Abstract
APOBEC3F and APOBEC3G are DNA cytosine deaminases that potently restrict Human Immunodeficiency Virus-type 1 replication when the virus is deprived of its accessory protein Vif. Vif counteracts these restriction factors by recruiting APOBEC3F and APOBEC3G to an E3 ubiquitin ligase complex that mediates their polyubiquitination and proteasomal degradation. While previous efforts have identified single amino acid residues in APOBEC3 proteins required for Vif recognition, less is known about the downstream ubiquitin acceptor sites that are targeted. One prior report identified a cluster of polyubiquitinated residues in APOBEC3G and proposed an antiparallel model of APOBEC3G interaction with the Vif-E3 ubiquitin ligase complex wherein Vif binding at one terminus of APOBEC3G orients the opposite terminus for polyubiquitination [Iwatani Y, et al. (2009) PNAS 106(46):19539–19544]. To test the generalizability of this model, we carried out a complete mutagenesis of the lysine residues in APOBEC3F and used a complementary, unbiased proteomic approach to identify ubiquitin acceptor sites targeted by Vif. Our data indicate that internal lysines are the dominant ubiquitin acceptor sites in both APOBEC3F and APOBEC3G. In contrast with the proposed antiparallel model, however, we find that the Vif-dependent polyubiquitination of APOBEC3F and APOBEC3G can occur at multiple acceptor sites dispersed along predicted lysine-enriched surfaces of both the N- and C-terminal deaminase domains. These data suggest an alternative model for binding of APOBEC3 proteins to the Vif-E3 ubiquitin ligase complex and diminish enthusiasm for the amenability of APOBEC3 ubiquitin acceptor sites to therapeutic intervention.
Keywords: APOBEC3F, APOBEC3G, HIV, Vif, ubiquitin
Introduction
Human APOBEC3F (A3F) and APOBEC3F (A3G) are DNA cytosine deaminases capable of inhibiting Human Immunodeficiency Virus-type 1 (HIV-1) reverse transcription and integration, most prominently through their active C-terminal deaminase domains, which introduce massive levels of G-to-A mutations in the nascent provirus that contribute to incomplete reverse transcription and render hypermutated genomes hypofunctional [reviewed 2; 3; 4]. To achieve this restriction activity, A3F and A3G must encapsidate in producer cells during viral budding, but HIV-1 typically circumvents this inhibitory APOBEC3 (A3) activity by utilizing its accessory protein Vif as a substrate receptor to link A3F and A3G to an E3 ubiquitin (Ub) ligase complex consisting of CBFβ, ELOB and ELOC, CUL5 and RBX2, which results in the polyubiquitination (polyUb) and subsequent proteasomal degradation of these restriction factors [see recent papers 5; 6 and references therein].
Much effort has been devoted to identifying the determinants of the Vif-A3 interaction critical for this degradative process, including a variety of changes in Vif that result in the functional inactivation of its anti-A3 activity [e.g. 7; 8; reviewed in 9; 4]. On the A3 side of this direct, host-pathogen interaction, initial efforts suggested that the Vif binding sites in A3F and A3G may be structurally distinct, occurring in the C-terminus of the former and in the N-terminus of the latter at unrelated residues 10; 11; 12; 13; 14; 15. However, independent studies indicated that these single amino acid determinants of Vif recognition typically occur within a common region of a given susceptible A3 deaminase domain centered on the predicted α4 helix, suggesting that a conserved structural determinant may be targeted by Vif [DPD128-130 in A3G 10; 11; 12; 16; 13, E324 and E289 in A3F 17; 18, and D/E121 in A3H 19, as well as multiple residues of a distinct region recently described in A3C, A3D and A3F 20].
Less is known about the downstream Ub acceptor sites targeted for polyUb during the degradation process. Iwatani et al. have previously reported that four lysines in the C-terminus of A3G are the Ub acceptor sites required for the Vif-mediated degradation of this enzyme 1. Combined with the aforementioned studies localizing Vif interaction to the N-terminal half of the A3G, this led to the proposal of an antiparallel model of A3 binding to the Vif-E3 Ub ligase complex wherein Vif interacts with one deaminase domain, thereby orienting the second domain for polyUb by an activated E2~Ub conjugate at the opposite end of the CUL5 scaffold.
To test this model and potentially enhance our understanding both of A3 binding to the Vif-E3 Ub ligase complex and of how these substrate acceptor sites might be utilized to block the degradation of A3 proteins, we set out to define the sites of polyUb in A3F. Consistent with results previously reported by Iwatani et al., we find that internal lysines are the dominant Ub acceptor sites in both A3F and A3G. Analysis of the specific residues available for functional polyUb, however, reveals that these are dispersed throughout both domains of A3F at lysine residues clustered on one side of predicted structural models of the A3F N-terminal and C-terminal deaminase domains, opposite the Vif interaction site in the latter case. Furthermore, mutation of the lysine residues determined by Iwatani et al. to be the sites of polyUb in A3G confers only partial Vif resistance. Consistent with these genetic data, mass spectrometric analysis demonstrates Vif-dependent Ub modification of at least 6 sites in A3F and 10 sites in A3G localized to both the N- and C-terminal deaminase domains of each. We conclude, therefore, that the lysine residues available for Vif-dependent polyUb in human A3 proteins are diverse and therefore unlikely to be leveraged by novel therapeutics. To explain this flexibility, we further propose an alternative model of A3 binding to the Vif-E3 Ub ligase complex.
Results
The sites of Vif-mediated polyUb in A3F are distributed throughout the protein
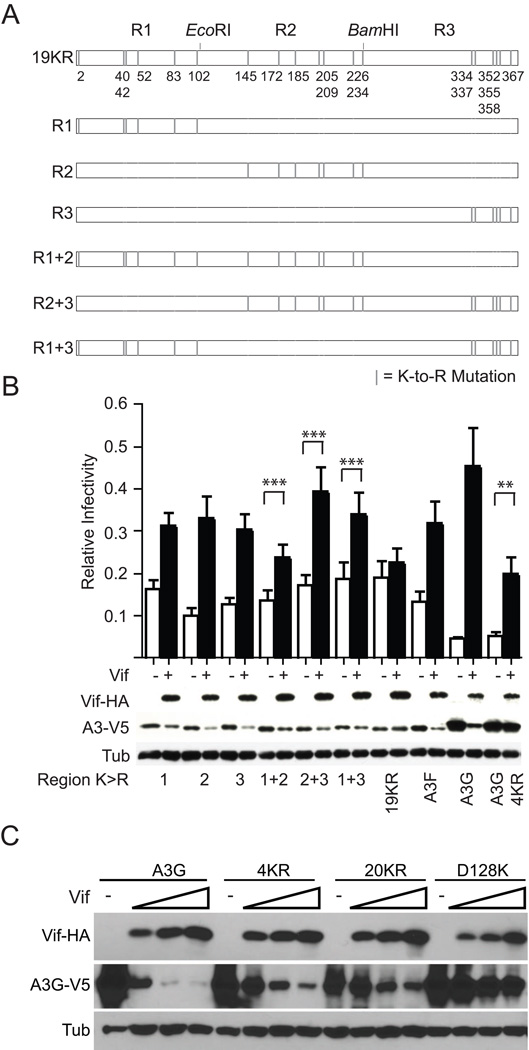
The lysine codons in A3F cluster into three linear groups separated by unique EcoRI and BamHI sites. We first used serial mutagenesis to convert each of these groups from K-to-R. Next, we joined the three regions together to make a panel of K-to-R mutants including a derivative completely devoid of lysines, A3F-19KR, and tested the restriction activity and Vif susceptibility of the resultant proteins in a single cycle of replication (Fig. 1A).
Fig. 1.
The sites of functional Vif-mediated polyUb in A3F are distributed throughout the protein. (A) A schematic showing the K-to-R variants of A3F tested in Fig. 1B. (B) Single-cycle infectivity of Vif-deficient viruses produced in the presence of the indicated A3F-V5 variant −/+ transcomplementation by Vif-HA. Infectivity data represent the mean and SEM of four independent experiments; here as in all singe-cycle infectivity experiments conducted, infectivity is normalized to the infectivities of virus produced in the presence of an A3 vector control −/+ Vif. Statistics were derived by carrying out one-tailed, paired T tests comparing the infectivity of virus produced in the presence of each A3 construct −/+ Vif. ** = p < 0.05; *** = p < 0.01. Western blots showing the steady state levels of Vif-HA and A3-V5 are derived from the producer cells from one of the experiments conducted. (C) A degradation experiment in which constant amounts of A3G or mutant derivatives thereof has been cotransfected with increasing amounts of Vif. Data are representative of two independent experiments.
The restriction activity of all A3F mutants was similar to that of the wild-type protein, and the mutation of all of the K-to-R conversion of any single linear region resulted in A3F variants with Vif susceptibility similar to that of the wildtype protein as evidenced both by the recovery of infectivity and of the decrease in steady state A3F levels in the presence of Vif (Fig. 1B). Similarly, the combination of any two linear regions of K-to-R mutants resulted in somewhat more resistant but still notably Vif-susceptible variants of A3F. Only the variant devoid of all lysines, A3F-19KR, was largely resistant to the effects of Vif. Taken together, these data indicate that at least one lysine residue in each linear region is available for Vif-mediated polyUb.
We also noted that the putative Vif-resistant control protein, A3G-4KR, in which the polyUb residues previously described by Iwatani et al. were changed to arginine, was only partially resistant to HIV-1 Vif (Fig. 1B). To confirm this observation, we cotransfected increasing amounts of Vif with a constant amount of A3G, A3G-4KR, a lysine-free variant of A3G (A3G-20KR) or A3G-D128K and assessed the resultant steady state levels of A3G. While A3G-4KR is indeed partially resistant to Vif as evidenced by enhanced steady state levels in the presence of Vif in comparison with the wildtype protein, the decrease in A3G-4KR steady state levels is greater than that observed for A3G-20KR, particularly considering the greater expression levels of the former in the absence of Vif (Fig. 1C). Furthermore, Vif susceptibility is not due to the epitope tag, as A3G and the lysine-to-arginine mutant derivatives reported here use a single, lysine-free V5 epitope. Thus, while the lysine residues identified by Iwatani et al. are important for the degradation of A3G, A3G appears to also contain alternative sites of polyUb that account for the residual sensitivity of A3G-4KR to Vif.
Multiple internal lysine residues in the A3F N- and C-termini are suitable substrates for Vif-dependent polyUb
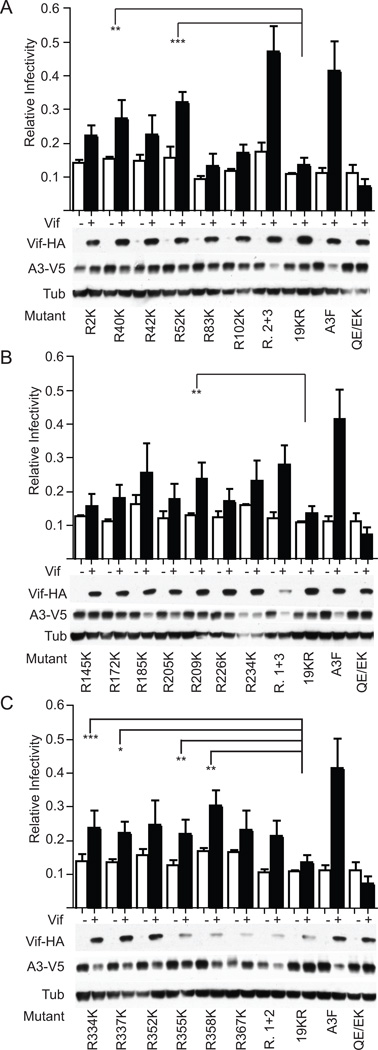
To map the individual lysines in A3F available for polyUb, we reverted each of the 19 R residues of A3F-19KR back to K and assessed the sensitivity of these mutants to Vif in comparison with the parent A3F-19KR in a single cycle of replication. Seven individual lysine reversions in A3F-19KR – R40K, R52K in Region 1; R209K in Region 2; and R334K, R337K, R355K, R358K in Region 3 – induced a statistically significant increase in infectivity in the presence of Vif over the parent lysine-free variant (Fig. 2A–C). In addition to these seven residues, it is possible that others in each region might also be functional targets since we noted some variability from experiment to experiment in which R-to-K changes appeared to sensitize A3F-19KR to Vif, the extent of which is indicated by the associated error bars. For example, A3F-19KR R185K appeared Vif-susceptible in the experiment from which the Western blots shown in Fig. 2B derived but failed to yield a statistically significant increase in infectivity over A3F-19KR when averaged over three, independent experiments. Further examples of this variability are shown in Supplementary Fig. 1, where we have provided the infectivity in the absence or presence of Vif for Region 1 mutants from each of the three experiments averaged in Fig. 2A. This implies that, despite the preferential targeting of certain residues in A3F, the process of Vif-mediated polyUb may be sufficiently promiscuous to find alternative sites with lower efficiency when preferred options are not available. Such a semi-stochastic character is consistent with the distribution of more consistently targeted residues throughout both the N- and C-termini of A3F as well as the generally more efficient degradation of control mutants in which the lysine residues in all but one linear region have been changed to arginine, yielding six or seven potential targets of polyUb in that region rather than the one found in associated individual revertants. The degradative Vif-mediated polyUb of A3F is therefore not a structurally fixed Ub event such as those associated with regulatory functions or that previously described for A3G [e.g. 21; 22; 23; 24; 1; 25]. Rather, it may be more accurately described as a flexible sampling of available substrates wherein at least seven residues throughout both the N- and C-termini of A3F can be polyubiquitinated despite binding of Vif at the protein C-terminus [26; 14; 17; 18; 20 and see Discussion].
Fig. 2.
Multiple internal lysines residues in A3F are suitable substrates for functional Vif-dependent polyUb. (A–C) Single-cycle infectivity of individual single lysine revertants in an A3F-19KR background in the absence or presence of Vif where data represent the mean and SEM of three independent experiments with associated Western blots showing producer cell steady state levels of Vif-HA and A3-V5 derived from one of these experiments. Statistics represent two-way ANOVA with Dunnett’s post-test comparing the infectivity recovery of each single amino acid revertant (Vif+/Vif−) to that of the A3F-19KR control. * = p < 0.1; ** = p < 0.05; *** = p < 0.01. Controls that appear in multiple panels of this figure (e.g. A3F in A, B and C) are regraphed and reblotted for visual comparison with the mutants of each individual region; all data shown in this figure are the result of one large experiment repeated three times and presented visually as three parts.
Mass spectrometric identification of lysine acceptor sites in A3F and A3G
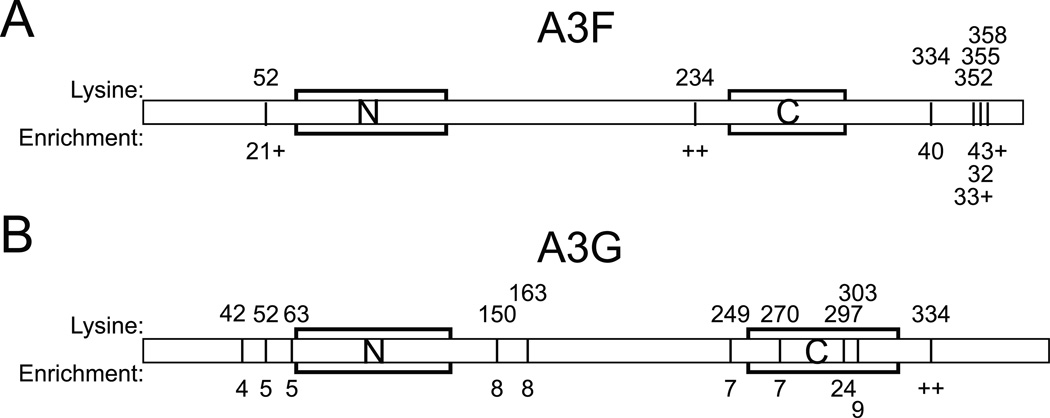
Previous analyses have relied exclusively on genetic and biochemical techniques for the determination of Ub sites in A3G 27; 1; 28; 29. To acquire direct, relatively unbiased biophysical evidence for Ub of the lysine residues identified as functional targets in A3F and to determine whether sites in A3G beyond those previously identified might be modified by Ub, we subjected A3F and A3G to Ub remnant profiling, a technique whereby a monoclonal antibody specific for the K-GG motif characteristic of trypsinized, Ub peptides is used to immunoprecipitate the Ub proteome in a cell lysate prior to mass spectrometric analysis 30. Peptides harboring Ub on residues 52, 234, 334, 352, 355 and 358 of A3F were highly enriched in the presence of Vif, indicating substantial modifications at the A3F C-terminus despite the binding of Vif to the same end of the protein (Fig. 3A and Supplementary File 1). Moreover, despite a few minor discrepancies in the specific acceptor sites implicated by the mass spectrometric and genetic datasets, the results of these approaches largely overlap and collectively reinforce the notion that the modification of A3 proteins is not restricted to a structurally rigid subset of lysine acceptor sites (see Discussion).
Fig. 3.
Mass spectrometry indicates Ub of multiple internal lysine residues in the N- and C-termini of both A3F and A3G. (A–B). Diagrams showing Ub-modified residues in A3F (A) or A3G (B), with residues numbered at the top and the fold-enrichment of associated peptides indicated on the bottom. ++ indicates that all of the peptides detected indicating modification of the associated residues were so rare in the absence of Vif as to preclude calculation of fold-enrichment. The combination of a number with a + reflects calculation of a fold enrichment for a mixed residue in which some associated peptides permitted calculation while others were so rare in the absence of Vif as to preclude a fold-enrichment calculation. Zinc-coordination motifs of N- and C-terminal deaminase domains are indicated by boxes.
For A3G, peptides corresponding to ubiquitination of five N-terminal and five C-terminal residues were enriched in the presence of Vif (Fig. 3B and Supplementary File 2). These data again implicate residues of both the N- and C-termini as Ub acceptor sites and are further consistent with the inability of K-to-R mutations at the residues previously identified by Iwatani et al. to render A3G fully resistant to Vif (Fig. 1B). Importantly, like the variation observed in the genetic analysis of single K-to-R revertants in A3F-19KR, we found that the exact repertoire of modified sites as determined by mass spectrometry differed slightly from experiment to experiment. For example, our previous, unpublished analysis of A3G by this same technique identified modification only at residues 297 and 303 among the four residues reported by Iwatani et al. while also finding no modification at residue 63 (Fig. 3B and see Discussion).
Lysine residues in A3F and A3G cluster at distinct predicted surfaces
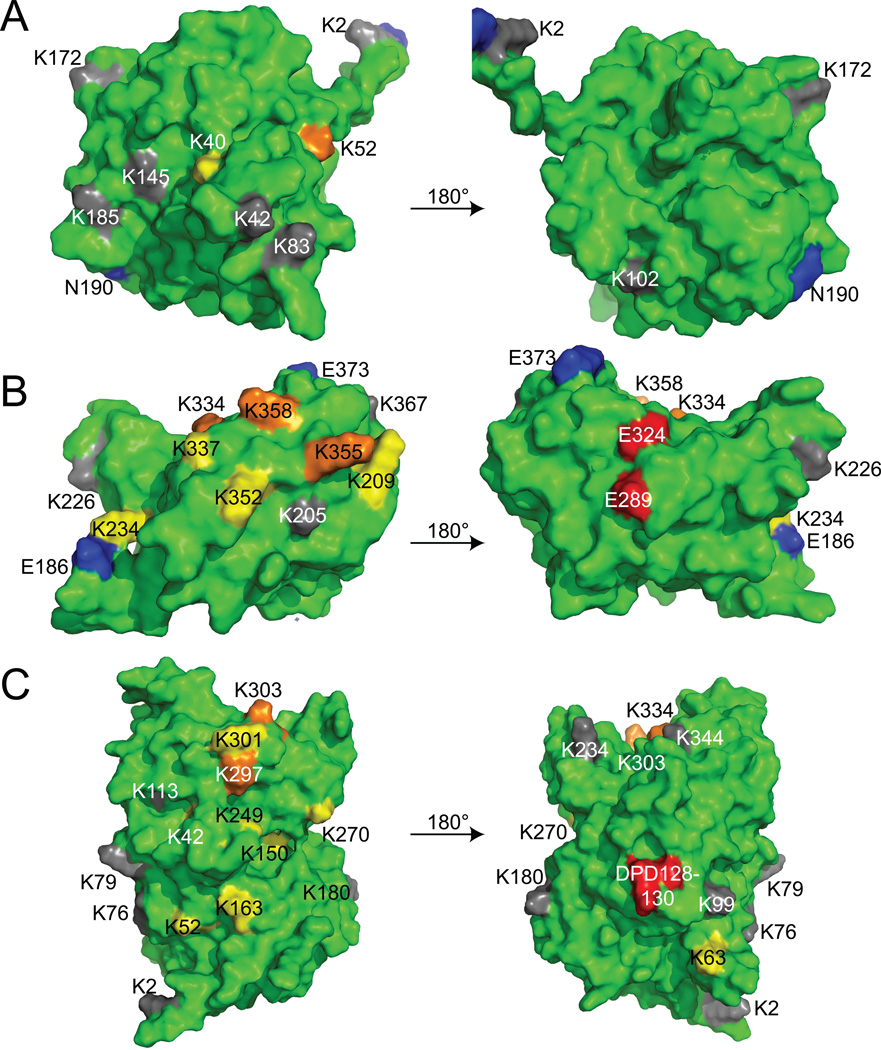
To determine whether the susceptibility patterns observed in Figs. 2 and 3 might have a structural basis, we created a predicted structural model of the A3F N-terminus to complement the predicted structural model of the C-terminus reported previously 17. This yielded the surprising finding that in both the N- and C-termini, the lysine residues in A3F typically cluster at one predicted surface, which in the case of the C-terminus is opposite the surface strongly implicated in the interaction with Vif (Fig. 4A–B). Thus, the placement of lysines within A3F is consistent with the promiscuous ability of the Vif-E3 ligase complex to promote polyUb at multiple residues in A3F since these residues are, broadly speaking, structurally comparable. It is important to note that structural data are not yet available to make a robust model of full-length A3F. When the Ub acceptor sites implicated in Fig. 3B are visualized on our previously-described full-length predicted structural model of A3G, however, we observe a similar clustering of lysine residues at a surface opposite that bound by Vif (Fig. 4C).
Fig. 4.
Lysines in the N- and C-termini of A3F and A3G cluster at distinct predicted surfaces. (A) A model of the A3F N-terminal deaminase domain is shown rotated 180° about the y-axis. (B) A model of the A3F C-terminal deaminase domain is shown rotated 180° about the y-axis. (C) A previously-described model of full-length A3G is shown rotated 180° about the y-axis 46. Orange indicates residues implicated by both genetic and biophysical experiments, while yellow indicates residues implicated by one or the other. A3G residues implicated by Iwatani et al. and also identified in Fig. 3B are orange in (C), while additional residues not implicated by Iwatani et al. are yellow. Lysine residues not significantly implicated by either data set are gray, while Vif interaction residues are colored red 17; 18.
Lack of evidence for polyUb at sites other than internal A3 lysines
Although our results above as well as those of Iwatani et al. provide positive evidence for internal lysines as the predominant sites of polyUb in A3 proteins, others have previously reported that either Vif itself or the N-terminal residue of A3G may be important sites of polyUb en route to A3G degradation 27; 28; 29. The results of Dang et al. implicating autoUb of Vif as the functionally relevant modification for A3G degradation may reflect the targeting of lysine residues in the C-terminal tag of the otherwise lysine-free A3G variant used [27; 1 and consistent with sequencing of constructs provided by Dr. Yong-Hui Zheng, Michigan State University]. To functionally address the possibility that A3F or A3G may be modified at their respective N-terminal residues, however, we sought to create variants that would be predicted to be resistant to N-terminal modification.
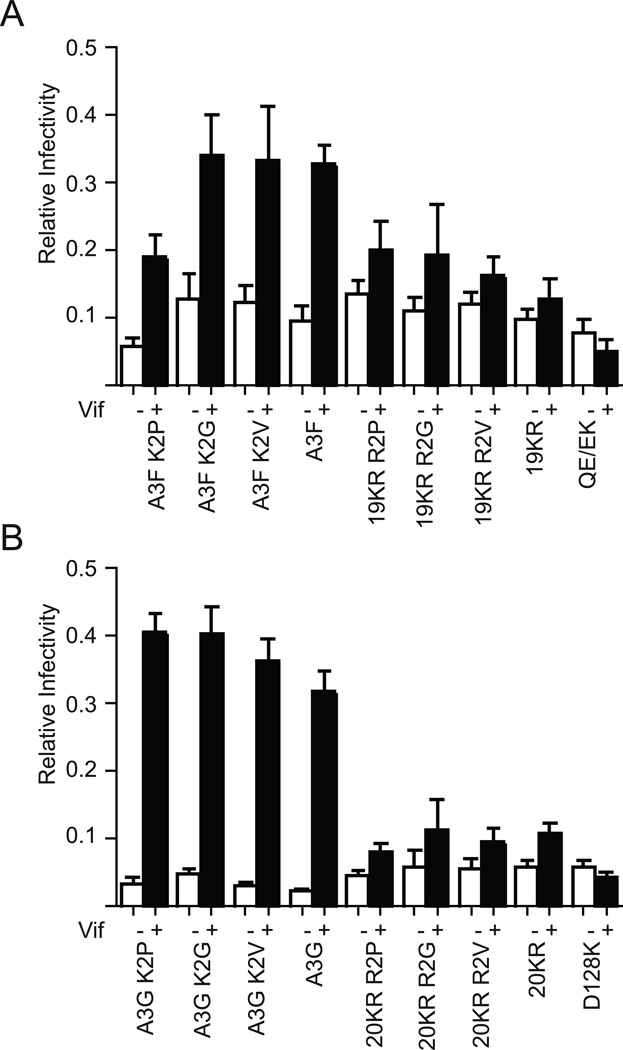
Changing residue two of A3F and A3G from K to P, G or V favors cleavage of the initiating methionine to create a new N-end [e.g. 31]. Among potential N-terminal residues, the imino group of P is biochemically distinct and likely resistant to N-terminal Ub since, unlike all other amino acids, artificial fusion of Ub to an N-terminal P yields a bond resistant to the action of the deubiquitinating enzymes present in yeast and mammalian cell lysates [e.g. 32; 33]. The resistance of P to N-terminal acetylation also obviates potential complications associated with the ability of this modification promote degradation 34, while G and V are subject to modification by N-terminal acetylases distinct from those predicted to act on wild-type A3F and A3G [e.g. 35]. We therefore predicted that changing the second A3 amino acid to P, G or V should alter the Vif sensitivity of the resultant variant if Vif regulates A3 proteins via modifications of the A3 N-terminus. We observed, however, that changes to the second amino acid of A3 proteins do not substantially alter their Vif susceptibility in a single cycle of replication (Fig. 5). Combined with positive evidence for the Ub of internal lysines, these data therefore indicate that the A3 N-terminal residue is not a major target of Vif function.
Fig. 5.
Changes at the A3 N-terminus do not alter Vif susceptibility. (A–B) Single-cycle infectivity experiments demonstrate the Vif sensitivity of A3F, A3G and lysine-free variants thereof to Vif when the second amino acid is altered as shown. Infectivity data represent the mean and SEM of four independent experiments.
Discussion
We present here genetic and biophysical evidence for the polyUb of multiple internal lysines distributed throughout both the N- and C-termini of A3F and A3G (Figs. 1–3). The flexibility of the Vif-E3 ligase complex to promote the polyUb of A3 proteins at a wide range of acceptor sites is consistent with an active sampling of A3 lysines by the associated E2~Ub conjugate. Such a sampling model is further consistent with the observation that single R-to-K reversions in A3F-19KR context are generally less efficient in sensitizing A3F-19KR to Vif relative to control mutants in which all the available substrates of a given region are lysines, particularly in Region 1 (Fig. 2A). This relatively stochastic and indiscriminate process makes intuitive sense since the default mode for Vif should be to rapidly degrade A3 proteins with no apparent need for tight regulation of the kind mediated by more selective Ub events [e.g. 21; 22; 23; 24; 1; 25].
Consistent with the promiscuous modification of A3F lysines, we observe that mutation of the residues previously reported by Iwatani et al. as the sole functional targets of Vif-mediated polyUb in A3G render the protein only partially Vif-resistant, while many N- and C-terminal lysines in A3G are modified in the presence of Vif (Figs. 1B, 1C and 3B; compare also the infectivity recovery in the presence of Vif associated with A3G-4KR in Fig. 1B versus that associated with A3G-20KR in Fig. 5B). We therefore think it likely that the antiparallel model for A3 polyUb previously proposed does not accommodate all of the data [1 and Fig. 6A]. It is possible that said model may be partially correct since the A3G-4KR mutant is notably less susceptible to Vif than wild-type A3G, and we do note the presence of some susceptible lysine residues in the A3F N-terminus (Figs. 1B and 2A–B). Nonetheless, the reversion of multiple lysines in the A3F C-terminus can also render A3F-19KR sensitive to Vif, demonstrating extensive flexibility in the ability of activated E2~Ub conjugates to reach dispersed sites.
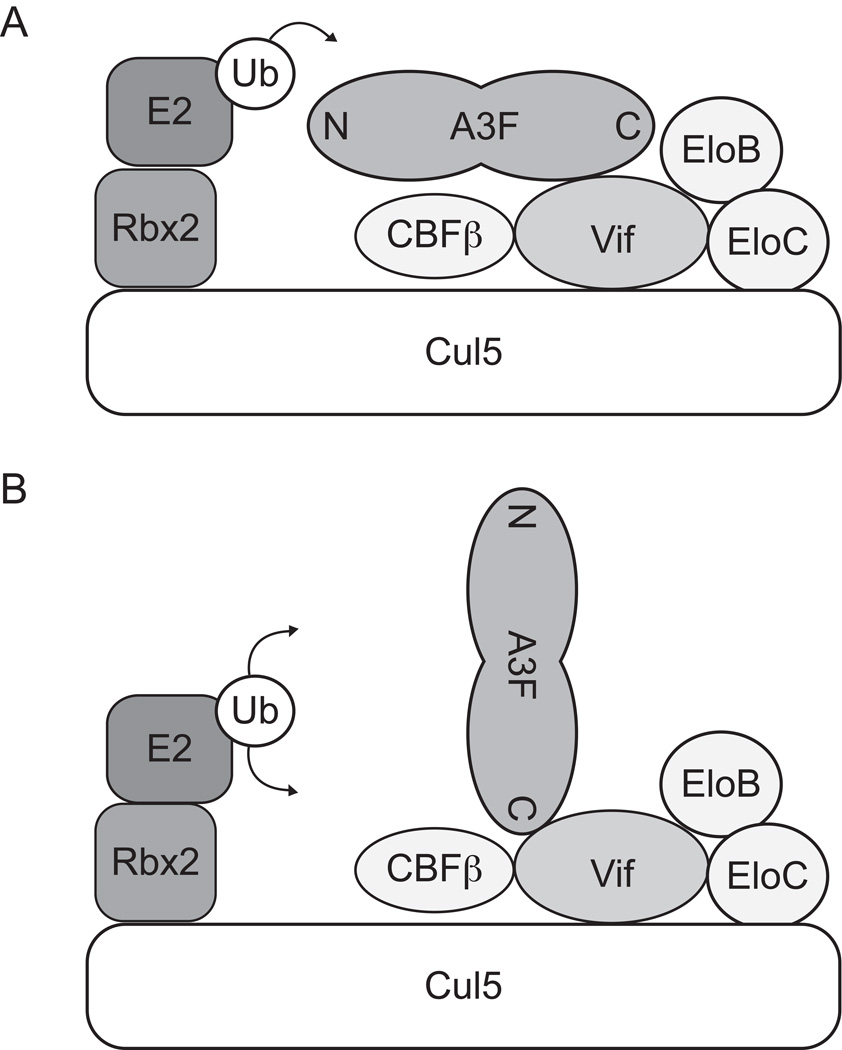
Fig. 6.
An alternative model of A3 binding to the Vif-E3 ligase complex. (A) The antiparallel model previously proposed by Iwatani et al. in which binding of Vif at one deaminase domain orients an A3 protein for polyUb at its second deaminase domain 1. (B) An alternative, angled/perpendicular model for A3F binding to the Vif-E3 ligase complex in which binding of Vif to a lysine-poor surface orients the lysine-rich surfaces of A3F for polyUb.
Rather than the rigid antiparallel model of A3 polyUb originally proposed and depicted schematically in Fig. 6A, it is more likely that A3 proteins bind Vif in an angled or perpendicular orientation that would expose to activated E2~Ub the entirety of the lysine-rich surfaces predicted to be opposite the Vif binding sites in A3F and A3G (Fig. 6B). It is also possible that multimeric forms of A3 proteins or of the E3 Ub ligase complex might yield results similar to ours, although we have no evidence to directly support such a model. Regardless, while we agree that the four lysines initially identified by Iwatani et al. are important substrates for polyUb in A3G, our data suggest that other sites can also be modified, including sites in the N-terminal half of A3G. Moreover, it is clear that both N- and C-terminal residues of A3F can be modified. If these observations are each accurate, then the binding model shown in Fig. 6A cannot account for all of the data, while that shown in Fig. 6B can.
Alternatively, the ease with which Vif can degrade A3F mutants with one or more linear regions available for polyUb contrasted with the prominent albeit incomplete Vif-resistance of A3G-4KR may imply differential binding of Vif to A3F versus A3G. By extension, this may lead to differential availability of acceptable lysine substrates for polyubiquitination. Such a view could be consistent with studies genetically mapping the A3F/G and Vif binding sites in each protein, which suggest that different regions of Vif interact with different regions of A3F versus A3G [reviewed in 9]. Whatever binding model one favors, then, the central question remains one of accessibility.
We noted that there was some variability in which specific R-to-K reversions in A3F-19KR sensitized the protein to Vif from experiment to experiment, while linear regional mutants with multiple sidechains available for polyUb were consistently Vif-sensitive (Figs. 1–2 and Supplementary Fig. 1). Similarly, independent, unbiased mass spectrometric analyses of A3G indicated some variation in the specific residues modified. For example, focusing just on the four residues functionally implicated by Iwatani et al. 1, residues 297, 303 and 334 were identified in one experiment (Fig. 3B), while only residues 297 and 303 were identified in a second and neither experiment produced any evidence for the modification of residue 301. We interpret these data to mean that the Vif-mediated polyUb of A3 proteins is a semi-stochastic affair in which the associated E2~Ub conjugate is able to sample multiple available sidechains for polyUb. As a result, changing the residues most efficiently targeted for modification may simply force the modification of less efficiently targeted sidechains, consistent with data from other fields indicating that components of the E3 Ub ligase complex may display substantial structural flexibility [e.g. 36; 37; 38; 39].
Similarly, there were several discrepancies between the specific residues likely modified according to genetic versus biophysical approaches (Figs. 2–3). Three A3F lysines – K40, K209 and K337 – were found to be significantly targeted in the genetic data but were not identified by mass spectrometry, while the opposite was true of K234. This may reflect the fact that the wild-type protein used for mass spectrometry simply has more residues available for polyUb and that, consequently, the wildtype protein is preferentially modified at those residues common to the genetic and mass spectrometric datasets. Alternatively, the structures of A3F and A3F-19KR may differ slightly despite their similar restriction activities (Figs. 1–2). It is also possible that further experiments may find additional residues modified that are not apparent in the data presented here, as we have observed with A3G. Regardless, the overarching message remains clear: many residues in both the N- and C-terminal domains of both A3F and A3G are suitable substrates for polyUb.
In summary, we report here that the Ub acceptor sites in A3F and A3G are distributed throughout each protein, suggesting that the conjugated E2 enzyme(s) recruited by the Vif-E3 ligase complex are more flexible in their substrate targeting potential than previously proposed and that blocking the modification sites themselves will not be a viable strategy for the therapeutic stabilization of A3 proteins. Of note, the crystal structure of A3C has recently been solved, an effort which further included extensive genetic identification of the Vif interaction surface in A3C 20. These authors further determined that the residues important for Vif interaction with A3C are conserved in A3F. With a handful of exceptions, mostly notably those residues corresponding to A3F K352, K355 and K358, there is little linear conservation of lysine residues between A3C and A3F (data not shown). On comparing the mapped Vif interaction surface with the available lysine residues, however, it appears that the theme of Vif binding at a lysine-poor surface distinct from available polyubiquitination sites holds true in this single-domain APOBEC3 protein. It will be interesting to find whether the apparent clustering of lysines at surfaces distinct from those at which Vif interacts is a general feature of the A3-Vif interaction. It may be that the positively-charged Vif protein achieves efficient degradation of A3 proteins, in part, by binding to negatively charged regions of susceptible deaminase domains such as the α4 helix, thereby positioning distinct, lysine-rich surfaces for efficient polyUb in the context of the fully assembled E3 Ub ligase complex.
Materials and Methods
Plasmids
Wildtype A3F and A3G coding sequences are identical to those found in GenBank entries NM_145298 and NM_021822 and previously reported [e.g. 40]. A3 expression constructs were expressed in pcDNA3.1-derived vectors with a C-terminal V5 tag 40; in all V5 constructs except the Vif interaction mutants A3F QE323-324EK and A3G D128K, however, the inherent tag lysine was mutated to arginine to prevent artifactual tag Ub such as that previously described 1. K-to-R mutants were made by sequential site-directed mutagenesis and subcloning; A3G-20KR was graciously provided by Dr. Yong-Hui Zheng 27 and cloned into our lysine-free V5 vector. The HIV-1 Vif-deficient IIIB Vif-XX proviral construct and IIIB Vif-HA expression construct were as previously described 40. All constructs were verified by restriction digestion and sequencing.
Cell lines
HEK293T cells were maintained in DMEM medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. CEM-GFP reporter cells were obtained from the AIDS Research and Reference Reagent Program 41 and maintained in RPMI medium supplemented with 10% fetal bovine serum, penicillin-streptomycin and β-mercaptoethanol.
Single-cycle infectivity protocol
Single-cycle infectivity experiments were carried out as previously described 40. 250,000 HEK293T cells were plated into 2 mL of supplemented DMEM in 6-well plates. The following day, cells were transfected with 200 ng of either a vector control or an A3-V5 expression construct, 100 ng of a Vif-HA expression construct, and 1.6 µg of replication competent, Vif deficient HIV-1IIIB provirus using TransIT transfection reagent (Mirus Bio). Two days after transfection, transfected cells were harvested and lysed as described below, and virus-containing supernatants were filtered through 0.45 µm filters and used to infect CEM-GFP cells in 96-well plates. Three days post-infection, target CEM-GFP cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry for viral infectivity on a FACSCanto flow cytometer (BD Biosciences).
Vif titration experiments
Cells were transfected as in single-cycle infectivity experiments above but with 200 ng A3-V5 expression construct, 50–200 ng Vif-HA expression construct balanced to 200 ng total with a Vif expression vector and 600 ng pcDNA3.1 (Invitrogen) vector DNA for a total of 1 µg per well. Two days post-transfection, cells were harvested as described above and analyzed by Western blot as described below.
Western blot analysis
HEK293T cells transfected as described above were washed in PBS and lysed in 250 µL of a lysis buffer composed of 25 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 50 µM ZnCl2, 10% glycerol and 1% Triton X-100 and supplemented with 50 µM MG132 and complete protease inhibitor (Roche). A 5× sample buffer consisting of 62.5 mM Tris pH 6.8, 20% glycerol, 2% sodium dodecyl sulfate, 5% β-mercaptoethanol, and 0.05% bromophenol blue was added to each cell lysate to a 2× concentration, and the mixture was boiled for 5–10 minutes and subjected to fractionation by 10% SDS-PAGE and transfer to PVDF membranes. Primary antibodies utilized include mouse anti-V5 (Invitrogen), mouse anti-HA.11 (Covance), and mouse anti-tubulin (Covance). Between probing with different primary antibodies, membranes were stripped with 62.5 mM Tris pH 6.8, 2% SDS, and 100 mM β-mercaptoethanol at 50°C and washed in PBS 0.01% Tween.
Mass spectrometry
HEK293T cells were labeled using a SILAC approach 42. Cells were cultured in either “light” SILAC media containing the normal complement of amino aids or “heavy” SILAC media wherein lysine was replaced with 13C6-lysine. FBS was dialyzed in both light and heavy media formulations to remove free unlabeled amino acids. Four 15 cm plates of light media each were transfected with 2.5 µg Vif-Strep and 2.5 µg of either A3F-V5 or A3G-FLAG. An additional four 15 cm plates of 293T cells cultured in heavy media each were also transfected with 2.5 µg pcDNA4 vector and 2.5 µg either A3F-V5 or A3G-FLAG. Cells were detached from the plate with 10 mM EDTA in PBS, spun down, and snap frozen. The frozen cell pellets were lysed in buffer containing 8M urea, 0.1M Tris pH 8.0, 150 mM NaCl, and a combination of protease inhibitors (Complete tablet, Roche). Protein concentration was measured by Bradford assay (QuickStart 1× Reagent, Bio-Rad). 50 mg protein lysate from equal portions of light and heavy culture conditions for A3F-V5 or A3G-FLAG, respectively, were combined and then subjected to reduction with 4 mM TCEP for 30 minutes at room temperature, alkylation with 10 mM iodoacetamide for 30 minutes and room temperature in the dark, and overnight digestion with 250 µg of trypsin (Promega) at room temperature. Treated lysates were subsequently desalted using SepPak C18 cartridges (Waters), lyophilized for two days and immunoprecipitated using an antibody specific for the K-GG motif characteristic of trypsinized, Ub-modified peptides (UbiScan, Cell Signaling Technology). Immunoprecipitates were then desalted using C18 STAGE tips (ThermoScientific), evaporated, and analyzed in duplicate with a two-hour gradient on an Orbitrap Elite Mass Spectrometer (Thermo Scientific). The reverse phase gradient was delivered by an Easy nLC 1000 liquid chromatography system (ThermoScientific) from 5% to 30% acetonitrile in 0.1% formic acid. Columns used were a 100 µm × 2 cm pre-column packed with 5 µm ReproSil Pur C18 particles and a 75 µm × 10 cm analytical column packed with 2 µm ReproSil Pur C18 particles (Thermo Scientific). The Orbitrap Elite continuously collected data in data-dependent acquisition mode, acquiring a full scan in the orbitrap at 120,000 resolution followed by collision-induced fragmentation of the top 20 most intense peaks from full scan in the ion trap. Dynamic exclusion was enabled to exclude repeated fragmentation of peaks for 30 seconds. Charge state screening was enabled to reject fragmentation of unassigned or singly charged species. Results were analyzed using the MaxQuant software package 43.
Homology modeling of the A3F N- and C-terminal deaminase domains
Modeling of the A3F CTD has been described previously 17. The A3F NTD model including residues 1–190 was generated using YASARA 44 with the crystal structure of A3G191–384 2K3A (PDB 3IR2) as a template 45. Alignment with the template sequence was iteratively optimized using SwissProt and TrEMBL sequences, the predicted secondary structure and the three dimensional structure of the template. Refinement of the model was carried out using knowledge-based and electrostatic interactions in unrestrained molecular dynamics with explicit solvent molecules. Insertions were accounted for by searching the Protein Data Bank for superimposable loop ends with model anchor points and optimized for energy minimization.
Supplementary Material
Supplementary Files 1 & 2. Raw mass spectrometry data. Raw mass spectrometry data used in making Fig. 3 are provided for A3F (Supplementary File 1) and A3G (Supplementary File 2). H/L ratios of NaN indicate that the background levels of the indicated peptide in the absence of Vif were too low to permit the calculation of fold-enrichment. Note also that lysines at the extreme C-terminus of a given peptide are not valid hits in the method used here. Residue numbers for the main lysine detected in a given peptide have been manually placed next to the gene name under the Gene Name column.
Supplementary Fig. 1. Variation in targeted lysine residues as exemplified in linear Region 1. (A) The infectivity of virus produced in the presence of the indicated A3F mutant −/+ Vif for Experiment 1 of 3 shown averaged in Fig. 2A. (B) As in (A) with Experiment 2 of 3. (C) As in (A) with Experiment 3 of 3. Error bars indicate the mean and standard deviation of the relative infectivity from duplicate infections in a given experiment.
Sites of Vif-mediated polyUb in APOBEC3 proteins are not well understood
Residues in the N- and C-termini of APOBEC3F can act as ubiquitin acceptor sites
Data suggest a flexible model for polyUb of A3 proteins by the Vif-E3 ligase complex
Acknowledgements
We thank A.K. Bielinsky, K. Walters and the U. of MN Statistical Consulting Service for helpful discussion, and Y.-H. Zheng and the AIDS Research and Reference Reagent Program for materials. This research was funded by the National Institute of Allergy and Infectious Disease (R01 AI064046 to RSH and PO1 AI090935 to NJK) and the National Institute of General Medical Sciences (P01 GM091743 to RSH and P50 GM082250 and P50 GM081879 to NJK). JSA was supported in part by the National Institute on Drug Abuse (F30 DA026310) and by the University of Minnesota Medical Scientist Training Program (T32 GM008244). NJK is a Searle Fellow and Keck Young Investigator.
Abbreviations
- A3F
APOBEC3F
- A3G
APOBEC3G
- HIV-1
Human Immunodeficiency Virus-type 1
- Vif
virion infectivity factor
- Ub
ubiquitin
- E2
ubiquitin conjugating enzyme
- E3
ubiquitin ligase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, Sugiura W. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc Natl Acad Sci U S A. 2009;106:19539–19544. doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 3.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Reviews in Molecular Medicine. 2010;12:1–26. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jäger S, Kim DY, Hultquist JF, Shindo K, Larue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBFβ to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBFβ regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2011;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 7.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JL, Bu W, Burdick RC, Pathak VK. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol Sci. 2009;30:638–646. doi: 10.1016/j.tips.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 12.Schröfelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavens D, Peelman F, Van der Heyden J, Uyttendaele I, Catteeuw D, Verhee A, Van Schoubroeck B, Kurth J, Hallenberger S, Clayton R, Tavernier J. Definition of the interacting interfaces of APOBEC3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 2010;38:1902–1912. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albin JS, LaRue RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem. 2010;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JL, Pathak VK. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol. 2010;84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J Virol. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 21.Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc Natl Acad Sci U S A. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 23.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 24.Batonnet S, Leibovitch MP, Tintignac L, Leibovitch SA. Critical role for lysine 133 in the nuclear ubiquitin-mediated degradation of MyoD. J Biol Chem. 2004;279:5413–5420. doi: 10.1074/jbc.M310315200. [DOI] [PubMed] [Google Scholar]
- 25.Das-Bradoo S, Nguyen HD, Wood JL, Ricke RM, Haworth JC, Bielinsky AK. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat Cell Biol. 2010;12:74–79. doi: 10.1038/ncb2007. sup pp 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Chen G, Niewiadomska AM, Xu R, Yu XF. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS ONE. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang Y, Siew LM, Zheng YH. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J Biol Chem. 2008;283:13124–13131. doi: 10.1074/jbc.M708728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao Q, Wang Y, Hildreth JE, Liu B. Polyubiquitination of APOBEC3G is essential for its degradation by HIV-1 Vif. J Virol. 2010;84:4840–4844. doi: 10.1128/JVI.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Shao Q, Yu X, Kong W, Hildreth JE, Liu B. N-terminal hemagglutinin tag renders lysine-deficient APOBEC3G resistant to HIV-1 Vif-induced degradation by reduced polyubiquitination. J Virol. 2011;85:4510–4519. doi: 10.1128/JVI.01925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 33.Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 34.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnesen T. Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 2011;9:e1001074. doi: 10.1371/journal.pbio.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Nussinov R. The mechanism of ubiquitination in the cullin-RING E3 ligase machinery: conformational control of substrate orientation. PLoS Comput Biol. 2009;5:e1000527. doi: 10.1371/journal.pcbi.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Nussinov R. Molecular dynamics reveal the essential role of linker motions in the function of cullin-RING E3 ligases. J Mol Biol. 2010;396:1508–1523. doi: 10.1016/j.jmb.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Nussinov R. Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. J Biol Chem. 2011;286:40934–40942. doi: 10.1074/jbc.M111.277236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albin JS, Haché G, Hultquist JF, Brown WL, Harris RS. Long-term Restriction by APOBEC3F Selects Human Immunodeficiency Virus Type 1 Variants with Restored Vif Function. Journal of Virology. 2010;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci U S A. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 45.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Files 1 & 2. Raw mass spectrometry data. Raw mass spectrometry data used in making Fig. 3 are provided for A3F (Supplementary File 1) and A3G (Supplementary File 2). H/L ratios of NaN indicate that the background levels of the indicated peptide in the absence of Vif were too low to permit the calculation of fold-enrichment. Note also that lysines at the extreme C-terminus of a given peptide are not valid hits in the method used here. Residue numbers for the main lysine detected in a given peptide have been manually placed next to the gene name under the Gene Name column.
Supplementary Fig. 1. Variation in targeted lysine residues as exemplified in linear Region 1. (A) The infectivity of virus produced in the presence of the indicated A3F mutant −/+ Vif for Experiment 1 of 3 shown averaged in Fig. 2A. (B) As in (A) with Experiment 2 of 3. (C) As in (A) with Experiment 3 of 3. Error bars indicate the mean and standard deviation of the relative infectivity from duplicate infections in a given experiment.