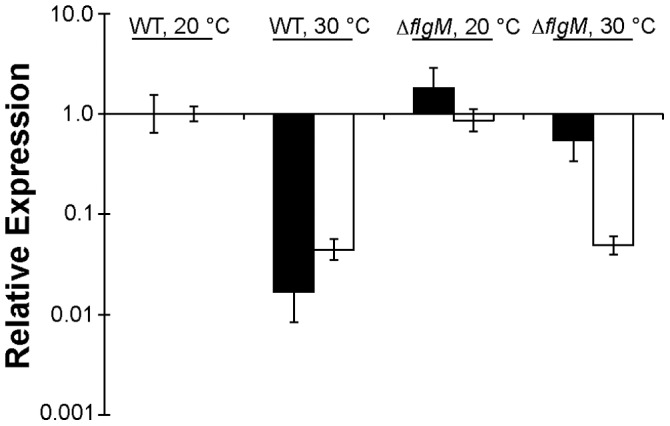
Relative expression of either fliC (black bars) or syfA (white bars) in wild type Pseudomonas syringae B728a or a ΔflgM mutant at either 20°C or 30°C. Expression normalized to WT, 20°C. Error bars represent the 95% confidence interval.
Thermo-regulation of the Flagellum Predominately Affects Stage III and IV Flagellar Genes
Using an RNA sequencing approach, we determined the global effect of temperature on the P. syringae transcriptome (see below for full description). We were particularly interested in assessing the effect of temperature on the flagellar cascade to determine if all genes in the cascade were thermo-regulated similar to fliC.
Assuming that regulation of P. syringae flagellum synthesis follows the same four-tier system of Pseudomonas aeruginosa [22], we found that thermo-regulation appeared to be prominent in class III genes, and to an even greater extent in class IV genes (Table 1). Analysis of the transcriptome suggested that the late stage (class III and IV) genes were the most strongly temperature-regulated genes in the flagellar cascade, with fliC being the most differentially expressed. Neither of the class I genes (fleQ and fliA) were temperature regulated. Interestingly, morA, a protein involved in c-di-GMP metabolism [23] (possessing both GGDEF and EAL domains) was the only gene directly involved in flagellar motility that was more highly expressed at 30°C.
Table 1. Thermo-regulation of flagellar genes.
| Class I | Class II | Class III | Class IV | |
| number ofthermo-responsivegenesa | 0/2 | 3/22 | 6/12 | 7/13 |
| mean foldtemperatureeffectb | n/a | 2.6 | 2.1 | 4.8 |
Genes thermo-regulated within class/total number of genes within class.
Determined only for the genes within class that are thermo-regulated.
flgM is Required for Thermo-repression of fliC but not syfA
The cascade of flagellum regulation has been elucidated in detail in both Salmonella typhimurium and E. coli, where the interaction of FlgM (an antisigma factor) and FliA (an alternative, flagellum-specific sigma factor) is required for the inhibition of expression of late stage genes (genes encoding the flagellar filament, cap, motor proteins, and chemotaxis components) until the hook and basal body has been completed [24]. Since analysis of the flagellar cascade revealed that the class IV flagellar genes were the most strongly affected by incubation temperature (fliC having the greatest difference in expression between warm and cool growth conditions), we hypothesized that the FlgM-FliA interaction might also mediate thermo-repression of class IV genes, and in particular, fliC expression in P. syringae. We compared fliC expression in a flgM deletion strain (ΔflgM) to that in the WT strain at both 20°C and 30°C. While the expression of fliC decreased with increasing growth temperature above 25°C in WT, its expression in ΔflgM was high and independent of growth temperature (Fig. 4A). Over-expression of flgM in trans suppressed the expression of fliC at both cool and warm temperatures in both the WT and ΔflgM strains (Fig. 4B). The transcript abundance of fliC measured by qRT-PCR also was much higher in ΔflgM compared to WT when cells were grown at 30°C (Fig. 3). These results indicate that flgM is required for thermo-repression of fliC and that its over-expression on a high copy number plasmid suppresses fliC at all temperatures.
Figure 4. Reduction of fliC expression in ΔflgM by complementation.
Cell-normalized GFP fluorescence of either wild type Pseudomonas syringae B728a (black squares) or a ΔflgM mutant (open diamonds) harboring a gfp reporter gene fusion with a promoter of fliC (A). Cell normalized GFP fluorescence of wild type P. syringae or a ΔflgM mutant harboring a gfp reporter gene fusion with the promoter of fliC as well as either a flgM complementing vector (p519Mcomp) (black and dark gray bar, respectively) or vector control (p519empty) (white and light gray bar, respectively) when grown at either 20°C or 30°C (B). The vertical bars represent the standard deviation of the mean.
To test whether flgM was also responsible for thermo-repression of syfA at 30°C, we assessed syfA expression in ΔflgM at 20°C and 30°C using qRT-PCR. ΔflgM exhibited thermo-repression of syfA similar to WT (Fig. 3). Additionally, ΔflgM produced less surfactant at 30°C than at 20°C, similar to WT (Fig. S1).
salA Positively Regulates syfR and syfA
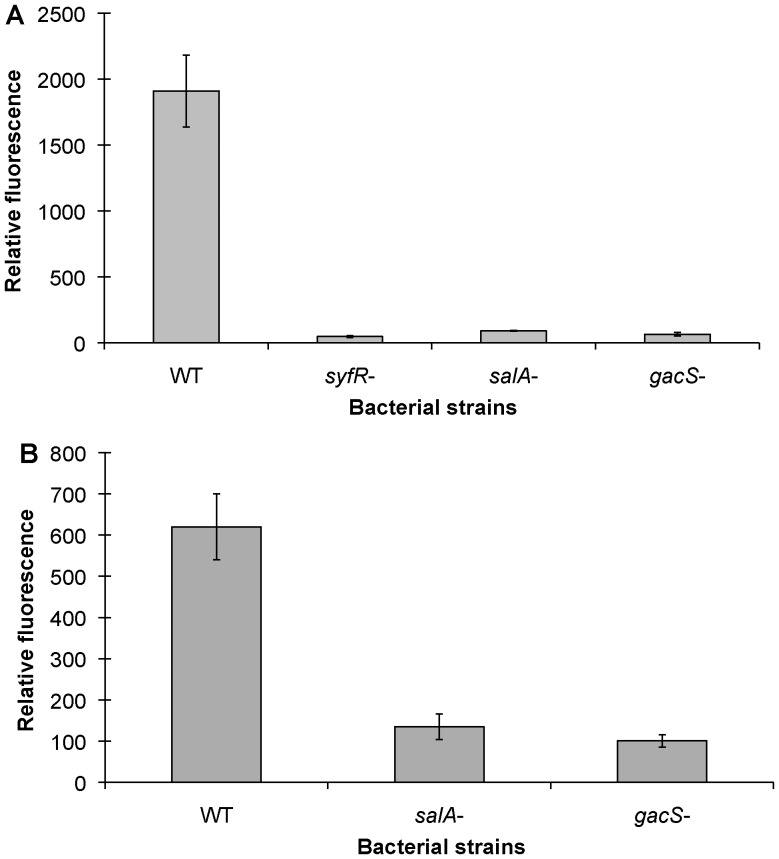
While we were unable to link flgM to the regulation of syfA, results from the transcriptome indicated that genes involved in the biosynthesis and transport of the phytotoxins syringomycin and syringopeptin, as well as their regulators, salA, syrG, and syrF were all more highly expressed at 20°C than at 30°C, similar to fliC and syfA. Because of the similarities in synthesis (lipopeptides synthesized by NRPSs) and temperature-dependent regulation between the former two phytotoxins and the latter biosurfactant, we posited that they shared a common regulator. We found that both syfA as well as syfR (a LuxR-type regulator required for syringafactin expression [20], [21]) expression was dependent on salA (Fig. 5). Additionally, as salA is positively regulated by gacS [25], gacS was also required for expression of syfR and syfA. Taken together, this data demonstrates that syringafactin expression is under gac-regulation via salA.
Figure 5. salA positively regulates syfA.
Cell-normalized GFP fluorescence exhibited by wild type and various mutants of Pseudomonas syringae B728a harboring a gfp reporter gene fused either to syfA (A) or syfR (B) following 24 hours of incubation at 20°C. Vertical bars represent the standard deviation of the mean.
Temperature Affects the Ability of P. syringae to Survive Desiccation Stress on Leaves
Given that motility is expected to be required for cells to access sites where environmental extremes such as desiccation stress could best be avoided, and that high temperatures appeared to strongly suppress the motility of P. syringae, we measured the temperature-dependent survival of P. syringae on leaves. We compared epiphytic populations of viable cells recovered from plants incubated under cool conditions (20°C, motility-inducing) and warm conditions (30°C, motility-suppressing) after inoculation. To minimize the confounding effect of temperature on the plant itself, as well as possible differences in population sizes of the bacterium on plants due to different growth rates at various temperatures, bacteria were only incubated on plants at the test temperature under moist conditions for 6 hours. After this brief period of incubation under near saturating humidity such that motility would be permitted, all plants were moved to a common area where leaves were allowed to dry with a constant temperature of 28°C. Bacterial cells incubated on cool, moist leaves survived the subsequent desiccating conditions an average of 3.2-fold better than cells that had been incubated on warm, moist leaves (Fig. 6). Importantly, a flgK mutant (which is defective in flagellar motility) survived poorly on both cool and warm leaves and exhibited similarly large declines in viable population sizes as observed for the WT strain incubated at 30°C. These results suggest that motility is required for stress survival on leaves and that the elevated motility of P. syringae on cool plants facilitated its acquisition of sites where desiccation survival was enhanced. In contrast, we found no evidence that syringafactin production contributed to enhanced survival on leaves incubated at cool temperatures under these conditions since the survival of a WT and a syfA mutant were similar on moist leaves incubated at a given temperature (data not shown). Together, these results suggest that bacterial cell behavior during the brief periods that leaves were moist modulated their ability to survive the subsequent desiccation stresses, and motility was a sufficient trait to account for this apparent stress avoidance.
Figure 6. Temperature-dependent epiphytic survival.
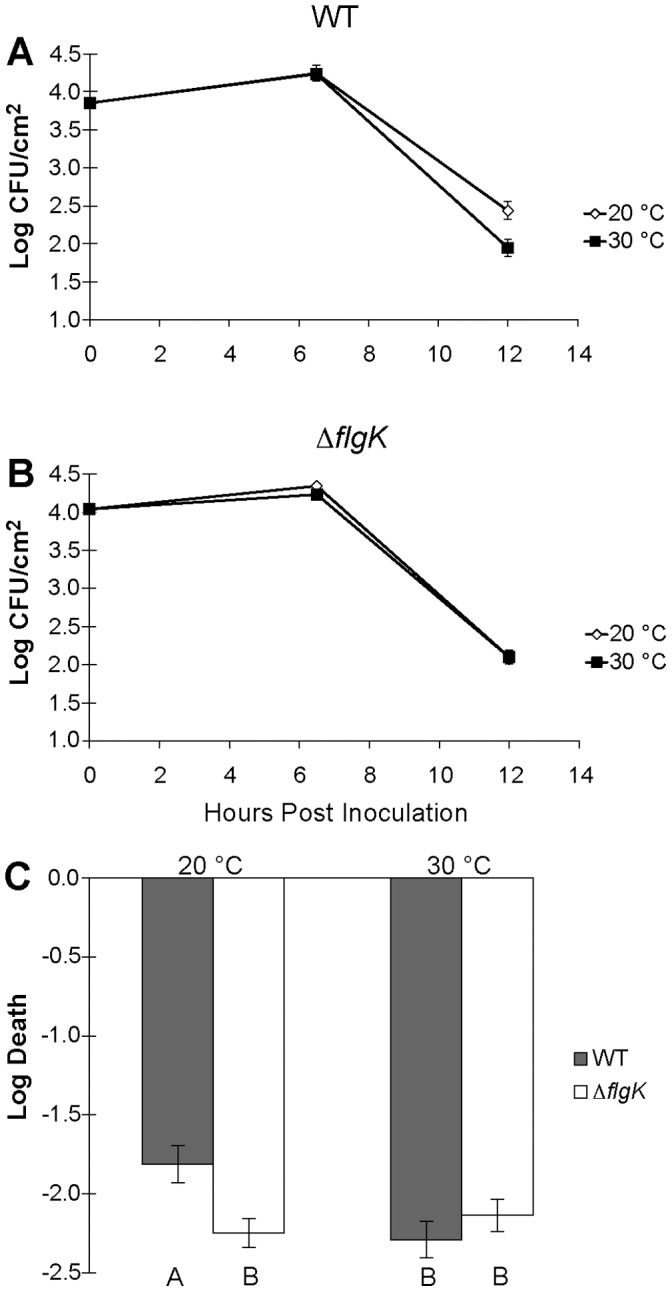
Epiphytic population size of wild type Pseudomonas syringae B728a (A) or a ΔflgK mutant (B) incubated at 20°C (open diamond) or 30°C (black square) for 6.5 hours at 100% RH prior to being exposed to desiccation at 26–28°C and 60–65% RH. Log-transformed population size decline was measured during the desiccation period that occurred between 6.5 and 12 hours after inoculation (C). This experiment was repeated several times with similar results. Treatments noted with the same letter do not differ significantly at a p-value ≤0.05 as determined by least significant difference test (LSD). The vertical bars represent the standard deviation of the mean. Error bars are present in panel B, but in some cases are small and obscured by the treatment symbol.
Global Transcriptome Analysis of P. syringae Incubated under Warm and Cool Conditions
As the effect of temperature on swimming and swarming motility was linked to a transcriptional response of genes encoding flagellin and syringafactin, we assessed the effect of temperature on the entire transcriptome of P. syringae by comparing the abundance of messages in cells incubated at 20°C and 30°C using Illumina next-generation sequencing of cDNAs. In total, 28.3% of the putative protein-encoding genes were differentially expressed between these two temperatures, with 338 genes more highly expressed at 20°C than at 30°C, while 1107 genes were more highly expressed at 30°C than at 20°C (Tables S1 and S2). Of the 338 genes that were up-regulated under cool conditions, 205 were induced more than 2-fold, while 133 were induced less than 2-fold. Of the 1107 genes that were up-regulated under warm conditions, 572 were induced more than 2-fold, while 535 were induced less than 2-fold. The differentially regulated genes were grouped into functional categories to determine if particular processes were more prominently temperature-dependent in P. syringae. Tables 2 and 3 list the functional categories in which genes preferentially expressed at 20°C and 30°C, respectively, were significantly over-represented. More functional categories were enriched at 20°C than at 30°C. The differential expression of genes in the following functional groups was particularly noteworthy:
Table 2. Functional gene categories preferentially up-regulated under cool growth conditions.
| Category | p-valuea |
| Polysaccharide synthesis and regulation | 8.03E−05 |
| Phage & IS elements | 5.70E−14 |
| Type VI secretion system | 1.17E−13 |
| Chemosensing & chemotaxis | 4.18E−09 |
| Translation | 0.02 |
| Flagellar synthesis and motility | 9.32E−06 |
| Phytotoxin synthesis andtransport | 4.77E−10 |
Significance of functional category enrichment assessed using the hypergeometric distribution [77].
Bonferroni corrected p-value.
Table 3. Functional gene categories preferentially up-regulated under warm growth conditions.
| Category | p-valuea |
| Hypothetical | 0.04 |
| Transcriptional regulation | 8.73E−04 |
| QAC metabolism and transport | 1.56E−04 |
| Chaperones/heat shock proteins | 0.01 |
Significance of functional category enrichment assessed using the hypergeometric distribution [77].
Bonferroni corrected p-value.
Polysaccharide synthesis and regulation
Genes in this functional category were predominantly induced at 20°C. Noteworthy were genes involved in alginate synthesis (algE, 1.9-fold; algK, 2.2-fold; alg44 2.4-fold; alg8, 2.1-fold; and algD, 3.0-fold), levan synthesis (lscC-1, 4.2-fold), and Psl polysaccharide synthesis (pslA, 3.7-fold; pslB, 3.5-fold; pslD, 2.1-fold; pslE, 2.8-fold; pslF, 3.4-fold; pslG, 2.6-fold; pslH-1, 2.3-fold; pslI, 3.0-fold; pslJ, 2.0-fold). Interestingly, there is a second copy of a gene putatively encoding levansucrase in the P. syringae genome (psyr_2103, lscC-2) that is induced by 2.1-fold at 30°C, exhibiting an opposite pattern of temperature-dependent expression compared to lscC-1. A few other genes in this functional category were also more highly expressed at 30°C, including 3 genes annotated as polysaccharide deacetylases with predicted functions as xylanases or chitin deacetylases (psyr_1809, 1.7-fold; psyr_2692, 2.2-fold; psyr_3937, 1.6-fold). mucD, encoding a negative regulator of AlgT (AlgU/σE) that controls alginate expression, also was more highly expressed at warm incubation temperatures (2.6-fold), which might explain the apparent induction of alginate at 20°C.
Phage and IS elements
Genes in this functional category were predominantly expressed at a higher level at 20°C. Prominent among such genes were those in two predicted prophage regions in the P. syringae genome corresponding to psyr_2761 to psyr_2832 (prophage region I) and psyr_4582 to psyr_4597 (prophage region II) identified with the Prophinder tool in ACLAME [26]. At least 22 genes in prophage region I were more highly expressed at 20°C than at 30°C, with a mean induction of 3.2-fold (range of 1.8 to 6.0-fold). However, 3 genes in prophage region I were more highly expressed at 30°C (psyr_2773, 2.0-fold; psyr_2791, 2.7-fold; psyr_2826, 1.7-fold). All 16 genes in prophage region II were more highly expressed at 20°C with a mean induction of 5.0-fold (range 1.7 to 7.3-fold).
Type VI secretion system
Most genes associated with the recently described type VI secretion system (T6SS) in P. syringae B728a [27], including genes encoding structural components of the secretion apparatus as well as ClpV, an ATPase putatively involved in driving export of secretion substrates, were more highly expressed at 20°C than at warmer temperatures. T6SS genes had a mean induction of 2.0-fold (range of 1.7- to 2.6-fold). However, 2 putative hcp homologs often associated with T6SS but not co-located with other T6SS genes, were both more highly expressed at 30°C (psyr_1935, 2.1-fold; psyr_4039, 2.9-fold).
Chemosensing and chemotaxis
Many genes in this functional category, including those encoding putative methyl-accepting chemotaxis proteins (15) as well as homologs of cheV (2), cheW (3), cheB (1), cheA (1), and an aerotaxis receptor, were more highly expressed at 20°C. The mean induction of these genes at 20°C was 3.3-fold (range of 1.7- to 11.9-fold). In contrast, 2 putative methyl-accepting chemotaxis proteins were more highly expressed at 30°C (psyr_0868 and psyr_1539, 2.0-fold and 1.8-fold, respectively).
Translation
Many genes encoding proteins associated with the 50S and 30S ribosomal subunits, as well as elongation factor Tu and G, were more highly expressed at 20°C than at 30°C (average of 2.0-fold induction with a range of 1.6- to 2.3-fold).
Transcriptional regulation
The genes encoding numerous DNA-binding transcriptional regulators belonging to the AraC, AsnC, DoeR, GntR, IclR, LacI, LuxR, LysR, MarR, TetR families, as well as putative response regulator partners of two component systems that also possess DNA binding domains were more highly expressed at 30°C (Table 4). Very few genes encoding putative transcriptional regulators were induced at 20°C, with the exception of 4 LuxR-type regulators that are involved in syringomycin, syringopeptin, and syringafactin regulation (salA, syrG, syrF, and syfR). Interestingly, those putative response regulator partner proteins that lack discernible DNA binding domains, were more likely to be induced at 20°C compared to 30°C (17.6% vs. 2.9%).
Table 4. Transcriptional regulators influenced by temperature.
| Regulator Type | Annotated in the genome | Induced at 30°C(%) | Induced at 20°C(%) |
| LysR | 74 | 21 (28.4) | 1 (1.4) |
| GntR | 27 | 12 (44.4) | 0 (0.0) |
| TetR | 27 | 9 (33.3) | 0 (0.0) |
| LuxR | 24 | 6 (25.0) | 4 (16.7) |
| AsnC | 7 | 2 (28.6) | 0 (0.0) |
| AraC | 19 | 7 (36.8) | 0 (0.0) |
| IclR | 5 | 1 (20.0) | 0 (0.0) |
| DoeR | 5 | 2 (40.0) | 0 (0.0) |
| LacI | 15 | 3 (20.0) | 1 (6.7) |
| MarR | 7 | 1 (14.3) | 0 (0.0) |
| Response Regulator with putative DNA binding domaina | 28 | 11 (39.3) | 0 (0.0) |
| Response Regulatorwith out putative DNA binding domain | 34 | 1 (2.9) | 6 (17.6) |
This contains 10 Response regulator domains that are paired with LuxR DNA binding domains, 4 of which are induced at 30°C, none of which are induced at 20°C.
Quaternary ammonium compound (QAC) metabolism and transport
Genes encoding enzyme systems involved in choline, glycine betaine (GB), and carnitine import and metabolism were generally up-regulated at 30°C, while the opuC transporter, which is associated with osmoprotection [28], was modestly induced at 20°C (range of 1.6- to 1.8-fold). The betT transporter (associated with osmoprotection), along with betX and caiX solute binding proteins (associated with catabolism) which import betaine and carnitine, respectively, were induced by 2.3- to 2.8-fold at 30°C [29], [30]. All genes necessary for conversion of choline and glycine betaine to glycine were induced at 30°C, including betAB (range of 4.3 to 4.9-fold), gbcAB (range of 1.8 to 2.1-fold), dgcAB (range of 2.0- to 2.3-fold), and soxBDAG (range of 1.7- to 2.3-fold) [31]. dhcA and dhcB, both predicted to encode proteins that catalyze the second catabolic step in carnitine breakdown, were more highly expressed at 20°C (range of 1.9- to 2.1-fold), while their regulator (dhcR) was expressed 2.8-fold higher at 30°C than at 20°C. As DhcR induces transcription of dhcAB in the presence of 3-dehydrocarnitine (an intermediate in the catabolism of carnitine), these results might suggest that elevated growth temperature increases carnitine catabolism, but that the inducing intermediate was not present in this experiment.
Hypothetical
Surprisingly, the proportion of genes with no known function that were more highly expressed at high temperatures than at cooler temperatures was much higher than their proportion in the genome. Such genes included both those encoding proteins that possess domains of unknown function, as well as those without such conserved domains.
In addition to the general categories noted above, genes involved in auxin biosynthesis were temperature regulated. Both homologs of iaaM (psyr_1536, psyr_4667) and iaaH (psyr_2208, psyr_4268), involved in biosynthesis of indole-acetic acid (IAA) were induced at 30°C, as were genes involved in the biosynthesis of tryptophan, a precursor for IAA.
Discussion
The motility of P. syringae, like some other bacteria, is very strongly influenced by temperature. Both swimming and swarming motility are inhibited by incubation at 30°C and this is apparently due to the suppression of expression of flagellar components required for both swimming and swarming motility, as well as reduced expression of syringafactin that contributes to swarming. In addition to syringafactin, P. syringae B728a produces a second biosurfactant, 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA), which is synthesized by RhlA. In contrast to syfA, rhlA (psyr_3129) was not thermo-regulated at the transcript level (Table S1), potentially indicating that regulation of this surfactant is independent of temperature. Temperature-regulated swarming motility and surfactant production has also been observed in Pseudomonas putida KT2440, a non plant-pathogenic, soil-associated Pseudomonad, which moves at 22°C but not 30°C [32]. Interestingly, biosurfactant production has not been reported for this strain and its swarming motility is not flagellar-dependent, but rather type IV pilus-dependent [32]. In contrast, another strain, P. putida PCL1445, produces the biosurfactants putisolvin I and II, required for swarming, in a temperature-dependent manner [33], [34], [35], but neither temperature-regulated flagellar expression nor swimming motility has been reported. The production of these biosurfactants is, however, dependent on the gacS/gacA two component signaling system, both of which are more highly expressed at 11°C than at 28°C or 32°C [34], [35]. In contrast to this strain, the expression of neither gacS nor gacA in P. syringae B728a was influenced by temperature under the conditions tested, as determined by the transcriptome results.
Inhibition of flagellar motility occurs at the optimal growth temperature of P. syringae as in many bacteria. This phenomenon has been studied most extensively in animal pathogens [36] but it seems likely that the biological relevance for such a phenotype differs in bacteria colonizing such different habitats. flgM, encoding an anti-sigma factor known to inhibit the activity of FliA in numerous organisms, also was required for thermo-repression of fliC in P. syringae. The patterns of thermo-regulation in P. syringae seem similar to that of flagellar thermo-regulation in Yersinia enterocolitica, whose motility is regulated in a manner that generally conforms to the three-tiered regulatory hierarchy established in E. coli and S. typhimurium [37]. In Y. enterocolitica flagellar-mediated motility is expressed at 25°C but repressed at 37°C, while plasmid-encoded virulence factors are oppositely regulated [38]. Thermo-regulation occurs at the level of fliA expression, while expression of other class I and class II flagellar genes are insensitive to incubation temperature [37], [39], [40]. Disruption of fliA leads to reduced or abolished thermo-regulation of plasmid-encoded virulence factors, and therefore is thought to be a key mediator of the temperature response [37]. In P. syringae, however, we found that fliA transcription itself was insensitive to incubation temperature, suggesting instead that FlgM mediates thermo-regulation. Indeed, the FlgM-FliA interaction in Campylobacter jejuni is temperature dependent, being destabilized at 42°C compared to 32°C or 37°C [41]. Campylobacter is unique in that it is one of the few genera where motility and flagellin expression is maximal at its growth temperature optimum (42°C) [41], [42]. It is not clear whether the FliA-FlgM interaction in P. syringae is itself temperature sensitive or if other factors mediate a temperature-sensitive interaction of these two components. If this interaction is itself temperature sensitive, our results would suggest that the interaction in P. syringae is stabilized with increasing temperature, opposite to that in Campylobacter. It is unclear if thermo-regulation of FliA activity in P. syringae contributes to thermo-regulation of non-flagellar genes similar to its role in Y. enterocolitica. However, the fact that flgM is not required for thermo-repression of syfA, coupled with the observation that the expression of over 1000 genes is significantly influenced by incubation temperature indicates that at least one, if not several, other genes contribute to thermo-regulation in P. syringae.
Syringomycin production was previously shown to be thermo-regulated [43], which is consistent with our results indicating that the genes involved in the biosynthesis, transport and regulation of syringomycin and syringopeptin were all thermo-regulated in the transcriptome. Both syfR and syfA are positively regulated by salA (Fig. 6), which is also a positive regulator of syringomycin and syringopeptin [44], [45]. While it is not clear at this point which gene(s) are required for thermo-regulation of these three lipopeptides, it seems likely that they may share a common thermo-regulator. Additionally, it appears that thermo-regulation of the flagellum and thermo-regulation of syringafactin are independent of each other because mutations affecting thermo-regulation of syfA do not influence thermo-regulation of fliC (Hockett and Lindow, unpublished).
The apparent temperature-dependent expression of a variety of EPS species in P. syringae suggests that such molecules play a very context-dependent role in the fitness of this bacterium. In addition to alginate production, genes involved in producing other EPS species such as Psl and levan were, in general, strongly suppressed at 30°C. In P. syringae pv. glycinea, levan production is also thermo-regulated, accumulating at 18°C but not 28°C [46]. Similar to our observation that lscC-2 was oppositely regulated compared to the majority of genes involved in EPS synthesis, being more highly expressed at 30°C than 20°C, LscC in P. syringae pv. glycinea is more abundant within the cell at 28°C compared to 18°C [46]. The enzyme is not, however, exported at the elevated temperature, which restricts levan formation to cool temperatures. Levansucrase enzyme location may be similarly regulated in P. syringae B728a. Psl, a recently discovered EPS involved in biofilm formation in Pseudomonas aeruginosa [47], [48], is hypothesized play a similar role in P. syringae [49], [50]. There may be a conflict in the roles of Psl and motility in biofilm formation as flagellar-mediated motility apparently contributes to initial attachment of cells to surfaces [51], while the expression of EPS could interfere with motility of P. syringae. Further investigation will be required to directly address conflicts arising from increased expression of genes mediating motility along with genes mediating EPS production. We presume that during our in planta assays, cells that became motile under the cool conditions accessed preferred sites of colonization (base of glandular trichomes, intercellular grooves between epidermal cells) that facilitated their subsequent desiccation tolerance or enabled them to escape such stresses. A non-mutually exclusive possibility is that such cool conditions stimulated biofilm formation due to both enhanced motility and EPS production and that survival was enhanced on the leaf surface itself [52], [53]. Further directed studies will be required to discern whether motility itself or its role in biofilm formation (or both) contribute to cool temperature induced desiccation tolerance.
It was noteworthy that nearly all of the known uptake systems for QAC compounds, whether for their accumulation for osmoprotection or for their catabolism were more highly expressed at 30°C. These results suggest that increased import capacity of these compounds would allow the cell to rapidly accumulate compatible solutes by repressing expression of the catabolic enzymes, should water limitation occur (conditions which did not occur in our experiment). Alternatively, P. syringae may anticipate the increased abundance of QACs in the plant environment using temperature as a cue as tomato plants were found to accumulate QACs at temperatures above 25°C [54]. Indeed, other researchers have observed that P. syringae does not maintain a measurable glycine betaine pool at 30°C, but does maintain such a pool at 22–25°C, which would be consistent with increased catabolism (M. Wargo, personal communication and [55]).
One of the most obvious effects of temperature on P. syringae that is linked to its behavior on plants is the thermo-regulation of motility. The lower survival of cells of P. syringae exposed to warm temperatures during moist incubation prior to desiccation stress is consistent with previous work that showed that non-motile mutants of this species were less resistant to desiccation and UV stress in planta [6]. While there have been numerous descriptions of thermo-regulated motility in animal pathogens, there have been only two studies to date that have linked temperature with the motility of plant pathogenic bacteria [7], [56]. Flagellar motility of Erwinia amylovora, which is suppressed at warm temperatures, appeared to play little role in the infection and disease process, as its virulence was independent of temperature. In addition to thermo-regulation of motility, E. amylovora expresses amylovoran, an EPS required for virulence, and type III secretion system-associated genes in a temperature dependent manner, with greater expression at cooler temperatures [57], [58], [59], [60]. In contrast to E. amylovora, symptom development in lilac inoculated with P. syringae isolates was lower on plants incubated at temperatures above 24°C than at cooler temperatures [61]. However, due to the invasive method of inoculation used in this study, motility may not have been required for infection and the effect of temperature on the motility of the pathogen was not assessed. While plant disease symptoms caused by other P. syringae strains also have been noted to be higher under cool conditions, such effects on virulence cannot be linked to the motility of the pathogen. For example, pre-incubation of P. syringae pv. glycinea PG4180 at 18°C enhanced subsequent in planta multiplication and disease symptoms compared to when preincubated at 28°C [62]. This temperature-dependent interaction was dependent on production of the toxin coronatine, which is produced only at cool temperatures. It has been hypothesized that the increased occurrence and severity of bacterial plant disease often seen under cool, wet weather in temperate climates is a result of increased opportunity for invasion due to increased water availability that coincides with cool temperatures [18]. Our results, in conjunction with previous results, suggest that temperate pathogens are also more invasive under these conditions because they more highly express motility-related traits under cool conditions.
This work supports the notion of P. syringae as a conservative pathogen that coordinates motility with environmental conditions in a way that will be optimally productive for the pathogen. It also suggests that the pathogen exhibits anticipatory behaviors, using high temperature as an indicator that water availability may soon be limited, and thus dictating that it express such a trait in a conservative manner. This strategy would maximize its fitness over the long run by avoiding risks associated with short-term exposure to stresses such as desiccation that could be avoided.
Materials and Methods
Bacterial Strains, Plasmids, Culture Media, and Growth Conditions
Pseudomonas syringae pv. syringae B728a [63] was routinely cultured in King’s medium B (KB) broth, or on KB plates supplemented with 1.5% (w/v) Difco agar technical (BD, Sparks, MD) at 28°C [64]. Escherichia strains TOP10 (Life technologies, Carlsbad, CA) and S17-1 [65] were cultured in Luria-Bertani (LB) medium broth, or on LB plates supplemented with 1.5% (w/v) Difco agar technical at 37°C. Antibiotics were used at the following final concentrations: rifampicin, 100 µg/mL; kanamyacin, 50 µg/mL; gentamicin, 15 µg/mL; spectinomycin 20 µg/mL; tetracycline, 15 µg/mL; nitrofurantoin (NFT), 30 µg/mL. Natamycin (antifungal) was used at 20 µg/mL. Strains and plasmids used in this work are listed in Table S3. Primers used in this work are listed in Table S4. The temperature of agar plates were routinely monitored using a CZ-IR thermometer (ThermoWorks, Lindon, UT). Incubator temperatures and relative humidity were routinely monitored using HOBO data loggers (Onset, Bourne, MA).
Detection of Biosurfactants
Biosurfactants were detected with an atomized oil assay similar to previously described [20]. Bacterial cultures were grown overnight and diluted into fresh medium the following morning and grown for 2–4 hours. Cultures were then washed once with 10 mM KPO4 and resuspended to a concentration of 2.0×108 CFU/mL, as determined by turbidity (OD600). 5 µL of resuspended culture was spotted onto KB plates and incubated for 20–30 hours at either 20°C or 30°C, then sprayed with a mist of mineral oil. The diameter of the visible halo of brighter oil drops was measured and the area of the producing bacterial colony was calculated and subtracted from that of the surfactant halo to yield the adjusted halo area.
Swimming and Swarming Assays
Swimming media (50% KB containing 0.25% agar) was allowed to dry overnight on the bench top (unstacked). Bacterial cultures were incubated overnight (28°C with shaking at 200 RPM) in KB broth amended with the proper antibiotics. Cultures were then diluted into fresh media and allowed to incubate for 2–4 hours and cells recovered by centrifugation and washed once with 10 mM KPO4 buffer before being resuspended to a final concentration of 2.0×108 CFU/mL and inoculated by stabbing with a sterile toothpick. Plates were incubated for 20–30 hours prior to observation. Swarming assays were performed similar to swimming assays except 5 ul of bacterial suspensions were spot inoculated singly onto the center of swarming plates (undiluted KB containing 0.4% agar).
Transcriptional Reporter Assays
Transcriptional reporter assays were performed similar to other studies [66]. Briefly, culture spots were resuspended in 10 mM KPO4 buffer and diluted to a final OD600 of 0.1–0.2. GFP fluorescence intensity was determined using a TD-700 fluorometer (Turner Designs, Sunnyvale, CA) with a 486-nm-band-pass excitation filter and a 510- to 700-nm combination emission filter. Relative fluorescence was determined by normalizing the arbitrary fluorescence units to optical density.
RNA Isolation and qRT-PCR
Bacterial cultures were harvested in 1.0 mL of RNA later (Life Technologies; Carlsbad, CA) and stored at 4°C for no longer than one week prior to RNA isolation. Total RNA was isolated using TRIzol® reagent (Life Technologies) similar to manufacturer’s protocol except differing in the following ways: homogenized cells were incubated at 65°C for 10 minutes in TRIzol® prior to addition of chloroform; after adding chloroform, samples were incubated at RT for 15 minutes; RNA was precipitated in isopropanol at −80°C for 20 minutes. Washed RNA was resuspended in 30–50 µl of RNAsecure™ (Life Technologies), according to manufacturer’s protocol. To remove contaminating genomic DNA, RNA was treated with TURBO DNA-free™ (Life Technologies) according to manufacturer’s protocol. DNase-treated samples were either cleaned using the DNase inactivating reagent included in the TURBO DNA-free™ kit, or were column purified using RNeasy Mini Kit (QIAGEN; Valencia, CA). The absence of contaminating DNA was confirmed by performing a PCR reaction using Phusion® High-Fidelity DNA Polymerase (New England Biolabs; Ipswich, MA) with either DNase treated or untreated samples using rpoD-RT-S and AS primers (Table S4). RNA purity and abundance was routinely assessed using an ND-1000 spectrophotometer (Thermo Scientific; Lafayette, CO). cDNA was generated from 0.5 or 10 µg of DNase-treated RNA, using SuperScript® II reverse transcriptase (Life Technologies) with random primers (Life Technologies). qPCR was performed using a 7300-Real-Time PCR System (Life Technologies) with QuantiTect SYBR Green I (QIAGEN) on 5 or 10 µl of 1∶1000 diluted cDNA. Samples not treated with reverse transcriptase were routinely included, which showed no significant increase in fluorescence following 35 cycles. Amplification efficiencies were determined using LinReg [67]. Amplification efficiencies were determined to be 95% of maximum, and were consistent between gene targets. Relative expression was calculated according to the ΔΔCt method with a base of 1.9 to account for amplification efficiency. rpoD and psyr_3981, encoding pseudouridine synthase which was found to be stably expressed under numerous conditions and in numerous mutant backgrounds (R. Scott and K. Hockett, unpublished data), were used as endogenous controls. qRT-PCR data normalized to either endogenous control gave similar results; results normalized to rpoD are shown.
mRNA Sequencing
Total RNA was harvested as described above. Following total RNA isolation, 16S and 23S rRNA was removed using Ribo-Zero™ rRNA removal Kit (Gram-Negative Bacteria) (Epicentre, Madison, WI) according to the manufacturer’s protocol. Enriched mRNA samples were assayed with a 2100 Bioanalyzer (Agilent; Santa Clara, CA) to confirm removal of rRNA. Quantification of mRNA and dsDNA was routinely performed with qBit RNA and dsDNA HS assays (Life Technologies), respectively, according to the manufacturer’s protocol. Ambion fragmentation reagent (Life Technologies) was used to generate 100–200 nucleotide fragments from enriched mRNA. Fragmented RNA was isolated by ethanol precipitation with glycogen amendment, followed by resuspension in RNase free water. First strand cDNA synthesis was performed using random primers (Life Technologies) and Superscript II reverse transcriptase (Life Technologies), followed by second strand synthesis using RNase H and DNA pol I (Life Technologies). dsDNA was routinely purified using AMPure XP beads (Beckman Coulter, Brae, CA) with PEG 6000 added to a final concentration of 6.5% (w/v). cDNA end repair was performed using a combination of Klenow DNA polymerase, T4 DNA polymerase, and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). An A tail was added to end-repaired fragments using Klenow exo minus with 250 µM ATP (final concentration) (New England Biolabs). Illumina adapters were ligated to A-tailed dsDNA fragments using T4 DNA ligase (New England Biolabs), with PEG 6000 addition to 5.0% (w/v) final concentration. Adapter-ligated fragments were amplified using Illumina barcoded primers with Phusion DNA polymerase (New England Biolabs). Amplified fragments were purified using AMPure XP beads without PEG addition. Fragment sizes were confirmed to be in the size range of 200–300 nucleotides using the 2100 Bioanalyzer. Sequencing was performed using the Illumina HiSeq 2000 for 50 base pair reads through the Vincent J. Coates Genomics Sequencing Laboratory. Three biological repeats were sequenced per temperature treatment on three separate flow cells. Reads were aligned to the P. syringae pv. syringae B728a genome (downloaded from the Integrated Microbial Genomes website) using bwa [68] allowing for a maximum of three mismatches between a given read and the reference genome. The number of reads that overlapped with a given gene was counted using a custom script. Differential expression of genes and statistical significance were assessed using edgeR [69]. Briefly, significance was established by comparing gene expression levels, normalized by the trimmed mean of M-values (TMM) [70], between three biological replicates incubated at either 20°C or 30°C (six samples total) using an empirical Bayes estimation and exact tests based on the negative binomial distribution. Genes were considered significantly differentially regulated if the p-value (after adjustment for multiple comparisons) for a difference in expression between the two treatments was less than or equal to 0.05. Expression data is available at the Integrated Microbial Genome website (http://img.jgi.doe.gov).
Deletion of flgK
A deletion mutant of flgK was constructed by cloning approximately 1 kb fragments upstream and downstream of flgK into pENTR/D-TOPO:MCS-Kan [71]. The region downstream of flgK was amplified using the primers flgKe-xhoF and flgKe-xbaR, digested with XhoI and XbaI and ligated into pENTR/D-TOPO:MCS-Kan. The region upstream of flgK was amplified using the primers flgKs-hindF and flgKs-speR, digested with HindIII and SpeI, and ligated into pENTR/D-TOPO:MCS-Kan. The resulting region containing both flanking sequences and npt2 driving kanamycin resistance were transferred to pLVC/D [72] via a clonase LR reaction (Invitrogen). The resulting plasmid was isolated and electroporated into E. coli S17-1 for conjugal transfer. Both E. coli and P. syringae were grown individually overnight on plates, then mated overnight. Initial transformants were isolated on KB plates containing rifampin, kanamycin and tetracycline. Deletion mutants were selected that were kanamycin resistant but tetracycline sensitive. Deletions were confirmed by PCR amplification, which verified that the kanamycin cassette had replaced flgK.
Deletion and Complementation of flgM
A markerless flgM deletion strain was constructed using an overlap extension PCR protocol similar to that of other studies [73]. Regions of approximately one kilobase of sequence upstream and downstream of flgM were amplified using 5′-S-flgM, 5′-AS-flgM, 3′-S-flgM, and 3′-AS-flgM. These fragments were combined with a kanamycin resistance cassette flanked by FLP recombinase target sites (kan-FRT) amplified from pKD13 [74] using primers pKD-13-site-4-AS and pKD4/13-site-1-S. The upstream, downstream, and kan-FRT fragments were combined into a single PCR reaction which was cycled for 15 amplification cycles in the absence of primers, followed by 20 amplification cycles with 5′-S-flgM and 3′-AS-flgM primers added. The resulting fragment was cloned into the suicide vector pTOK2T [29], creating pflgM-KO, which was transformed into the E. coli mating strain, S17-1. S17-1 harboring pflgM-KO was mated with P. syringae B728a overnight on KB, followed by transfer onto KB amended with kanamycin and NFT (counter selection for E. coli). Single colonies were streaked onto KB amended with either rifampicin and kanamycin or rifampacin and tetracyline. Colonies displaying kanamycin resistance and tetracycline sensitivity were screened for replacement of flgM with the kanamycin resistance cassette using PCR with the external and internal primer sets, 5′-S-flgM +3′-AS-flgM and flgM-coding-S+flgM-coding-AS, respectively. A spectinomycin-resistant derivative of pFLP2 [66], [75] was electroporated into the kanamycin resistant, ΔflgM. pFLP2-containing colonies were screened for kanamycin sensitivity. A single kanamycin sensitive isolate was grown in KB broth over night in the absence of selection, yielding a markerless ΔflgM strain.
To construct p519Mcomp, p519nGFP was digested with XbaI and EcoRI, followed by treatment with T4 DNA polymerase (to create blunt ends). The digested/blunted vector was ligated to a PCR fragment containing flgM with its promoter region, amplified with flgM-5′-S-complement and flgM-3′-AS-complement. p519empty was constructed by self-ligating the digested/blunted vector.
Plant Assays
Three week old bean plants (Phaseolus vulgaris cv. Blue Lake 274) were acclimated to 30°C for 48 hours on a long day (16 hours light/8 hours dark) light regime prior to inoculation. Bacterial strains were grown over night at 30°C on KB to suppress motility. Cultures were resuspended in 10 mM KPO4 and diluted to a final concentration of 106 CFU/mL in sterile, distilled water. Cultures were spray inoculated onto plants until dripping, and bagged to maintain high relative humidity. Inoculated plants were incubated at either 20°C or 30°C for 6 hours, prior to being unbagged and moved to a common incubation area (28°C, 50–60% relative humidity) for an additional six hours of incubation. At the time of transfer, condensation was present on the interior surfaces of plant bags, indicating near saturating humidity under both temperature treatments. For each treatment three primary leaves were sampled immediately after inoculation, and ten primary leaves were sampled immediately after the plants were un-bagged and again six hours later. Harvested leaves were sonicated in washing buffer (100 mM KPO4 buffer, 0.1% peptone) for 5 minutes, followed by dilution plating onto KB plates amended with rifampicin and natamycin. Bacterial population size was normalized for leaf surface area determined by analysis of digital images of leaves using ImageJ software [76]. The log death value was calculated as the difference between the log-transformed, leaf-area normalized population size determined immediately after the plants were un-bagged and those at the conclusion of the experiment.
Supporting Information
Temperature-dependent biosurfactant production. Area of biosurfactant-coverage on agar plates produced by either wild type Pseudomonas syringae B728a (grey bars) or a ΔflgM mutant (white bars) when grown at either 20°C or 30°C. The vertical bars represent the standard deviation of the mean.
(TIF)
Thermo-regulated genes in P. syringae.
(CSV)
Functional categories of thermo-regulated genes.
(DOCX)
Bacterial strains and plasmids.
(DOCX)
Primers used in cloning and qRT-PCR.
(DOCX)
Acknowledgments
We are grateful to Gwyn Beattie for providing the gene functional category assignments for P. syringae. We thank Ali Irani for technical assistance with the construction, complementation, and analysis of the flgM mutant. We thank Konstantinos Billis at the Joint Genome Institute for determining gene read counts.
Funding Statement
Support for KH provided by the Hildebrand-Laumeister fund for research in plant pathology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rouse DI, Nordheim EV, Hirano SS, Upper CD (1985) A Model relating the probability of foliar disease incidence to the population frequencies of bacterial plant-pathogens. Phytopathology 75: 505–509. [Google Scholar]
- 2. Hirano SS, Upper CD (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae - a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64: 624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monier JM, Lindow SE (2004) Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl Environ Microbiol 70: 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schonherr J (2006) Characterization of aqueous pores in plant cuticles and permeation of ionic solutes. J Exp Bot 57: 2471–2491. [DOI] [PubMed] [Google Scholar]
- 6. Haefele DM, Lindow SE (1987) Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae . Appl Environ Microbiol 53: 2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hattermann DR, Ries SM (1989) Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology 79: 284–289. [Google Scholar]
- 8. Panopoulos NJ, Schroth MN (1974) Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola . Phytopathology 64: 1389–1397. [Google Scholar]
- 9. Schreiber KJ, Desveaux D (2011) AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol 80: 364–377. [DOI] [PubMed] [Google Scholar]
- 10. Quinones B, Dulla G, Lindow SE (2005) Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae . Mol Plant Microbe Interact 18: 682–693. [DOI] [PubMed] [Google Scholar]
- 11. Dechesne A, Wang G, Gulez G, Or D, Smets BF (2010) Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci USA 107: 14369–14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macnab RM (1996) Flagella and Motility. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd ed. Washington, D.C.: ASM Press. 123–145.
- 13. Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S (2007) Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa . J Bacteriol 189: 3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tart AH, Blanks MJ, Wozniak DJ (2006) The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188: 6483–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caiazza NC, Merritt JH, Brothers KM, O’Toole GA (2007) Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189: 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindeberg M, Cunnac S, Collmer A (2009) The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol 10: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danhorn T, Fuqua C (2007) Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61: 401–422. [DOI] [PubMed] [Google Scholar]
- 18. Smirnova A, Li HQ, Weingart H, Aufhammer S, Burse A, et al. (2001) Thermoregulated expression of virulence factors in plant-associated bacteria. Arch Microbiol 176: 393–399. [DOI] [PubMed] [Google Scholar]
- 19. Kearns DB (2010) A field guide to bacterial swarming motility. Nat Rev Microbiol 8: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burch AY, Shimada BK, Browne PJ, Lindow SE (2010) Novel High-Throughput Detection Method To Assess Bacterial Surfactant Production. Appl Environ Microbiol 76: 5363–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berti AD, Greve NJ, Christensen QH, Thomas MG (2007) Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J Bacteriol 189: 6312–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, et al. (2003) A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa . Mol Microbiol 50: 809–824. [DOI] [PubMed] [Google Scholar]
- 23. Choy W-K, Zhou L, Syn CK-C, Zhang L-H, Swarup S (2004) MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J Bacteriol 186: 7221–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aldridge P, Hughes KT (2002) Regulation of flagellar assembly. Curr Opin Microbiol 5: 160–165. [DOI] [PubMed] [Google Scholar]
- 25. Kitten T, Kinscherf TG, McEvoy JL, Willis DK (1998) A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae . Mol Microbiol 28: 917–929. [DOI] [PubMed] [Google Scholar]
- 26. Leplae R, Lima-Mendez G, Toussaint A (2010) ACLAME: A CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res 38: D57–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Records AR, Gross DC (2010) Sensor Kinases RetS and LadS Regulate Pseudomonas syringae Type VI Secretion and Virulence Factors. J Bacteriol 192: 3584–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen CL, Beattie GA (2007) Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J Bacteriol 189: 6901–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen CL, Malek AA, Wargo MJ, Hogan DA, Beattie GA (2010) The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol 75: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C, Beattie GA (2008) Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J Bacteriol 190: 2717–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wargo MJ, Szwergold BS, Hogan DA (2008) Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190: 2690–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matilla MA, Ramos JL, Duque E, Alche JD, Espinosa-Urgel M, et al. (2007) Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida . Environ Microbiol 9: 1842–1850. [DOI] [PubMed] [Google Scholar]
- 33. Kuiper I, Lagendijk EL, Pickford R, Derrick JP, Lamers GEM, et al. (2004) Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol 51: 97–113. [DOI] [PubMed] [Google Scholar]
- 34. Dubern JF, Bloemberg GV (2006) Influence of environmental conditions on putisolvins I and II production in Pseudomonas putida strain PCL1445. FEMS Microbiol Lett 263: 169–175. [DOI] [PubMed] [Google Scholar]
- 35. Dubern JF, Lagendijk EL, Lugtenberg BJJ, Bloemberg GV (2005) The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J Bacteriol 187: 5967–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soutourina OA, Bertin PN (2003) Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27: 505–523. [DOI] [PubMed] [Google Scholar]
- 37. Horne SM, Pruss BM (2006) Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch Microbiol 185: 115–126. [DOI] [PubMed] [Google Scholar]
- 38. Rohde JR, Fox JM, Minnich SA (1994) Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol 12: 187–199. [DOI] [PubMed] [Google Scholar]
- 39. Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA (1996) Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol 19: 1061–1071. [DOI] [PubMed] [Google Scholar]
- 40. Kapatral V, Campbell JW, Minnich SA, Thomson NR, Matsumura P, et al. (2004) Gene array analysis of Yersinia enterocolitica FlhD and FlhC: regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiology 150: 2289–2300. [DOI] [PubMed] [Google Scholar]
- 41. Wosten M, van Dijk L, Veenendaal AKJ, de Zoete MR, Bleumink-Pluijm NMC, et al. (2010) Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol Microbiol 75: 1577–1591. [DOI] [PubMed] [Google Scholar]
- 42. Alm RA, Guerry P, Trust TJ (1993) The Campylobacter sigma 54 flaB flagellin promoter is subject to environmental regulation. J Bacteriol 175: 4448–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinden SL, Devay JE, Backman PA (1971) Properties of syringomycin, a wide spectrum antibiotic and phytotoxin produced by Pseudomonas syringae, and its role in bacterial canker disease of peach trees. Physiological Plant Pathology 1: 199-&.
- 44. Lu SE, Wang N, Wang JL, Chen ZJ, Gross DC (2005) Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol Plant Microbe Interact 18: 324–333. [DOI] [PubMed] [Google Scholar]
- 45. Lu SE, Scholz-Schroeder BK, Gross DC (2002) Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol Plant Microbe Interact 15: 43–53. [DOI] [PubMed] [Google Scholar]
- 46. Li HQ, Schenk A, Srivastava A, Zhurina D, Ullrich MS (2006) Thermo-responsive expression and differential secretion of the extracellular enzyme levansucrase in the plant pathogenic bacterium Pseudomonas syringae pv. glycinea. FEMS Microbiol Lett 265: 178–185. [DOI] [PubMed] [Google Scholar]
- 47. Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ (2004) Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186: 4466–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ (2006) Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188: 8213–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lindeberg M, Myers CR, Collmer A, Schneider DJ (2008) Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol Plant Microbe Interact 21: 685–700. [DOI] [PubMed] [Google Scholar]
- 50. Laue H, Schenk A, Li H, Lambertsen L, Neu TR, et al. (2006) Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae . Microbiology 152: 2909–2918. [DOI] [PubMed] [Google Scholar]
- 51. Karatan E, Watnick P (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73: 310–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jacques MA, Josi K, Darrasse A, Samson R (2005) Xanthomonas axonopodis pv. phaseoli var. fuscans is aggregated in stable biofilm population sizes in the phyllosphere of field-grown beans. Appl Environ Microbiol 71: 2008–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Monier JM, Lindow SE (2003) Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc Natl Acad Sci USA 100: 15977–15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rivero RM, Ruiz JM, Romero LM (2004) Importance of N source on heat stress tolerance due to the accumulation of proline and quaternary ammonium compounds in tomato plants. Plant Biol (Stuttg) 6: 702–707. [DOI] [PubMed] [Google Scholar]
- 55. Fitzsimmons LF, Hampel KJ, Wargo MJ (2012) Cellular choline and glycine betaine pools impact osmoprotection and phospholipase C production in Pseudomonas aeruginosa. J Bacteriol 194: 4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raymundo AK, Ries SM (1981) Motility of Erwinia amylovora . Phytopathology 71: 45–49. [Google Scholar]
- 57. Wei ZM, Sneath BJ, Beer SV (1992) EXPRESSION OF ERWINIA-AMYLOVORA HRP GENES IN RESPONSE TO ENVIRONMENTAL STIMULI. J Bacteriol 174: 1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gaudriault S, Malandrin L, Paulin JP, Barny MA (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol 26: 1057–1069. [DOI] [PubMed] [Google Scholar]
- 59. Kelm O, Kiecker C, Geider K, Bernhard F (1997) Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol Gen Genet 256: 72–83. [DOI] [PubMed] [Google Scholar]
- 60. Bereswill S, Jock S, Aldridge P, Janse JD, Geider K (1997) Molecular characterization of natural Erwinia amylovora strains deficient in levan synthesis. Physiological and Molecular Plant Pathology 51: 215–225. [Google Scholar]
- 61. Young JM (1991) Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. syringae Van Hall 1902. Ann Appl Biol 118: 283–298. [Google Scholar]
- 62. Budde IP, Ullrich MS (2000) Interactions of Pseudomonas syringae pv. glycinea with host and nonhost plants in relation to temperature and phytotoxin synthesis. Mol Plant Microbe Interact 13: 951–961. [DOI] [PubMed] [Google Scholar]
- 63. Loper JE, Lindow SE (1987) Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77: 1449–1454. [Google Scholar]
- 64. King EO, Ward MK, Raney DE (1954) 2 simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- 65. Simon R, Priefer U, Puhler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio-Technology 1: 784–791. [Google Scholar]
- 66. Burch AY, Shimada BK, Mullin SWA, Dunlap CA, Bowman MJ, et al. (2012) Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J Bacteriol 194: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rutledge RG, Stewart D (2008) A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dulla G (2007) Bacterial babel: breaking down quorum sensing cross-talk in the phyllosphere; analysis of the contributions of abiotic and biotic factors on AHL-mediated quorum sensing to epiphytic growth and virulence in Pseudomonas syringae. Berkeley: University of California, Berkeley, CA.
- 72. Marco ML, Legac J, Lindow SE (2005) Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ Microbiol 7: 1379–1391. [DOI] [PubMed] [Google Scholar]
- 73. Choi KH, Schweizer HP (2005) An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86. [DOI] [PubMed] [Google Scholar]
- 76. Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image Processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- 77. Castillo-Davis CI, Hartl DL (2003) GeneMerge - post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891–892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Temperature-dependent biosurfactant production. Area of biosurfactant-coverage on agar plates produced by either wild type Pseudomonas syringae B728a (grey bars) or a ΔflgM mutant (white bars) when grown at either 20°C or 30°C. The vertical bars represent the standard deviation of the mean.
(TIF)
Thermo-regulated genes in P. syringae.
(CSV)
Functional categories of thermo-regulated genes.
(DOCX)
Bacterial strains and plasmids.
(DOCX)
Primers used in cloning and qRT-PCR.
(DOCX)