Abstract
Activation of KIT, by its ligand, stem cell factor (SCF), results in the initiation of signal transduction pathways that influence mast cell survival and proliferation. Activating mutations in KIT have thus been linked to clonal MC proliferation associated with systemic mastocytosis. SCF also modulates MC function by inducing MC chemotaxis and by potentiating antigen (Ag)/IgE-mediated MC degranulation. Thus, mutations in KIT also have the potential to affect these processes in allergic and other mast cell-related diseases. Studies to determine how native and mutated KIT may modulate MC chemotaxis and activation have, however, been limited due to the lack of availability of a suitable functional MC line lacking native KIT which would allow transduction of KIT constructs. Here we describe a novel mouse MC line which allows the study of normal and mutated KIT constructs. These cells originated from a bone marrow-derived mouse MC culture out of which a rapidly dividing mast cell sub-population spontaneously arose. Over time, these cells lost KIT expression while continuing to express functional high affinity receptors for IgE (FcεRI). As a consequence, these cells degranulated in response to Ag/IgE but did not migrate nor show any evidence of potentiation of Ag/IgE degranulation in response to SCF. Retroviral transduction of the cells with a human (hu)KIT construct resulted in surface expression of huKIT which responded to huSCF by potentiation of Ag/IgE-induced degranulation and chemotaxis. This cell line thus presents a novel system to delineate how MC function is modulated by native and mutated KIT and for the identification of novel inhibitors of these processes.
Keywords: Degranulation, FcεRI, KIT, Mast Cells, KIT, SCF
1. Introduction
MCs are terminally differentiated cells of hematopoietic origin that contribute to both innate and acquired immune responses (Galli and Tsai, 2010). Mature MCs express a wide array of surface receptors which recognize exogenous and endogenous factors that regulate MC growth, development, homing, homeostasis, and function (Rivera and Gilfillan, 2006; Bischoff, 2007; Brown et al., 2008). The major receptor regulating MC responses associated with acquired immune reactions is the FcεRI, whereas TLRs and NOD receptors are likely the major regulators of MC-dependent innate responses (Gilfillan and Beaven, 2011). Initiation of signaling following Ag/IgE-induced FcεRI aggregation results in degranulation, arachidonic acid metabolism and cytokine/chemokine gene expression leading to the generation and/or release of inflammatory mediators (Galli and Tsai, 2012). In contrast, TLRs and NOD receptors respond to bacterial and viral components to induce cytokine production in the absence of degranulation (Marshall, 2004). Hence, MC responses elicited through the various classes of receptors are both discrete and overlapping (Gilfillan and Beaven, 2011).
The receptor with tyrosine kinase activity family member KIT, is the major receptor regulating the growth, development and homeostasis of MCs (Okayama and Kawakami, 2006). However, following cross linking with its ligand, stem cell factor (SCF), KIT may additionally play a role in MC homing (Jensen et al., 2008), and potentiation of Ag-mediated MC activation. Activating mutations in KIT are associated with specific disease states such as mastocytosis (D816V, and potentially others) and gastrointestinal stromal cell tumors (GISTS) (Antonescu, 2008). In the case of mastocytosis, this is linked to dysregulated MC growth and/or survival and an increased incidence of anaphylaxis (Brockow and Metcalfe, 2010). Therefore, understanding how native and mutated KIT modifies MC processes has been, and remains, an active area of research. These efforts, however, have been hampered by the lack of functional MCs which lend themselves to transduction with genetically modified KIT. For example, the manifestations of mutations in KIT have often been examined in KIT-transduced non-mast cell lines (Gleixner et al., 2007; Xiang et al., 2007; Bougherara et al., 2009; Sun et al., 2009) or in cells bearing KIT mutations (Aichberger et al., 2009; Bai et al., 2012) which do not degranulate nor produce other MC-associated responses. Additionally, these cells may not express all of the signaling molecules required for the ability of KIT to modulate MC responses. Examining the manifestations of ectopically expressed KIT in MCs may also be complicated by the expression of endogenous KIT that may dominantly compete for resident signaling molecules. Thus, there is a need for functional MC lines which do not express native KIT and which would facilitate the examination of manifestations of human (hu)KIT.
Here we describe a novel mouse MC line which possesses such characteristics. A rapidly dividing mouse MC population expanded from a differentiated bone marrow-derived mouse MC (BMMC) culture. Over time, these cells lost the ability to express endogenous mouse (m)KIT while still expressing functional FcεRI. These cells were readily transducible with huKIT and the transduced cells responded in a predicted manner to huSCF. Thus, this cell line represents a unique system in which to examine the role of wild type (WT) and mutated KIT on MC function and furthermore may be useful for screening potential novel therapeutic KIT inhibitors.
2. Material and Methods
2.1. Mice and BMMCs
C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME). The mTOR knock-in mice, from which the parental BMMC culture was derived, were generated as described (Zhang et al., 2011). All mice were treated in compliance with protocols approved by the Animal Care and Use Committee of the National Institutes of Health (NIH). BMMCs were developed from mouse bone marrows as described (Kuehn et al., 2008). CD34+ peripheral blood progenitors, isolated from healthy donors, were used to prepare huMCs as described (Kirshenbaum et al., 1999). The donors provided an informed consent, and cells were obtained under a protocol (NCT00001756) approved by the Institutional Review Board of The National Institute of Allergy and Infectious Diseases.
2.2. Antibodies and reagents
Anti-DNP-IgE (clone SPE-7) and DNP-HSA were obtained from Sigma-Aldrich (St. Louis, MO). HuSCF, mouse (m)SCF and mIL-3 were from Peprotech (Rocky Hill, NJ). The protein-specific antibodies were: anti-KIT (Cell Signaling Technology, Beverley, MA), anti-MC tryptase (Abcam, Cambridge, MA), and anti-β-actin (Sigma-Aldrich, St. Louis, MO; Cell Signaling Technology). The conjugated protein-specific antibodies were: fluorophore-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), horseradish peroxidase-conjugated rabbit IgG (Amersham Biosciences, Piscataway, NJ) and mouse IgG Fc-specific antibody (Sigma-Aldrich). The phosphoprotein-specific antibodies: anti-phospho-KIT(Tyr823) and anti-phospho-PLCγ1(Tyr783) antibodies were from Invitrogen (Carlsbad, CA). Other phosho-specific antibodies were from Cell Signaling Technology. Other reagents used were: donkey serum (Santa Cruz Biotechnology, Santa Cruz, CA), imatinib (LC Laboratories, Woburn, MA), Fura2-AM (Invirogen), and all other chemicals were from Sigma-Aldrich.
2.3. HuKIT cloning and expression
The WT KIT gene was cloned from CD34+ peripheral blood mononuclear cells and inserted into the pcDNATM3.1 expression vector (Invitrogen, Carlsbad CA) using standard molecular biology techniques. After verifying the correct sequence, the KIT gene was then subcloned into the retroviral vector pMX-puro (Cell BioLabs, San Diego, CA). The empty or huKIT gene-containing pMX-puro, was transfected into PLAT-E cells (Cell Biolabs, San Diego, CA) and virus produced as described (Ito et al., 2012). MCs were transduced by culturing the filtered virus-containing medium (1:1) with a suspension of the target cells (0.03–2 × 106) for 3 days. The transduced cells were selected by culturing in medium containing puromycin (Sigma Aldrich) at 3.0–4.0 μg/ml, a concentration we found to be lethal for non-transduced MCBS1 MCs. The experiments were then conducted after 2 weeks in culture.
2.4. Quantitative real-time PCR
Total RNA was isolated using QIAshredder in combination with RNase-Free DNase Set and RNeasy Mini Kit (Qiagen, Valencia, CA). The isolated RNA was transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen) and gene expression was determined as described (Smrž et al., 2011).
2.5. Cell activation, degranulation and cytokine production
The cells were sensitized with mouse DNP-specific IgE (100 ng/ml) overnight in cytokine-free media. For degranulation studies, the cells were harvested, then activated as described (Smrž et al., 2010). The percentage of β-hexosaminidase released into the supernatant (Choi et al., 1993) after 30 min activation was used to determine cell degranulation. To determine cytokine production in activated cells, the sensitized and starved cells were washed with cytokine-free media then stimulated with 2× concentrated activator added in 1:1 ratio to the cell suspension. The concentration of cytokines released by 1.0 × 106/ml cells into the supernatant after 6 h stimulation at 37 °C / 5 % CO2 was determined by Quantikine ELISA kits (R&D Systems, Minneapolis, MN).
2.6. Immunoblotting, flow cytometry and calcium response
Cell lysates were prepared, analyzed by immunoblotting, and immunoblots evaluated as described (Smrž et al., 2010). To determine cell viability, the cells were stained with annexin V and propidium iodide as described (Smrž et al., 2007). To determine surface expression of huKIT, mKIT, FcεRI or FcεRI-bound IgE, the cells were stained with the corresponding PE-labeled (huKIT or mKIT-specific) or FITC-labeled (mFcεRI and mIgE-specific) antibodies. Cell staining was determined by flow cytometry using FACSCalibur with CellQuest 3.3 software (BD Biosciences). The acquired data were analyzed and processed by FlowJo 7.6 (TreeStar). To determine changes in levels of intracellular calcium during activation, the sensitized cells were treated and analyzed as described (Tkaczyk et al., 2003).
2.7. Chemotactic purification
Chemotactic purification of the huKIT-transduced cells, driven by huSCF-elicited migration, was performed using Transwell® permeable supports with 5.0 μm pore size membranes in 24-well plates (Costar®) (Corning, Corning, NY). Cells were washed with culture medium and 100 μl cell suspensions (0.5–4 × 106 cells/ml) were transferred to the upper chambers and these chambers placed over the bottom chambers containing 600 μl culture medium. Following 30 min incubation at 37 °C / 5 % CO2, the upper chambers were replaced over the bottom chambers containing 600 μl culture medium with or without huSCF (10 ng/ml). After 20 min incubation at 37 °C / 5 % CO2, the upper chambers were removed and the migrated cells in the bottom chambers harvested. The cells were washed to remove huSCF before expansion in culture.
2.8. Toluidine blue staining and confocal microscopy
To visualize MC granules by metachromatic staining, cytospins were prepared, fixed and stained with toluidine blue as described (Kirshenbaum and Metcalfe, 2006). To visualize MC tryptase by confocal microscopy, the cells were washed with PBS, mixed 1:1 with [40 mM EDTA, 40 mM EGTA, PBS], then transferred onto poly-lysine (100 μg/ml)-coated slides positioned in a 24-well plate and the cells allowed to attach for 5 min. The cells were then fixed with 1 ml [4% (w/v) paraformaldehyde, 5 mM EDTA, 5 mM EGTA, PBS] for 30 min, washed with PBS and permeabilized [with 0.1 % (v/v) Triton-X100, 5 % (w/v) BSA, 2 mM EGTA, PBS] for 5 min, washed with PBS, then incubated with [5 % (w/v) BSA, 2 mM EGTA, PBS] for 30 min. The solution was removed and the cells stained with mouse MC tryptase-specific Ab in [5 % (w/v) BSA, 2 mM EGTA, PBS supplemented with 200 x diluted donkey serum] for 1 h. The cells were extensively rinsed with PBS and [5 % (w/v) BSA, 2 mM EGTA, PBS], then stained with donkey mouse IgG-specific Alexa488-conjugated secondary antibody [in 5 % (w/v) BSA, 2 mM EGTA, PBS supplemented with 200 x diluted donkey serum] for 1 h. The cells were washed with [5 % (w/v) BSA, 2 mM EGTA, PBS], then PBS, and stained with DAPI in PBS for 10–20 min. The slides were extensively washed with PBS and then mounted on support slides using Prolong® Gold anti-fade reagent (Invitrogen). The images were acquired on Leica SP5 confocal microscope (Leica Microsystems, Exton, PA) using 63× oil immersion objective with numerical aperture 1.4. The images were deconvolved using Huygens Essential software (Scientific Volume Imaging BV, Hilversum, The Netherlands). The Imaris software (Bitplane AG, Zurich, Switzerland) was used to process the deconvolved images and to create 3D volumetric surface models via a semiautomatic surface rendering module.
2.9. Statistical analysis
Means and SEM were computed from the number of experiments performed or the number of independent transfectants. The statistical significance between 2 groups was determined by unpaired Student t test. One-way ANOVA with the Tukey test was used to determine statistical significance among multiple groups. When P<0.05, the data were considered significant.
3. Results
3.1. Development and characteristics of a KIT-negative mouse bone marrow-derived MC line
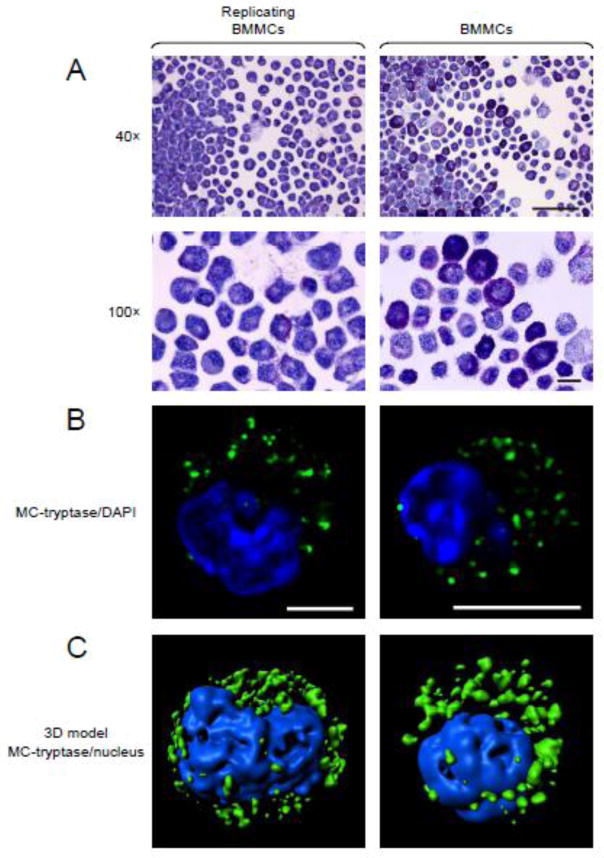
We detected a rapidly proliferating MC population that arose from one of many BMMC cultures derived from mTOR knock-in mice (Zhang et al., 2011) which had partially disrupted mTOR transcription and which were previously utilized to examine the role of mTOR complexes on MC homeostasis (Smrž et al., 2011). This population has now been maintained in culture for more than 30 months and has gone through multiple cryopreservation/reconstitution procedures. The doubling time of the cells was less than 24 h (2.57 ± 0.09 fold increase in 24 h; SEM, n=3) and the appearance of the cells following toluidine blue staining was comparable to that of regular 4–6 week old terminally differentiated, non-dividing BMMCs (Figure 1A). The similarity to BMMCs was also evident from the presence and distribution of MC tryptase as demonstrated on optical sections of deconvolved confocal images (Figure 1B) or the corresponding 3D volumetric surface models created out of these images (Figure 1C).
Fig. 1.
Morphology of the replicating BMMCs (subsequently named MCBS1 MCs). (A) Cytospins of these cells BMMCs (right) and regular 4–6 week old terminally differentiated, non-dividing BMMCs (left) were stained with toluidine blue. Scale bars represent 50 μm (40×) or 10 μm (100×). (B) Replicating BMMCs (right) and 4–6 week old terminally differentiated, non-dividing BMMCs (left) were stained with MC-tryptase-specific, then with Alexa488-labeled secondary, antibodies (green) and cell sections were visualized by confocal microscopy and acquired images deconvolved. The nucleus was visualized by DAPI (blue) and scale bars represent 5 μm. (C) Deconvolved images from multiple sections acquired in B were used to create 3D volumetric surface models of the stained cells. In A-C, representative images from 3 sample preparations (replicating BMMCs) or cells from 3 mice (regular non-dividing BMMCs) are shown.
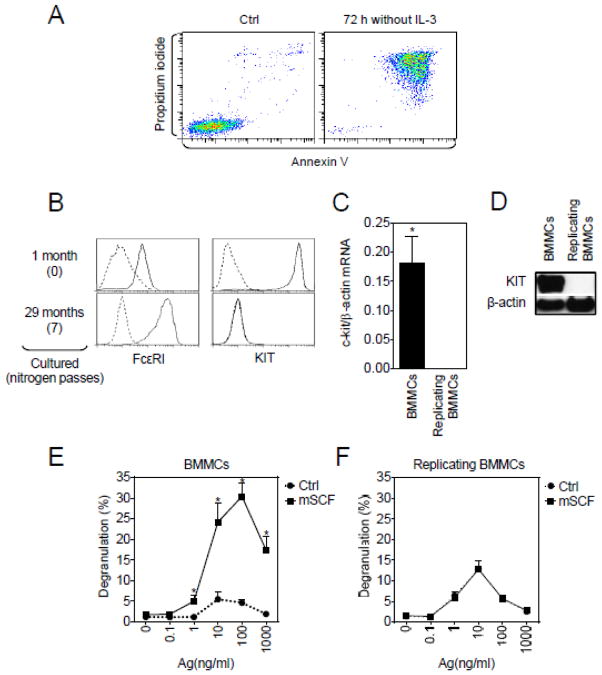
Regardless of the ability to continue to divide and survive in culture, as for normal non-replicating BMMCs, such survival was IL-3-dependent in that removal of IL-3 from the culture medium resulted in a significant increase in annexin V/propidium iodide positive cells (Figure 2A). Although initially the replicating BMMCs expressed both native KIT and FcεRI, long term culture and repeated cryopreservation/reconstitution procedures resulted in a loss of KIT on the cell surface, whereas the expression of FcεRI was greater in these cells (Figure 2B). The loss of surface expression of KIT from these cells was not a consequence of internalization or retention in cytoplasmic compartments, as quantitative real-time PCR and immunoblot analysis revealed lack of total cellular KIT expression (Figure 2C and 2D).
Fig. 2.
Functional analysis of the replicating BMMCs (MCBS1 MCs). (A) Cells were starved, or not (Ctrl), of IL-3 for 72 h, then labeled with annexin V and propidium iodide and analyzed by flow cytometry. (B) Surface expressions of FcεRI and mouse KIT in the initial 1 month and subsequent cultures these cells were determined by flow cytometry. Nitrogen passes indicates number of nitrogen cryopreservation cycles. (C) Expression of KIT mRNA (c-kit) was determined and compared with expression of β-actin. Regular 4–6 week old terminally differentiated, non-dividing BMMCs were used as a positive control. (D) Immunoblot analysis of the replicating BMMCs compared to the regular non-dividing BMMCs. Non dividing (E) or replicating BMMCs (F) were sensitized with anti-DNP-IgE overnight, then activated with DNP-HSA (Ag) alone or in combination with mSCF (100 ng/ml) for 30 min and degranulation was determined. In A, B and D, data of representative experiments performed from 3 independent sample preparations (replicating BMMCs) or cells from 3 mice (regular non-dividing BMMCs) are shown. In C E and F, the data represent means and SEM (replicating BMMCs: n=3 sample preparations; regular non-dividing BMMCs: n=cell preparations from 3 mice) and differences between the cell types (C) or cells co-stimulated with or without mSCF (E, F) are indicated (*; P<0.05, Student’s t-test).
Although SCF on its own does not induce degranulation, when added in conjunction with Ag, it results in a markedly enhanced degranulation (Hundley et al., 2004; Tkaczyk et al., 2004) and Figure 2E. Consistent with the lack of native KIT expression, we observed that mSCF had no effect on degranulation either in the absence or presence of Ag (Figure 2F).
As a whole, these results describe an immortal functional mouse IL-3 dependent MC line that does not express native KIT. From here on we have termed these cells MCBS1 to define the laboratory of origin (Mast Cell Biology Section within the Laboratory of Allergic Diseases, NIAID,NIH).
3.2. Reconstitution of MCBS1 MCs with huKIT
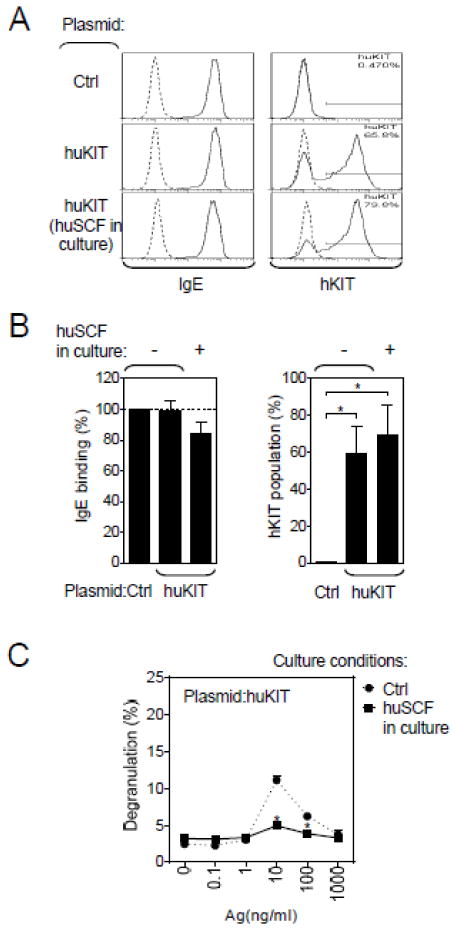
Having established a replicating mouse MC line that does not express native KIT, we next determined whether we could use the cell line to examine the functional consequences of huKIT expression. We therefore cloned the gene for huKIT into a retroviral expression system, transduced MCBS1 MCs with the corresponding vector and then selected the transduced cells by culturing in puromycin. MCBS1 MCs transduced with the empty vector were used as a control. Utilizing this approach, we determined that we could achieve, on average, 60% efficiency (range: 30–80%) after two weeks puromycin selection (Figure 3A and 3B) and that transduction of huKIT had no effect on FcεRI expression. However, huKIT expression was slightly increased when the culture medium was subsequently supplemented with huSCF for 2 weeks (Figure 3A and 3B).
Fig. 3.
Transduction of MCBS1 MCs with huKIT. (A) MCBS1 MCs were transfected with a retroviral plasmid containing huKIT (Plasmid:huKIT), selected by puromycin resistance, sensitized with anti-DNP-IgE overnight, and surface binding of IgE and huKIT surface expression was determined by flow cytometry. (B) Normalized IgE binding and the content of huKIT-expressing cell population in A (Plasmid:huKIT) was evaluated alongside with cells transfected with empty plasmid (Plasmid:Ctrl) or the huKIT transfected cells subsequently cultured in the presence of huSCF (100 ng/ml) for over 2 weeks. (C) The IgE-sensitized huKIT-transduced MCBS1 MCs cultured in the presence or absence (Ctrl) of huSCF (100 ng/ml) for over 2 weeks were activated with DNP-HSA (Ag) for 30 min and degranulation was determined. In A, data are representative experiments performed from 3 independent transfections. In B and C, data represents means and SEM (n=3 transfections) and differences among cells transfected with empty plasmid, huKIT-containing plasmid, or huKIT-containing plasmid then cultured in the presence of huSCF (B;1-way ANOVA) or between cells cultured in the presence or absence of huSCF (C; Student’s t-test) are indicated (*; P<0.05).
We have shown that prolonged exposure of mouse MCs to mSCF induces a hypo-responsive phenotype with regards to Ag-mediated degranulation (Ito et al., 2012). To determine whether the same phenotype is induced via the transduced huKIT in MCBS1 MCs, we analyzed the cells after 2 week culture in the presence or absence of huSCF. As shown in Figure 3C, the huKIT-transduced MCBS1 MCs were similarly rendered hyporesponsive to Ag following prolonged exposure to huSCF. Taken together, these results showed that the MCBS1 MC line transduced with huKIT produced the predicted outcome when challenged through the ectopically expressed receptor.
3.3. Chemotactic purification
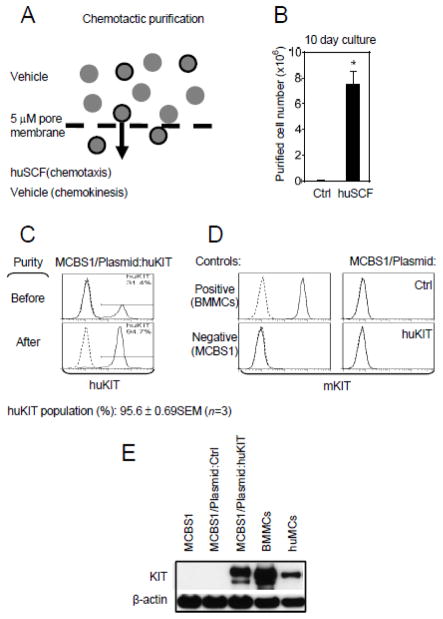
In view of the 60–70 % transduction efficiency of huKIT in MCBS1 MCs as noted above, further purification of huKIT-positve cells seemed desirable to reliably analyze the performance of huKIT in batch assays such as immunoblot analyses and assays for degranulation, calcium response and cytokine production. To achieve this goal, a strategy was devised based on the functional properties of huKIT. Accordingly, we examined whether the ability of SCF to induce MC chemotaxis (Halova et al., 2012) could be used for this purpose (Figure 4A). We utilized a two chamber-based system routinely used in migration assays (Kuehn et al., 2010; Rådinger et al., 2010) with huSCF as the chemoattractant. To avoid the SCF-mediated induction of hyporesponsive phenotype described above, we shortened migration to 20 min and washed the migrated cells to remove huSCF prior to expansion in culture. As shown in Figure 4B, numerous cells migrated in response to huSCF and expanded in culture thereafter whereas only a minimum number of cells were recovered in the chamber without chemoattractant. The fraction of cells expressing huKIT on the cell surface following purification by this procedure was found to be substantially enhanced, reaching over 95 % (Figure 4C). As for the non-transduced cells, the huKIT-transduced and chemotactically-purified MCBS1 MCs were negative for the presence of endogenous mKIT as determined by its absence on cell surface (Figure 4D) and in total cell lysate (Figure 4E). For all subsequent studies we utilized cells derived from cultures purified through chemotaxis. The selected, transduced cells have been used for up to a month by keeping them under puromycin selection to prevent expansion of the cells with low/no KIT expression. Furthermore, the transduced cells can be frozen and reconstituted providing that the cells are maintained in puromycin. Re-purification by chemotaxis is however recommended following expansion of the cells from frozen stocks.
Fig. 4.
Chemotactic purification of huKIT-transduced MCBS1 MCs. (A) Schematic diagram. (B) Purified huKIT-transfectants, derived as shown in A, were cultured for 10 days and the number of cells recovered was determined. The cells subjected to chemotactic purification driven towards vehicle alone (chemokinesis) were used as a negative control (Ctrl). (C) The content of huKIT-expressing populations of the purified transfectants in A was evaluated by flow cytometry. (D) Surface expression of mKIT in non-transduced MCBS1 MCs (MCBS1), empty plasmid transfected (MCBS1/Plasmid:Ctrl), or chemotactically purified huKIT-transduced cells from A (MCBS1/Plasmid:huKIT) was determined by flow cytometry. Four to six week old cultured regular non-dividing mouse BMMCs were used as a positive control. (E) Immunoblot analysis for the presence of KIT (both mouse and human) in cells analyzed in D. Four to six week old cultured regular non-dividing mouse BMMCs were used as a positive control. In C-E, data of representative experiments performed from 3 independent sample preparations, transfections, or cells from 3 mice (regular non-dividing BMMCs) or donors (huMC) are shown. In B, data represents means and SEM (n=3 huKIT transfectants purified chemotactically) and a difference between the huSCF- and vehicle-driven purifications is indicated (*; P<0.05, Student’s t-test).
3.4. Functional analysis of human KIT transduced in MCBS1 MCs
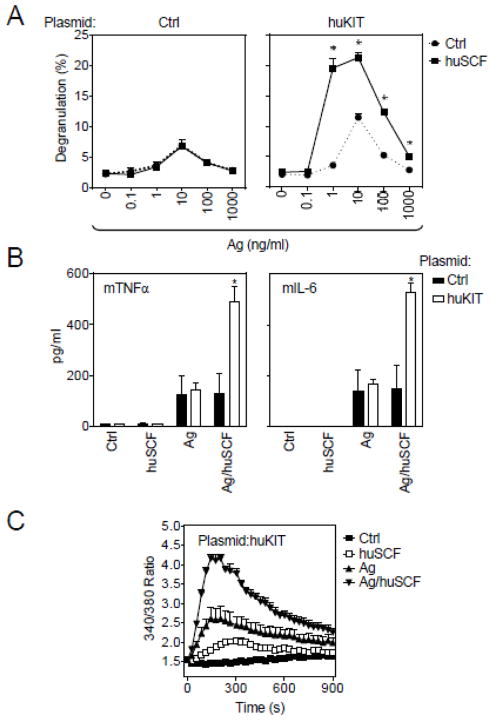
In addition to allowing purification of huKIT-transduced cells, the chemotaxis data demonstrated that transduced huKIT was functional with respect to huSCF-induced cell migration. To examine other KIT-dependent activities we tested the ability of huSCF to potentiate Ag/IgE-mediated degranulation in these cells. As for the parental cell line, the MCBS1 MCs transduced with control plasmid showed no response to huSCF in either the presence or absence of Ag challenge (Figure 5A, left panel). In contrast, in cells transduced with huKIT, huSCF markedly potentiated Ag-mediated degranulation (Figure 5A, right panel).
Fig. 5.
Degranulation, cytokine release and calcium response in purified huKIT-transduced MCBS1 MCs. (A) Three purified huKIT-transduced MCBS1 MC (Plasmid:huKIT) cultures were sensitized with anti-DNP-IgE overnight and then activated with DNP-HSA (Ag) alone or in the presence of huSCF (100 ng/ml) and degranulation after 30 min activation was determined. The cells transduced with empty plasmid (Plasmid:Ctrl) were used as a control. (B) The IgE-sensitized cells in A were activated with huSCF (100 ng/ml), DNP-HSA (Ag; 10 ng/ml), or with their combination (Ag/huSCF; 10 and 100 ng/ml, respectively), and cytokine release after 6 h activation was determined. (C) The IgE-sensitized cells in A were loaded with Fura-2, then activated with huSCF (100 ng/ml), DNP-HSA (Ag; 10 ng/ml), or with their combination (Ag/huSCF; 10 and 100 ng/ml, respectively), and calcium response was determined. In A-C, data represent means and SEM (n=3 transfections). In A, differences between cells co-stimulated with or without huSCF, and in B, between cells transduced with empty plasmid (Plasmid:Ctrl) or huKIT (Plasmid:huKIT) are indicated (*; P<0.05, Student’s t-test).
In addition to promoting chemotaxis and enhancing MC degranulation, SCF potentiates Ag-mediated MC cytokine production and calcium signaling (Hundley et al., 2004). Consistent with this report, and as shown in Figure 5B, huSCF enhanced Ag-mediated TNF-α and IL-6 production in huKIT-transfected MCBS1 MCs. Similarly, the Ag-mediated calcium signal was significantly enhanced by huSCF in these cells (Figure 5C).
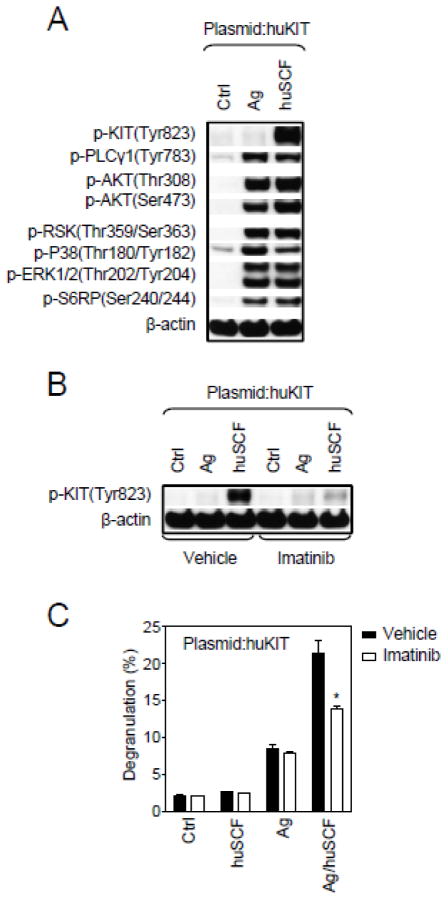
Following engagement with SCF and the initial autophosphorylation of KIT, KIT elicits a number of other critical signaling events in addition to the calcium signal (Gilfillan and Tkaczyk, 2006; Gilfillan and Rivera, 2009). From Figure 6A it can be seen that huSCF induced autophosphorylation of huKIT in the transduced MCBS1 MCs. Other critical signals including phosphorylation of PLCγ1, AKT (a surrogate marker for PI3K activation), RSK, p38, ERK1/2, and S6RP were also induced by huSCF, and to a comparable extent as Ag, in these cells (Figure 6A). Similar responses were observed in non-transduced MCBS1 cells when triggered through the FcεRI, but not when challenged with huSCF (data not shown). Taken together these data illustrate that MCBS1 MCs respond in the appropriate manner to huSCF when transduced with huKIT.
Fig. 6.
Effect of huSCF on cellular signaling and a KIT inhibitor on huSCF-mediated responses in huKIT-transduced MCBS1 MCs. (A) Purified huKIT-transduced MCBS1 MCs (Plasmid:huKIT) were sensitized with anti-DNP-IgE overnight and then activated with DNP-HSA (Ag; 10 ng/ml) or huSCF (100 ng/ml) for 2 min and protein phosphorylation assessed by immunoblot analysis of the selected signaling molecules. (B) The IgE-sensitized purified huKIT-transduced MCBS1 MCs (Plasmid:huKIT) were pre-treated with imatinib (1 μM) or vehicle alone for 20 min and then activated with DNP-HSA (Ag;10 ng/ml) or huSCF (100 ng/ml) for 2 min and tyrosine phosphorylation of KIT was analyzed by immunoblotting. (C) Sensitized purified huKIT-transduced MCBS1 MCs (Plasmid:huKIT) were again pre-treated with imatinib (1 μM) or vehicle alone for 20 min and then activated with huSCF (100 ng/ml), DNP-HSA (Ag; 10 ng/ml), or with both in combination and degranulation determined 30 min later. In A and B, representative experiments performed with 3 independent huKIT transfectants are shown. In C, data represent means and SEM (n=3 transfections) and differences between treatments are indicated (*; P<0.05, Student’s t-test).
3.5. Effect of a KIT inhibitor on huSCF-mediated responses in huKIT-transduced MCBS1 MCs
Finally, to determine the utility of this cell line in identifying potential inhibitors of huKIT, we examined the ability of imatinib (Gleevec), a recognized inhibitor of KIT catalytic activity, to inhibit huSCF-mediated huKIT phosphorylation and huSCF-enhancement of Ag-mediated degranulation. Indeed, as shown in Figure 6B and 6C, imatinib effectively inhibited these responses in the huKIT-transfected cells, thus demonstrating that these cells can be used to screen for potential inhibitors of huKIT phosphorylation and function.
4. Discussion
In this paper, we describe a novel transformed functional mouse MC line that is totally deficient in native KIT. The cells retain the characteristic morphological features of mouse BMMCs having non-lobate nucleii and abundant cytosolic granules; features which distinguish these cells from basophils which also express FcεRI (Stone et al., 2010). We demonstrate utility of this system for the determination of the functional consequences of human KIT activation and, furthermore, demonstrate that this system lends itself to the determination of the efficacy of potential inhibitors of human KIT activity. The parental cell line, and subsequent transfectants, are rapidly dividing and may be cryopreserved with effective recovery, thus providing a suitable inexhaustible source of material for more extensive study.
The parental cell line spontaneously expanded from a MC culture derived from the bone marrow of an mTOR-deficient mouse (Zhang et al., 2011). The replicative nature of the cell line, however, was independent of this defect as other cultures derived from this lineage failed to continue to expand or to constitute a stable cell population with regard to expression of FcεRI and/or KIT (data not shown). Currently, the underlying defect that allows expansion is unknown. However, it is clear that the cell line remains growth factor dependent as they failed to expand in the absence of IL-3, and when transfected with huKIT did not further proliferate and survive when cultured with huSCF alone (data not shown). Therefore, SCF cannot substitute for IL-3 as a growth factor with this cell line. However, unlike other cell lines that have been utilized to examine KIT function, as for example the IC2 and BAF3 cell lines (Koyasu et al., 1985; Ueda et al., 2002; Mayerhofer et al., 2008), the MCBS MCs retained granularity, expressed comparable levels of FcεRI to non-dividing mouse BMMCs and responded to Ag/IgE stimulation to elicit degranulation while retaining the ability to rapidly divide. Thus, the MCBS MCs have distinct advantages over other cell models.
The lack of expression of endogenous KIT is perhaps the most advantageous feature of the MCBS MCs. This deficiency of KIT expression was not evident when the culture was originally initiated but became apparent after several months in culture and a number of liquid nitrogen cryopreservations. It is possible that the loss of endogenous KIT was a result of a spontaneous change in the cell line but may also reflect a preferential expansion of a KIT negative clone over time. Nevertheless, this feature allowed transduction of the cells with a huKIT construct, and, in combination with chemotactic purification, to generate proliferating and highly uniform huKIT expressing MCs. The functionality of these transduced cells was demonstrated by the ability of huSCF to acutely enhance Ag-mediated degranulation, cytokine production, induce chemotaxis, and when chronically administered, downregulate the ability of Ag to induce degranulation. These responses recapitulate those previously demonstrated in mouse BMMCs when stimulated through endogenous mouse KIT (Hundley et al., 2004; Iwaki et al., 2005; Kuehn et al., 2010; Ito et al., 2012). Together with the demonstrated ability of human KIT to engage the necessary signaling machinery for the above responses, this shows that huKIT interacts with the endogenous signaling molecules in the identical manner as endogenous mouse KIT.
The ability of this cell model to function in the above manner again contrasts previous models used to examine KIT function in that we can now explore the structural requirements of huKIT for modulating MC degranulation, cytokine production and chemotaxis. For example, by mutational analysis, the importance of regulating residues that interact with critical signaling molecules for these responses can be determined. Similarly, it should also be possible to investigate the functional consequences of KIT mutations associated with specific disease states in this model.
Finally, in addition to facilitating studies for the investigation of the regulation of MC function by wild type and mutated KIT, this model system would have great utility for screening and identifying potential novel inhibitors of KIT and potentially mutated KIT signaling capacity. Proof of principal for this conclusion was provided by our demonstration that the enhancement of Ag-mediated degranulation by huSCF in the huKIT-transduced MCBS1 MCs was inhibited by the recognized KIT inhibitor imatinib.
Conclusions
In conclusion, we have described a novel rapidly dividing KIT-deficient mouse mast cell line that now permits transduction with human KIT constructs. This model will have great utility for determining the regulation of mast cell activation responses by non-transformed and mutated human KIT. Furthermore, this model will be most useful in screening for novel KIT inhibitors.
Highlights.
We describe a unique KIT-deficient functional mouse mast cell line
Readily transducible with functional human KIT
Utility for determining functional consequences of activated human WT and mutated KIT
Useful for screening for potential inhibitors of human KIT activity
Acknowledgments
Research was supported by funding from the Intramural Research programs within NIAID (D.S., A.M.G., D.D.M.), and NHLBI (M.A.B) and NCI (S.Z. and B.A.M.). We would like to thank Biological Imaging Facility (RTB/NIAID/NIH) for technical support in confocal image acquisition including Juraj Kabat for assistance in image post processing and analysis.
Abbreviations
- Ag
antigen
- BMMCs
bone marrow-derived mast cells
- FcεRI
high affinity IgE receptor
- hu
human
- MC
mast cell
- m
mouse
- SCF
stem cell factor
Footnotes
The authors declare no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aichberger KJ, Gleixner KV, Mirkina I, Cerny-Reiterer S, Peter B, Ferenc V, Kneidinger M, Baumgartner C, Mayerhofer M, Gruze A, Pickl WF, Sillaber C, Valent P. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood. 2009;114:5342–51. doi: 10.1182/blood-2008-08-175190. [DOI] [PubMed] [Google Scholar]
- Antonescu CR. Targeted therapy of cancer: new roles for pathologists in identifying GISTs and other sarcomas. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21(Suppl 2):S31–6. doi: 10.1038/modpathol.2008.9. [DOI] [PubMed] [Google Scholar]
- Bai Y, Bandara G, Ching Chan E, Maric I, Simakova O, Bandara SN, Lu WP, Wise SC, Flynn DL, Metcalfe DD, Gilfillan AM, Wilson TM. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund; U.K: 2012. Targeting the KIT activating switch control pocket: a novel mechanism to inhibit neoplastic mast cell proliferation and mast cell activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- Bougherara H, Subra F, Crepin R, Tauc P, Auclair C, Poul MA. The aberrant localization of oncogenic kit tyrosine kinase receptor mutants is reversed on specific inhibitory treatment. Mol Cancer Res. 2009;7:1525–33. doi: 10.1158/1541-7786.MCR-09-0138. [DOI] [PubMed] [Google Scholar]
- Brockow K, Metcalfe DD. Mastocytosis. Chem Immunol Allergy. 2010;95:110–24. doi: 10.1159/000315946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Wilson TM, Metcalfe DD. The mast cell and allergic diseases: role in pathogenesis and implications for therapy. Clin Exp Allergy. 2008;38:4–18. doi: 10.1111/j.1365-2222.2007.02886.x. [DOI] [PubMed] [Google Scholar]
- Galli SJ, Tsai M. Mast cells in allergy and infection: versatile effector and regulatory cells in innate and adaptive immunity. Eur J Immunol. 2010;40:1843–51. doi: 10.1002/eji.201040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Beaven MA. Regulation of mast cell responses in health and disease. Critical reviews in immunology. 2011;31:475–529. doi: 10.1615/critrevimmunol.v31.i6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol Rev. 2009;228:149–69. doi: 10.1111/j.1600-065X.2008.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nat Rev Immunol. 2006;6:218–30. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- Gleixner KV, Mayerhofer M, Sonneck K, Gruze A, Samorapoompichit P, Baumgartner C, Lee FY, Aichberger KJ, Manley PW, Fabbro D, Pickl WF, Sillaber C, Valent P. Synergistic growth-inhibitory effects of two tyrosine kinase inhibitors, dasatinib and PKC412, on neoplastic mast cells expressing the D816V-mutated oncogenic variant of KIT. Haematologica. 2007;92:1451–9. doi: 10.3324/haematol.11339. [DOI] [PubMed] [Google Scholar]
- Halova I, Draberova L, Draber P. Mast cell chemotaxis - chemoattractants and signaling pathways. Frontiers in immunology. 2012;3:119. doi: 10.3389/fimmu.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley TR, Gilfillan AM, Tkaczyk C, Andrade MV, Metcalfe DD, Beaven MA. Kit and FcεRI mediate unique and convergent signals for release of inflammatory mediators from human mast cells. Blood. 2004;104:2410–7. doi: 10.1182/blood-2004-02-0631. [DOI] [PubMed] [Google Scholar]
- Choi OH, Lee JH, Kassessinoff T, Cunha-Melo JR, Jones SVP, Beaven MA. Antigen and carbachol mobilize calcium by similar mechanisms in a transfected mast cell line (RBL-2H3 cells) that expresses ml muscarinic receptors. J Immunol. 1993;151:5586–95. [PubMed] [Google Scholar]
- Ito T, Smrž D, Jung MY, Bandara G, Desai A, Smržová Š, Kuehn HS, Beaven MA, Metcalfe DD, Gilfillan AM. Stem cell factor programs the mast cell activation phenotype. J Immunol. 2012;188:5428–37. doi: 10.4049/jimmunol.1103366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki S, Tkaczyk C, Satterthwaite AB, Halcomb K, Beaven MA, Metcalfe DD, Gilfillan AM. Btk plays a crucial role in the amplification of FcεRI-mediated mast cell activation by kit. J Biol Chem. 2005;280:40261–70. doi: 10.1074/jbc.M506063200. [DOI] [PubMed] [Google Scholar]
- Jensen BM, Akin C, Gilfillan AM. Pharmacological targeting of the KIT growth factor receptor: a therapeutic consideration for mast cell disorders. Br J Pharmacol. 2008;154:1572–82. doi: 10.1038/bjp.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13) Blood. 1999;94:2333–42. [PubMed] [Google Scholar]
- Kirshenbaum AS, Metcalfe DD. Growth of human mast cells from bone marrow and peripheral blood-derived CD34+ pluripotent progenitor cells. Methods Mol Biol. 2006;315:105–12. doi: 10.1385/1-59259-967-2:105. [DOI] [PubMed] [Google Scholar]
- Koyasu S, Nakauchi H, Kitamura K, Yonehara S, Okumura K, Tada T, Yahara I. Production of interleukin 3 and gamma-interferon by an antigen-specific mouse suppressor T cell clone. J Immunol. 1985;134:3130–6. [PubMed] [Google Scholar]
- Kuehn HS, Beaven MA, Ma HT, Kim MS, Metcalfe DD, Gilfillan AM. Synergistic activation of phospholipases Cγ and Cβ: a novel mechanism for PI3K-independent enhancement of FcεRI-induced mast cell mediator release. Cell Signal. 2008;20:625–36. doi: 10.1016/j.cellsig.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn HS, Rådinger M, Brown JM, Ali K, Vanhaesebroeck B, Beaven MA, Metcalfe DD, Gilfillan AM. Btk-dependent Rac activation and actin rearrangement following FcεRI aggregation promotes enhanced chemotactic responses of mast cells. J Cell Sci. 2010;123:2576–85. doi: 10.1242/jcs.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS. Mast-cell responses to pathogens. Nat Rev Immunol. 2004;4:787–99. doi: 10.1038/nri1460. [DOI] [PubMed] [Google Scholar]
- Mayerhofer M, Gleixner KV, Hoelbl A, Florian S, Hoermann G, Aichberger KJ, Bilban M, Esterbauer H, Krauth MT, Sperr WR, Longley JB, Kralovics R, Moriggl R, Zappulla J, Liblau RS, Schwarzinger I, Sexl V, Sillaber C, Valent P. Unique effects of KIT D816V in BaF3 cells: induction of cluster formation, histamine synthesis, and early mast cell differentiation antigens. J Immunol. 2008;180:5466–76. doi: 10.4049/jimmunol.180.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama Y, Kawakami T. Development, migration, and survival of mast cells. Immunol Res. 2006;34:97–115. doi: 10.1385/IR:34:2:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rådinger M, Kuehn HS, Kim MS, Metcalfe DD, Gilfillan AM. Glycogen synthase kinase 3β activation is a prerequisite signal for cytokine production and chemotaxis in human mast cells. J Immunol. 2010;184:564–72. doi: 10.4049/jimmunol.0902931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Gilfillan AM. Molecular regulation of mast cell activation. J Allergy Clin Immunol. 2006;117:1214–25. doi: 10.1016/j.jaci.2006.04.015. quiz 1226. [DOI] [PubMed] [Google Scholar]
- Smrž D, Dráberová L, Dráber P. Non-apoptotic phosphatidylserine externalization induced by engagement of glycosylphosphatidylinositol-anchored proteins. J Biol Chem. 2007;282:10487–97. doi: 10.1074/jbc.M611090200. [DOI] [PubMed] [Google Scholar]
- Smrž D, Iwaki S, McVicar DW, Metcalfe DD, Gilfillan AM. TLR-mediated signaling pathways circumvent the requirement for DAP12 in mast cells for the induction of inflammatory mediator release. Eur J Immunol. 2010;40:3557–69. doi: 10.1002/eji.201040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrž D, Kim MS, Zhang S, Mock BA, Smržová Š, DuBois W, Simakova O, Maric I, Wilson TM, Metcalfe DD, Gilfillan AM. mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and non-neoplastic human mast cells. Blood. 2011;118:6803–13. doi: 10.1182/blood-2011-06-359984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010;125:S73–80. doi: 10.1016/j.jaci.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Pedersen M, Ronnstrand L. The D816V mutation of c-Kit circumvents a requirement for Src family kinases in c-Kit signal transduction. J Biol Chem. 2009;284:11039–47. doi: 10.1074/jbc.M808058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkaczyk C, Beaven MA, Brachman SM, Metcalfe DD, Gilfillan AM. The phospholipase Cγ1 -dependent pathway of FcεRI-mediated mast cell activation is regulated independently of phosphatidylinositol 3-kinase. J Biol Chem. 2003;278:48474–84. doi: 10.1074/jbc.M301350200. [DOI] [PubMed] [Google Scholar]
- Tkaczyk C, Horejsi V, Iwaki S, Draber P, Samelson LE, Satterthwaite AB, Nahm DH, Metcalfe DD, Gilfillan AM. NTAL phosphorylation is a pivotal link between the signaling cascades leading to human mast cell degranulation following Kit activation and FcεRI aggregation. Blood. 2004;104:207–14. doi: 10.1182/blood-2003-08-2769. [DOI] [PubMed] [Google Scholar]
- Ueda S, Mizuki M, Ikeda H, Tsujimura T, Matsumura I, Nakano K, Daino H, Honda Zi Z, Sonoyama J, Shibayama H, Sugahara H, Machii T, Kanakura Y. Critical roles of c-Kit tyrosine residues 567 and 719 in stem cell factor-induced chemotaxis: contribution of src family kinase and PI3-kinase on calcium mobilization and cell migration. Blood. 2002;99:3342–9. doi: 10.1182/blood.v99.9.3342. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Kreisel F, Cain J, Colson A, Tomasson MH. Neoplasia driven by mutant c-KIT is mediated by intracellular, not plasma membrane, receptor signaling. Mol Cell Biol. 2007;27:267–82. doi: 10.1128/MCB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Readinger JA, DuBois W, Janka-Junttila M, Robinson R, Pruitt M, Bliskovsky V, Wu JZ, Sakakibara K, Patel J, Parent CA, Tessarollo L, Schwartzberg PL, Mock BA. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 2011;117:1228–38. doi: 10.1182/blood-2010-05-287821. [DOI] [PMC free article] [PubMed] [Google Scholar]