Abstract
P-glycoprotein (Pgp) is an important contributor to multidrug resistance of cancer. Pgp contains eleven native tryptophans (Trps) that are highly conserved among orthologs. We replaced each Trp by a conservative substitution to determine which Trps are important for function. Individual Trp mutants W44R, W208Y, W132Y, W704Y and W851Y, situated at the membrane surface, revealed significantly reduced Pgp induced drug resistance against one or more fungicides and/or reduced mating efficiencies in Saccharomyces cerevisiae. W158F and W799F, located in the intracellular coupling helices, abolished mating but retained resistance against most drugs. In contrast, W228F and W311Y, located within the membrane, W694L, at the cytoplasmic membrane interface, and W1104Y in NBD2 retained high levels of drug resistance and mating efficiencies similar to wild-type Pgp. Those were combined into pair (W228F/W311Y and W694L/W1104Y) and quadruple (W228F/W311Y/W694L/W1104Y) mutants that were fully active in yeast, and could be purified to homogeneity. Purified pair and quad mutants exhibited drug-stimulated ATPase activity with binding affinities very similar to wild-type Pgp. The combined mutations reduced Trp fluorescence by 35%, but drug induced fluorescence quenching was unchanged from wild-type Pgp suggesting that several membrane-bound Trps are sensitive to drug binding. Overall, we conclude that Trps at the membrane surface are critical for maintaining the integrity of the drug binding sites, while Trps in the coupling helices are important for proper interdomain communication. We also demonstrate that functional single Trp mutants can be combined to form a fully active Pgp that maintains drug polyspecificity, while significantly reducing intrinsic fluorescence.
Keywords: P-glycoprotein, native tryptophans, membrane bilayer, conservative tryptophan substitutions, drug binding sites, polyspecificity
1. Introduction
P-glycoprotein (Pgp, also known as ABCB1) is an integral membrane protein that functions as a multidrug exporter [1]. Pgp binds a wide array of structurally unrelated compounds, including many commonly used drugs, and transports them to the extracellular matrix, preventing intracellular drug accumulation [2,3]. Pgp mediated drug transport is a well-known contributor to multidrug resistance of cancer cells, and Pgp expression in tumors is associated with poor treatment outcome and patient relapse [4,5,6]. Pgp is also expressed at the blood brain barrier, intestinal epithelium and other sites where it can cause resistance to chemotherapeutic treatment for a variety of diseases including HIV infection and epilepsy [7,8,9,10]. Despite nearly four decades of work and multiple generations of clinical trials, a clinically viable Pgp inhibitor has remained elusive, substantiating the need for more in depth studies of the Pgp drug transport mechanism [11,12].
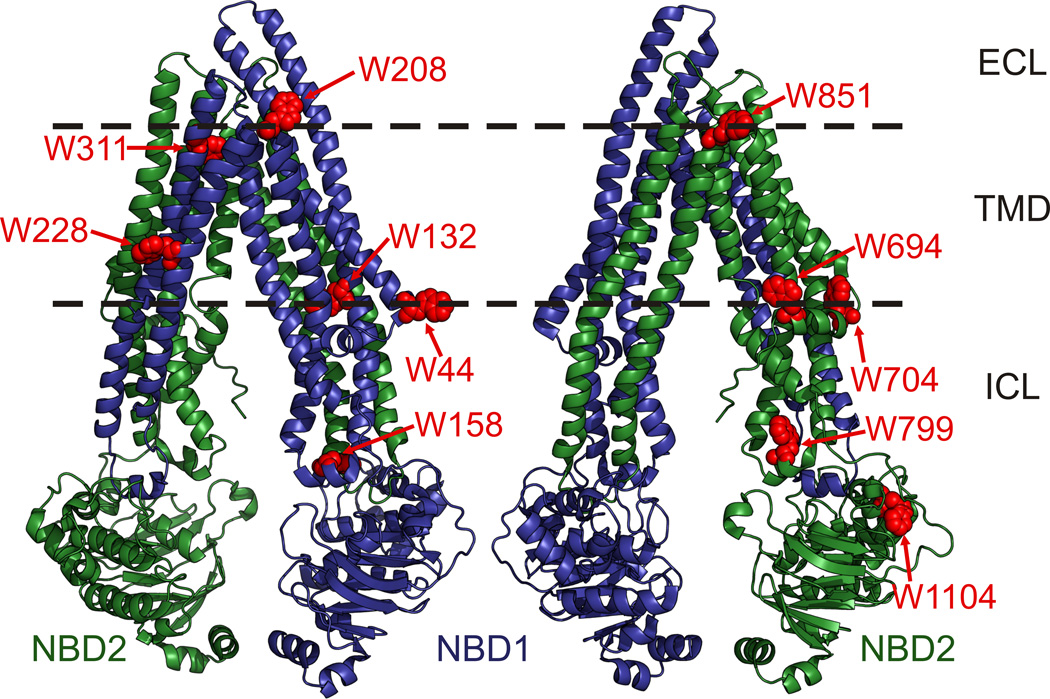
Pgp is a member of the ATP binding cassette (ABC) transporter superfamily [13]. It consists of two transmembrane domains (TMDs) with six α-helices and two nucleotide binding domains (NBDs) that bind and hydrolyze ATP [14]. The Pgp crystal structure (PDB: 3G5U, Figure 1) reveals that the two TMDs combine to form a voluminous central cavity with multiple sites for polyspecific substrate binding, which is likely mediated through varying interactions with amino acid side chains lining the central cavity [15,16,17,18]. The NBDs can dimerize to form two nucleotide binding sites that are catalytically active [19,20]. Nucleotide binding induced NBD dimerization likely provides the power to rearrange the TMDs from an inward-facing conformation, where transport substrates are able to enter the central cavity from the lipid bilayer or cytoplasm, to an outward-facing conformation that exposes the central cavity and bound substrates to the extracellular environment [21]. However, details of how substrate binding and transport are achieved through coordinated movements of the NBDs and TMDs remain poorly understood for Pgp and ABC transporters in general [22,23]. Site-specific tryptophan fluorescence spectroscopy, with probes placed at strategic sites, has been previously used with a variety of membrane proteins to monitor substrate binding and conformational changes that occur during the catalytic cycle [24,25,26]. Tryptophan (Trp) is considered to be the least invasive when compared to extrinsic fluorophores that must be covalently attached to the protein. Trp, as a fluorophore, eliminates labeling stoichiometry uncertainty (as it can be inserted genetically), and minimizes the risk of affecting protein structure and/or function, due to its relatively small size. However, site-specific Trp fluorescence has been precluded from use with Pgp due to the presence of eleven endogenous Trps that contribute to a high intrinsic fluorescence emission (Figure 1). Two Trps (W208 and W851) are located in extracellular loops close to the membrane bilayer interface, two (W311 and W228) are within the hydrophobic core of the membrane, W132 and three Trps in the elbow helices (W44, W694 and W704) are located at the cytoplasmic membrane interface, two (W158 and W799) are in the coupling helices that connect the TMDs to the NBDs, and W1104 is in NBD2. Sharom and colleagues have previously reported that intrinsic Pgp fluorescence is quenched by 20 to 50% upon binding of various cancer drugs [27]. However, eight Pgp Trps are located in the membrane domains (Figure 1), close enough to respond to drug binding via FRET, making it difficult to dissect which and how many Trps were responsible for the observed quenching. Intrinsic Pgp fluorescence must be reduced or eliminated to allow the use of newly-inserted Trps to localize the discrete drug binding sites within the drug binding cavity and gain insights into how the NBDs and TMDs communicate with one another to transport so many different compounds.
Figure 1. Location of the eleven endogenous Trps in Pgp.
Pgp crystal structure with the N-terminal and C-terminal homologous protein halves shaded in blue and green, respectively, and the Trps highlighted in red spheres. Dashed lines indicate the approximate boundaries of the lipid bilayer; ICL, intracellular loops; TMD, transmembrane domain; NBD1 and NBD1, N-terminal and C-terminal nucleotide binding domains, respectively. W44 and W694/W704 are located in the elbow helices that precede the N- and C-terminal TMDs, respectively. W158 and W799 are located in the ICL 2 and ICL 4 coupling helices, respectively, that connect the TMDs to the NBDs. Images were rendered in PyMol (www.pymol.org) using PDB: 3G5U.
Trp is generally the least abundant, but most conserved amino acid in a protein [28]. The aromatic amino acids, tyrosine and phenylalanine, have often been used to replace Trps while maintaining interactions that may be necessary for function, but in some situations another amino acid may be a better replacement depending on the local environment of the Trp residue [29,30]. A Trp-less Pgp was previously constructed by replacing all eleven Trps with phenylalanine. However, this protein exhibited low expression and minimal activity rendering it unsuitable for biochemical and biophysical studies [31]. In this study, we systematically analyzed the eleven endogenous Pgp Trps to identify which Trps could be replaced while maintaining protein expression and function. At each Trp position, we tested aromatic substitutions except where alignment with orthologous sequences suggested another amino acid substitution. The mutants with a single Trp removed were expressed in Saccharomyces cerevisiae to determine which mutants maintained Pgp function based on their ability to convey resistance against multiple fungicidal compounds, and complement for Ste6, a Pgp homolog required for yeast mating. Mutants that were fully active in these assays were then combined into multi-Trp mutant proteins, purified, and catalytic ability of these proteins was measured through drug-stimulated ATPase activities. Overall, we found that multiple Trps could be removed from Pgp while maintaining protein function and drug polyspecificity, but significantly reducing intrinsic fluorescence.
2. Materials and methods
2.1 Materials
FK506 and valinomycin were purchased from A.G. Scientific (San Diego, CA). Fluconazole was from LKT Laboratories (Saint Paul, MN). Doxorubicin, verapamil, cyclosporin A, and ATP were from Sigma Aldrich (Saint Louis, MO). E. coli lipids (Polar Extract) and PMPC were purchased from Avanti (Alabaster, AL), n-Dodecyl-β-D-maltopyranoside (DDM) was from Inalco (Italy).
2.2 Mutant construction and analysis in S. cerevisiae
Wt mouse Pgp (mdr3, abcb1a) in the pVT expression vector (pVT-mdr3.5) [32,33] served as a template throughout this study. Individual Trp mutations were introduced by QuickChange (Stratagene) site-directed mutagenesis using oligonucleotides containing the Trp mutation and silent mutations to identify the mutants by restriction enzyme digestion (all mutagenic oligonucleotides are listed in Supplementary Table 1). To generate the two Trp pair mutants, W311Y was introduced into the W228F ORF and W694L was introduced into the W1104Y ORF by site-directed mutagenesis. Then W228F/W311Y were combined with W694L/W1104Y by subcloning with SacI sites that flank W228F and W311Y to form the quadruple mutant W228F/W311Y/W694L/W1104Y (Quad). Mutant constructs were confirmed by DNA sequencing of the entire Pgp open reading frame. Wild-type abcb1 (Wt) and Trp mutants were then transformed into S. cerevisiae, JPY201(MATa ura3 Δste6::HIS3), cells for expression and functional assays that were performed essentially as previously described [33,34,35,36]. Briefly, 10 ml yeast cultures were grown overnight in uracil deficient medium containing 7.5% glycerol, diluted to OD600= 0.05 in YPD medium, and seeded into 96 well plates containing YPD alone or YPD plus 50 µM FK506, 100 µM valinomycin or 40 µM doxorubicin. Samples were grown in triplicate at 30°C for up to 30 hours, and yeast cell growth was monitored by measuring the OD600 at two hour increments in a microplate reader (Benchmark Plus, BioRad). The concentrations of drugs were fine-tuned to give maximum growth differences between Wt and pVT vector controls [35]. The remainder of the 10 ml cultures were used for microsomal membrane preparations to assess Pgp expression by Western blot analysis as described [35]. Statistical analyses of the functional assays were done with the SigmaPlot 11 software using a two-tail t-test.
2.3 Large scale Trp mutant expression and purification
For protein purification, Wt and mutant plasmids were transformed into strain BY4743 (relevant phenotype: MAT a/α ura3Δ0) [37], for increased biomass production versus JPY201 cells (provided by Dr. Brandt L. Schneider), and grown in 15 L fermentor cultures (BioFlow IV, New Brunswick) containing uracil deficient medium supplemented with 7.5% glycerol, as a chemical chaperone, to enhance Pgp expression [38]. Cells were harvested during log-phase growth (A280= 6–7) yielding 60–90 g cells per fermentor culture. Microsomal membranes were prepared as described for Pichia pastoris cells [36]. Wt and mutant Pgp proteins were extracted in 0.6% DDM and purified with successive nickel affinity, DE-52 anion exchange, and size exclusion chromatography steps, as previously described for P. pastoris expressed Pgp [35,36]. Protein concentration of Pgp containing fractions was determined by UV spectroscopy at 280 nm using a calculated molar extinction coefficient of 109,750 M−1cm−1 (1 A280 unit = 1.29 mg/ml). Wt and mutant proteins were further quantitated by resolving increasing protein amounts on Coomassie stained SDS-PAGE gels and compared to a BSA standard using ImageJ (http://rsbweb.nih.gov/).
2.4 ATPase Assays
For ATPase activity measurements, we modified the malachite green assay for detection of inorganic phosphate (Pi) release [39,40] for use with small volumes in a 96 well plate format as follows: Wt and mutant proteins in 0.1% DDM were activated with 10 mM DTT for 10 min on ice, and then mixed with an equal volume of 2% E. coli lipids and incubated at RT for 15 minutes [36]. Then, 0.1 µg activated protein was added to wells of a 96 well plate containing 10 µl of 10 mM ATP cocktail (10 mM ATP, 10 mM MgSO4 in 50 mM Tris-Cl buffer pH 7.5) with a 2-fold serial dilution of verapamil, FK506, valinomycin, or cyclosporin A with 150 µM verapamil, pre-warmed to 37°C. ATPase reactions were stopped at times ranging from 0 to 30 minutes by the addition of 100 µl cold 0.4 N H2SO4 containing 0.05 % DDM, and the 96 well plates were transferred to ice. Pi was measured by adding 100 µl of a stock color development solution to give a final concentration of 0.73% ammonium molybdate, 0.9 N H2SO4, 0.056% polyvinyl alcohol, and 0.011% malachite green in each sample well. Samples were then incubated at RT for 20 min and the absorbance of each sample was measure at 610 nm in a microplate reader (Benchmark Plus, BioRad). Mock samples containing buffer and lipids without any added Pgp were subtracted as background values, and inorganic phosphate standards (from 0.5 to 10 nmol) served as internal controls.
2.5 Statistical analysis
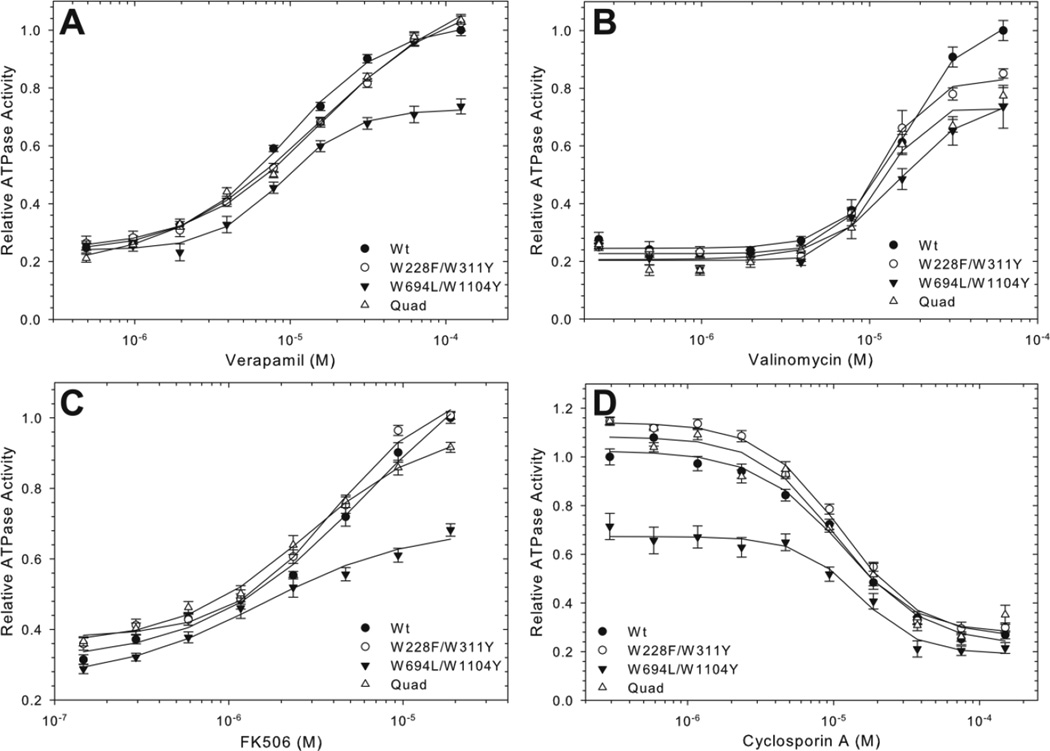
Significant differences in the yeast functional assays were determined using Student’s t-test with p=0.05 as the rejection limit. The ATPase activities for stimulatory compounds were analyzed by nonlinear regression analysis using the equation V= Vbas+ (Vmax×Sb)/(Sb+Ksb), where S is the concentration of stimulatory drug, V is the rate of ATP hydrolysis, Vbas is the ATPase activity in the absence of drug, Vmax is the maximum drug stimulated ATPase activity, Ks is the concentration of drug required for half-maximal stimulation, and b is the Hill coefficient. Cyclosporin A inhibition was analyzed using the equation V= Vbas−((Vbas−Imax)×Ib)/(Kib+Ib) where I is the inhibitor concentration, Imax is the maximal inhibited ATPase activity, and Ki is the inhibitor concentration required for half-maximal inhibition. The Hill coefficient for a given drug was similar between Wt and mutant proteins and was between ~1 for verapamil, FK506, and cyclosporin A, and ~2 for valinomycin. For each data fit, R2 was greater than 0.97 and each of the parameters was statistically significant (p<0.05). Wt and mutant Ks values were compared by Student’s t-test. All statistical analyses were performed with Sigmaplot 11.
2.6 Tryptophan fluorescence
Pgp intrinsic Trp fluorescence was measured by diluting purified proteins to 100 nM with purification buffer (20 mM HEPES, 50 mM NaCl, 10% glycerol, 0.1% DDM, pH 7.4) and measuring the steady state fluorescence emission from 310–410 nm with an excitation wavelength of 295 nm in a Fluorolog-3 spectrofluorometer (Horiba) at room temperature. Emission spectra were corrected by comparing the technical spectra of tyrosine and tryptophan with the corrected spectra as given by Eftink [41]. Rhodamine binding studies were performed in 20 mM HEPES, pH 7.4, 150 mM NaCl, 0.125% CHAPS, 0.5 mg/ml PMPC. Drug was added from concentrated stock in DMSO such that the final DMSO concentration was <1%, buffer controls containing the same amount of rhodamine/DMSO were subtracted from each sample. Inner filter corrections were carried out using NATA. Data were analyzed by nonlinear regression analysis using the equation f=d−(a×Sb)/(Sb+Kqb)), where S is the concentration of drug (rhodamine 123), a is the maximum quench, d is the Trp fluorescence in the absence of drug, Kq is the concentration of drug required for half-maximal quenching, and b is the Hill coefficient.
3. Results
3.1 Single Trp mutant expression
Our first objective was to mutate each of the eleven Pgp Trps, and determine how removing individual Trps affects protein expression and function. First, we compared Pgp amino acid sequences from different species to identify Trp substitutions that may preserve function. Human and mouse Pgp were aligned with orthologous Pgp sequences, having greater than 70% sequence identity, from the Ensembl database (http://ensembl.org) [42]. Nine of the eleven Trps were completely conserved across the 45 aligned orthologs. At W44, we found that Trp was replaced by arginine in guinea pig (Cavia porcellus), cysteine in chicken (Gallus gallus), and threonine in frog (Xenopus laevis) Pgp (Supplementary Figure 1A). We chose arginine to replace W44 since guinea pig is the closest relative to mouse and human. At W694, phenylalanine occurs in opossum (Monodelphis domestica) and frog, and leucine is present in chicken and turkey (Meleagris gallopavo) (Supplementary Figure 1B). We chose leucine to replace this Trp because ABCB4, a Pgp paralog, in mouse and several of its orthologous sequences also have leucine instead of Trp at W694. For the remaining six Trps located in the lipid bilayer or at the membrane interface (Figure 1), we chose tyrosine to replace Trp because membrane snorkeling preferences predict that it interacts more favorably with the lipid bilayer than phenylalanine [43,44,45]. The two Trps in the coupling helices (W158 and W799) were replaced by phenylalanine, found in the related bacterial ABC transporters MsbA and Sav1866 [46,47], and W1104 in NBD2 was replaced by a tyrosine.
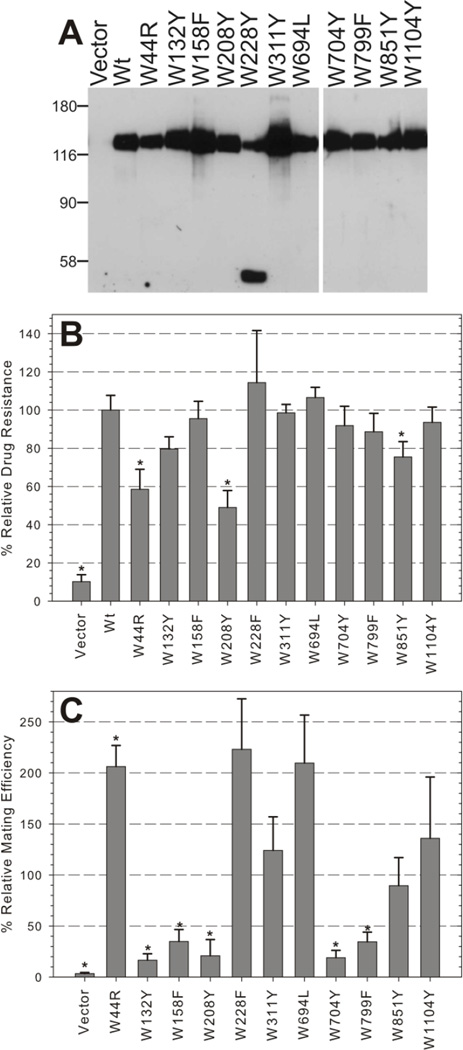
Each of these single mutants was transformed into S. cerevisiae for analysis of protein expression and function. Western blots revealed that the mutants were abundantly expressed as a full-length protein, at levels similar to or greater than Wt Pgp, except W228Y (Figure 2A). W228Y exhibited severely diminished levels of full-length protein and accumulation of a truncated mutant with an approximate molecular weight of 50 kDa. This result was reproducible with multiple clones of the mutant suggesting that tyrosine at this position adversely affects protein synthesis, folding, or trafficking. Subsequently, we replaced this membrane bound Trp (Figure 1) with phenylalanine, and found that the more hydrophobic W228F mutation is well expressed when compared to Wt Pgp (Supplemental Figure 2). Together, these data suggests that no single endogenous Trp was critical for Pgp expression.
Figure 2. Expression and function of single Trp mutants in yeast.
A, 15 µg of crude microsomal membrane protein from yeast expressing Wt Pgp, empty vector (negative control), and each Trp mutant was analyzed by Western blot using the C219 anti-Pgp antibody. A representative of at least three experiments is shown; protein MW marker positions are indicated in kDa. B, Fungicidal resistance was measured by growing mutant expressing yeast and control strains in the absence or presence of 50 µM FK506 and measuring the OD600 at 24 to 25 hours. Relative growth is the OD600 of cells grown in drug/the OD600 of cells grown in drug-free medium. Bars represent the mean relative growth ± SEM of triplicate samples from three or more independent experiments normalized to the relative growth of yeast expressing Wt Pgp. C, Mating efficiency of Trp mutants was determined by mating the a-type Trp mutant expressing cells with an α-type tester strain and plating on minimal media selective for diploids. Relative mating efficiency is the number of diploid cells divided by the total number of cells, then divided by the mating efficiency of Wt expressing yeast. Bars indicate the mean relative drug resistance ± SEM from four independent experiments. (*) indicate samples that were significantly different than Wt using a two tail t-test with p≤0.02.
3.2 Single Trp mutant function
Previous work has demonstrated that when Pgp is expressed in S. cerevisiae, it can convey fungicidal resistance to the yeast cells [34]. Furthermore, Pgp can complement for Ste6, the a-factor pheromone transporter required for mating, and restore mating in otherwise sterile Ste6 null yeast [33]. When the yeast strains expressing mutant Pgp were exposed to FK506, only three mutants (W44R, W208Y, and W851Y) had relative growths rates that were significantly less than strains expressing Wt (* in Figure 2B, p=0.0004, 0.001, 0.05, respectively). However, the mating efficiencies were severely impaired (<40% of Wt) in five of the mutants (Figure 2C) with W208Y showing low functionality in both assays. W311Y and W1104Y performed well at levels similar to Wt in both the mating and FK506 resistance assays while W228F and W694L outperformed Wt in mating efficiency by approximately 2 fold and maintained high levels of FK506 resistance. Interestingly, W44R had opposing effects in the two assays. This mutant significantly outperformed Wt in mating efficiency but had significantly impaired FK506 resistance. Overall, the combined results of these two functional assays identified a set of single Trp mutants (W228F, W311Y, W694L, and W1104Y) that likely preserve Pgp function at the plasma membrane based on FK506 transport (a macrocyclic lactone, MW of 804 g/mol) and export of yeast a-factor (a farnesylated dodecapeptide), compounds with vastly different sizes and structures [48].
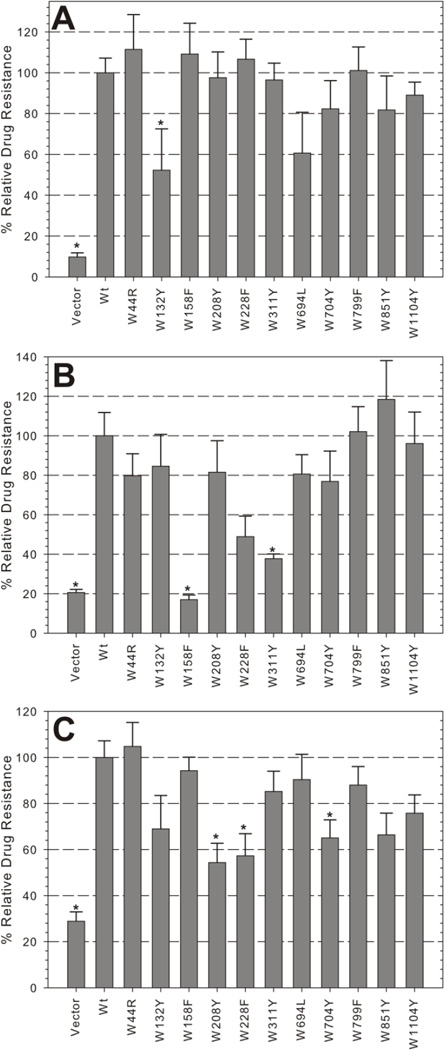
The differing performance of mutants with the two transport substrates suggested that some Trp mutations may not simply impair the protein, but may alter its specificity for some substrates. We further explored this possibility by subjecting the yeast strains to growth in three additional fungicidal drugs, valinomycin (1,111 g/mol), fluconazole (306 g/mol), and doxorubicin (544 g/mol), that represent a structurally diverse set of compounds. Most mutants retained some level of resistance (>50%) against these drugs except for W158F, which behaved similar to the vector control in fluconazole (<20% growth, Figure 3). Again, W694L and W1104Y, maintained a high level of activity (>70%) across all transport substrates assayed (Figure 3, see also Figure 2B, C). W228F and W311Y retained significant levels of drug resistance in all but one of the drugs (doxorubicin or fluconazole, respectively). Interestingly, W44R and W851Y, which were significantly impaired in FK506 (Figure 2B) but mated efficiently (Figure 2C), also maintained high levels of resistance against all three additional drugs. Together, these data indicate that the ability to mate (Figure 2C) is a stringent first test for function. However, drug resistance profiles may be altered in individual mutants and should be tested to confirm that polyspecificity, a hallmark of Pgp function, is maintained.
Figure 3. Fungicidal resistance profiles of single Trp mutant expressing yeast strains.
Mutant and control strains were analyzed for resistance to 75 µM (A) valinomycin, 65 µM fluconazole (B), and 25 µM doxorubicin (C) as in Figure 2B. Bars indicate the mean relative growth ± SEM of duplicate samples from four independent experiments, normalized to Wt relative growth. (*) indicate mutants that were significantly different than Wt (p<0.05).
3.3 Functionality of combined Trp mutations
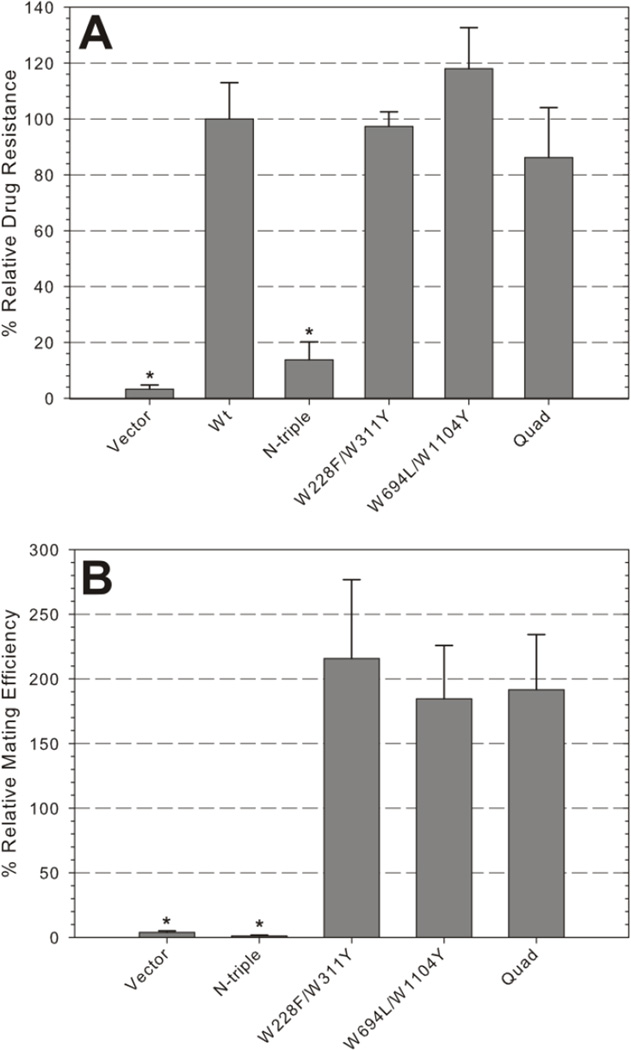
Our second objective was to determine how Pgp would be affected when multiple single Trp substitutions were combined into the same protein. Based on the functional data we first paired W228F/W311Y as well as W694L/W1104, because each of these single mutants were statistically equivalent to Wt in both the FK506 assay and the mating assay (Figures 2B and C). Coincidentally, these four Trps have previously been predicted to be major contributors to the Trp fluorescence lifetimes of Pgp [49]. Then W228F/W311Y was added to W694L/W1104Y to form the quadruple mutant W228F/W311Y/W694L/W1104Y (Quad). The pair and quad mutants exhibited expression levels similar to Wt (data not shown), maintained high levels of FK506 resistance (Figure 4A), and outperformed Wt in mating efficiency, similar to the respective single mutants (Figure 4B).These data suggest that the double and quad mutants combine the properties of the single mutants, or in other words the combinations were successful because we started with fully active single mutants.
Figure 4. Functionality of combined Trp mutations.
A, Relative resistance of mutant and control strains to 60 µM FK506. Bars indicate the mean relative growth ± SEM of triplicate samples, normalized to Wt, from five independent experiments. B, Relative mating efficiency ± SEM of five independent experiments. (*) indicate samples that are significantly different than Wt (p<0.01). N-triple mutant: W132Y/W158Y/W311Y; Quad mutant: W228F/W311Y/W694L/W1104Y.
As proof of principle, we also combined W132Y, W158F, and W311Y (N-triple mutant). All three individual mutants were highly resistant to FK506 (Figure 2B), but W132Y and W158F had significantly lower mating efficiencies than Wt. Additionally, each mutant showed significantly reduced resistance to one of the other drugs (W132Y to valinomycin, W158F to fluconazole, and W311Y to doxorubicin, see Figure 3). Combining these mutations completely ablated FK506 resistance (Figure 4A), even though protein expression was not impaired (data not shown). The N-triple mutant strain was also sterile in the yeast mating assay (Figure 4B) as expected from the low mating efficiencies of the respective W132Y and W158F mutants. The stark functional contrast between the N-triple mutant and Quad mutant demonstrates the importance of thoroughly testing individual Trp mutations before attempting to use them to construct a low-tryptophan Pgp protein that is functional.
3.4 Functionality of purified double and quad Trp mutations
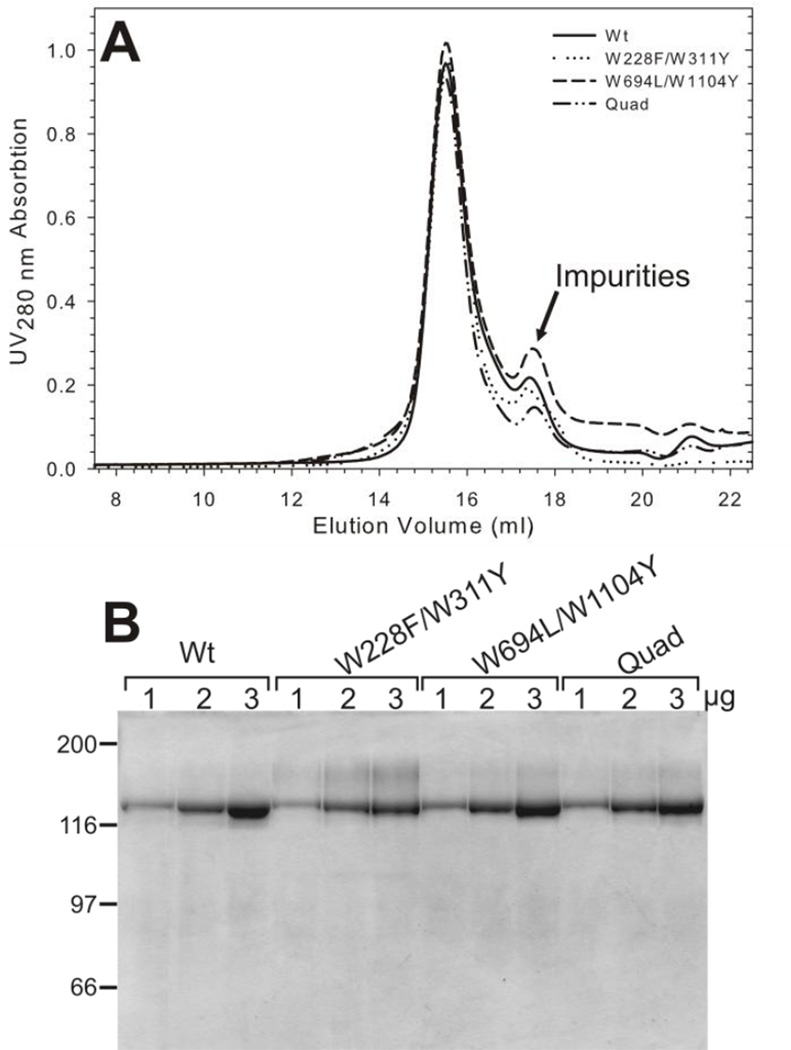
Our third objective was to examine the function of the combined pair and Quad mutants and their interaction with drug substrates at the purified protein level. Wt and the combination mutants were purified from S. cerevisiae fermentor cultures following essentially the same procedures we previously described for purification of Pgp from Pichia pastoris yeast (see Experimental Procedures). The last step involved size exclusion chromatography to remove residual contaminants, and exchange the proteins into a defined buffer for further biochemical and biophysical studies. Wt and mutant proteins all eluted as a sharp peak consistent with monomeric protein [35], well separated from smaller molecular weight proteins (labeled “impurities” in Figure 5A). Coomassie-stained SDS-gels revealed that Wt and each mutant protein were purified to near homogeneity (Figure 5B). Mutant protein yields were consistent with the Wt Pgp yield of 1.3 mg Pgp/fermentor culture (Table 1), which is somewhat lower than from Pichia pastoris [35]. However, the protein purity was excellent and the protein quantity is sufficient for biochemical and biophysical studies.
Figure 5. Purification of Wt and Trp mutant Pgp from S. cerevisiae.
A, Two milligrams (500 µl) of purified, detergent soluble proteins were loaded on a Superose 6B column (10×300 mm, GE Healthcare) and resolved in buffers containing 20 mM Hepes-NaOH pH 7.4, 10% glycerol, 50 mM NaCl, 1 mM DTT and 0.1% DDM as described [35]. A representative of four independent runs is shown for WT-Pgp and the mutant proteins. The Pgp peak at ~15.3 ml indicates an apparent size of ~200 kDa when compared to molecular mass markers resolved under identical buffer conditions [35]. The calculated molecular mass of monomeric Pgp (including the His6-tag) is 142 kDa, the predicted detergent micelle size for DDM is about 70 kDa. B) 1, 2, and 3 µg of Wt and mutant proteins were resolved on a 10% SDS-PAGE and stained with Coomassie blue. Protein MW marker positions are shown in kDa.
Table 1.
Yield and ATPase activity of Wt and Trp mutant proteins
| Yield (mg/fermentor culture) a |
ATPase Activity b (µmole Pi mg−1 min−1) |
|
|---|---|---|
| Wt | 1.3 | 0.49 ± 0.02 |
| W228F/W311Y | 1.5 | 0.54 ± 0.03 |
| W694L/W1104Y | 1.4 | 0.30 ± 0.01 |
| Quad | 1.1 | 0.63 ± 0.08 |
A 15 liter fermentor culture routinely gives between 80 to 90 g of wet cells.
Maximum drug-stimulated ATPase activity in the presence of 125 µM verapamil (mean ± SEM)
The drug stimulated ATPase activity of the purified proteins was evaluated, as this has long been a standard measure of in vitro Pgp function [36,50]. The maximum verapamil-stimulated ATPase activity of the purified pair and quad mutants was very similar to Wt Pgp, with verapamil providing a 5-fold stimulation in ATPase activity (Table 1). Half-maximal stimulatory concentration for verapamil ranged from 9 to 16 µM for Wt and the mutants (Fig. 6A, Table 2), not significantly different in a two tail t-test (p=0.05), and was in the same range as previously published [35]. Maximum stimulation of ATPase activity by valinomycin and FK506 was lower than with verapamil, about 4-fold and 3-fold over basal activity (in the absence of drug), and occurred at a concentration of 70 µM and 20 µM, respectively (Figures 6B and C). Again, the half-maximal stimulatory concentrations for valinomycin and FK506 of Wt and the mutants proteins were not significantly different in the two tail test (p=0.05, Table 2). Inhibition of verapamil-stimulated ATPase activity by the immunosuppressant cyclosporin A was also comparable for the Wt and mutant proteins, with half-maximal inhibition seen at 10–15 µM (p= 0.05 Figure 6D). Together these data indicate that the mutants were very similar to Wt Pgp in terms of yield, purity and their affinities for substrates and inhibitors in the purified proteins.
Figure 6. Drug stimulate/inhibited ATPase activity of purified Trp mutant protein.
Wt and mutant proteins were assayed for ATPase activity in increasing concentrations of verapamil (A), valinomycin (B), FK506 (C), and cyclosporin A with 150 µM verapamil (D). Data points indicate the average specific activity from multiple time points ± SEM from at least four independent experiments, relative to the maximum Wt activity. Where not visible, error bars are smaller than the symbols. Lines represent non-linear regression analysis of the data points with a Hill equation. R2 values for the data fits were between 0.97 and 0.99. Kinetic parameters of the data fits are given in Table 1.
Table 2.
Drug-stimulated ATPase activity of Wt and Trp Mutant proteins
| Verapamil a | Ks (µM) b | Valinomycin a | Ks (µM) b | FK506 a | Ks (µM)b | Cyclosporin A Ki (µM) c |
|
|---|---|---|---|---|---|---|---|
| Wt | 5.1 | 9.7 ± 0.5 | 5.5 | 16 ± 0.9 | 4.0 | 4.7 ± 1.3 | 12 ± 1.5 |
| W228F/W311Y | 5.2 | 16 ± 1.2 | 5.5 | 12 ± 0.7 | 4.2 | 4.0 ± 0.7 | 11 ± 1.0 |
| W694L/W1104Y | 2.7 | 9.0 ± 0.7 | 3.7 | 15 ± 2.7 | 2.1 | 1.6 ± 0.4 | 15 ± 2.0 |
| Quad | 4.0 | 16 ± 3.1 | 4.4 | 12 ± 1.5 | 3.3 | 2.8 ± 0.3 | 10 ± 2.0 |
Fold maximum stimulation of ATPase activity at 150 µM verapamil, 70 µM valinomycin and 20 µM FK506.
Concentration required for half-maximal ATPase activity derived from plots of V vs. [drug] in Figure 6A, B, and C, fitted for a single exponential (mean± SEM).
Cyclosporin A concentration required for half maximal inhibition of verapamil-stimulated ATPase (Figure 6D) (mean± SEM).
3.5 Intrinsic tryptophan fluorescence of Wt and mutant proteins
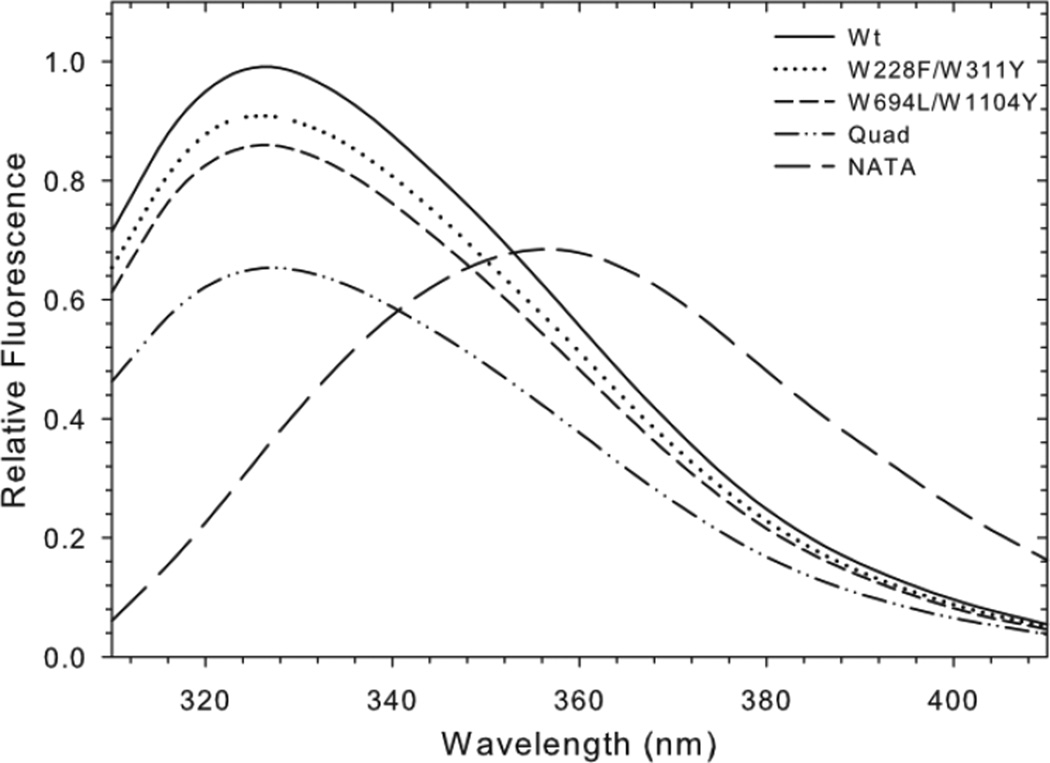
The goal of removing Pgp Trps was to minimize intrinsic protein fluorescence, but also to determine the contribution of certain Trps to the Pgp fluorescence signal. Trp fluorescence spectra of Wt and mutant Pgp proteins are shown in Figure 7. The Wt emission spectrum was shifted to shorter wavelengths (maximum at 327 nm) when compared to the tryptophan analog NATA (356 nm), as previously reported, suggesting that the native tryptophans are mostly in a hydrophobic environment [27]. Assuming a quantum yield of 0.13 for NATA [51], the average quantum yield of the eleven Wt Trps was estimated to be 0.17. The Wt emission maximum at 327 nm did not shift significantly when two or four Trps were removed (Figure 7). However, the emission intensity did decrease with subsequent removal of Trps. The W228F/W311Y and W694L/W1104Y mutations contributed to approximately 11 ± 7% and 14% ± 11% decreases in emission, respectively, while the Quad mutant emission was approximately 34% ± 7% less than Wt. Overall, the intrinsic fluorescent signal decreased proportionately with the number of Trps removed (Supplemental Figure 3). Removing four of the eleven endogenous Trps resulted in a 1/3 reduction in intrinsic fluorescence. Taking into account the rather high average quantum yield, the data suggest that most, if not all Trp residues contribute to the intrinsic Pgp fluorescence, in contrast to a previous report concluding that the intrinsic fluorescence arises from a few emitters [49].
Figure 7. Intrinsic Trp fluorescence of Wt and mutant Pgp.
100 nM purified Pgp in 0.1% DDM solutions were excited at 295 nm and the fluorescent emission spectra were collected from 310–410 nm. Lines represent the average corrected emission spectra from three independent experiments normalized to the maximum emission of Wt Pgp. The emission maxima of all four proteins occurred between 325 and 228 nm. For comparison, the emission spectra of a 1µM solution of the Trp analog NATA is also shown.
3.6 Drug binding to Wt and quad mutant proteins
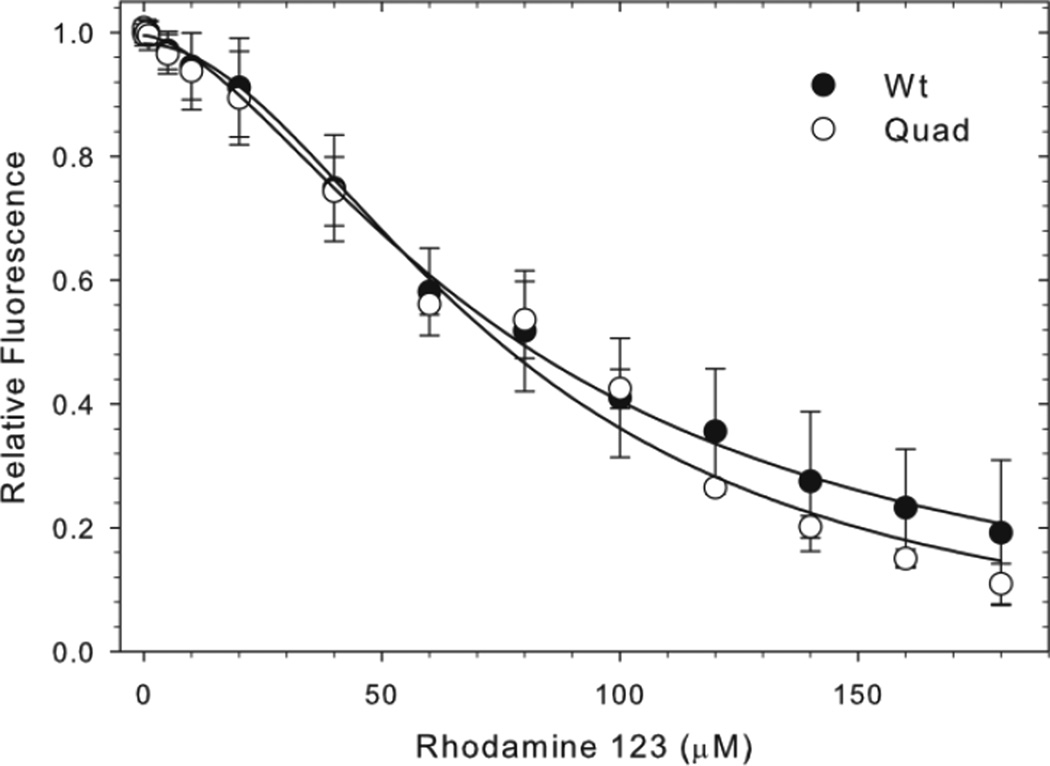
To compare the drug binding properties of Wt and quad mutant proteins we used the fluorescent transport substrate rhodamine 123 that produced strong Trp fluorescence quenching upon binding to Wt Pgp [27]. Experiments were carried out in the presence of phospholipid PMPC dissolved in 0.125% (2 mM) CHAPS, which gave low background fluorescence. Addition of rhodamine 123 resulted in a concentration-dependent Trp fluorescence quench that was similar in the Wt and quad mutant. Half-maximal quenching occurred at 80 ± 7 µM and 77 ± 11 µM in Wt and the quad mutant, respectively (Figure 8), indicating unaltered binding affinity of the quad mutant. Hill coefficients of 1.6 and 1.9 suggest positive cooperativity between two binding sites in both proteins. Wt Pgp contains eight Trps in the TMDs (Figure 1), close enough to the drug binding sites to serve as a FRET donor to Rhodamine 123 assuming a characteristic FRET transfer distance R0 of 17 Å of this donor/acceptor pair. The Quad mutant has W228F/W311Y/W694L/W1104Y substituted, three of which are located in the membrane. The similar quenching characteristics between Wt and quad mutant suggest that most or all of the eight transmembrane Trps contribute to the quench seen upon binding of rhodamine 123 to Wt Pgp.
Figure 8. Effect of binding of rhodamine 123 on the intrinsic Trp fluorescence of Wt and mutant Pgp.
100 nM purified Wt, 150 nM quad mutant Pgp or 1.5 µM NATA in 0.5 mg/ml PMPC/0.125 CHAPS solutions were excited at 295 nm and fluorescent emission spectra were collected after successive additions of rhodamine 123 (0.01 to 180 µM). Data points indicate the average Trp quenching (at 327 nm) of Wt and quad mutant ± SEM from four independent experiments, relative to the maximum Trp fluorescence, corrected for inner filter effects (NATA). Lines represent non-linear regression analyses of the data points; R squared values were 0.998 and 0.990 for Wt and quad mutants, respectively.
4. Discussion
Most Trps are highly conserved among Pgp species suggesting a role of Trps in maintaining the structural stability of the protein, the architecture of the drug binding sites and/or specific interactions with hydrophobic drug substrates [52,53]. This study addresses two basic questions: A) Can every one of the eleven native Trps in Pgp be substituted while retaining function? B) Can multiple Trp substitutions be combined into a low-Trp Pgp (several Trps replaced) that is fully functional and expressed at wild-type levels? To answer these questions we chose “traditional” substitutions (tyrosine and phenylalanine) with respect to the particular location within the membrane and in intracellular domains (Figure 1), or amino acids found in Pgp orthologs (W44R and W694L). Only four single Trp mutants fully retained their ability to convey resistance against a diverse set of antifungal drugs and to complement for Ste6, the yeast a-factor pheromone transporter required for mating (Figures 2 and 3). Among them were W228F and W311Y located within the membrane bilayer, W694L at the beginning of the elbow helix situated at the intracellular membrane interface, and W1104Y in NBD2. Those could be combined into pair (W228F/W311Y and W694L/W1104Y) and quadruple (W228F/W311Y/W694L/W1104Y) mutants that were fully active in yeast assays (Figure 4). Furthermore the pair and quad mutants could be purified to homogeneity in yields similar to Wt protein, and displayed drug-stimulated ATPase activity with binding affinities very similar to Wt Pgp (Tables 1 and 2). The intrinsic Trp fluorescence of the pair and quad mutants was reduced approximately proportionally to the number of Trps removed with ~35% reduction achieved in the quad mutant. Together these findings provide convincing evidence that polyspecificity for diverse transport substrates was preserved when fully active single Trp mutants were combined into a low-Trp Pgp.
This study also shed some light into those Trp residues that cannot be simply replaced by an aromatic amino acid without adversely affecting protein expression, folding and/or function. Strikingly, several single Trp mutants retained high levels of drug resistance against FK506 (and other drugs) but showed severely impaired mating efficiency (Figure 2A vs. B). Examples are W132Y and W704Y, situated at the cytoplasmic membrane interface, as well as W159F and W799F, located in the coupling helices that connect the TMDs to the NBDs. It is worth mentioning that in drug resistance assays, yeast cells are continuously exposed over a prolonged time (>24h) to the drug that may act in the manner of chemical chaperones and enhance folding and trafficking of the mutant proteins to the cell surface [54,55,56]. In contrast, mating is dependent on secretion of a-factor pheromone and only mutants that traffic normally to the cell surface and retain their ability to export the pheromone will attract α-tester cells and mate efficiently. The latter may account for the more stringent discrimination of mutant function seen in mating assays. Biophysical studies suggest tyrosine is a good substitute at the membrane surface because, like Trp, it can orient its polar side chain towards the polar phospholipid head groups of the membrane bilayer (‘snorkeling’ effect) and anchor helices in the membrane [43,44,45,52]. However for some tyrosine substitutions, interactions with the membrane interface alone may not fully account for the diverse phenotypes observed. For example, in the inside-facing conformation of the Pgp crystal structure the indole side chain of W132 points to the inside of the drug binding cavity and may thereby interact not only with the lipid interface but also directly contact some substrates by participating in H-bonding, cation-bonding, and aromatic stacking interactions. Effects on substrate specificity similar to W132Y were previously observed for tyrosine as well as phenylalanine substitutions of Trps located in the TMDs of the related multidrug ABC transporters MRP1, MRP2 and MRP3 [53,57,58,59]. Other examples are W704 that is situated at the turn of the elbow helix and the first α-helix of TMD2, or W208 and W851 located in the two very short extracellular loops. Possibly, a tyrosine substitution at these positions may experience additional stereochemical constraints leading to slight folding defects that can be ‘rescued’ by some drugs (in resistance assays) but are detectable in mating assays. Notably, W208Y was not only severely impaired in mating but also showed significantly reduced resistance to two of the drugs, FK506 and doxorubicin, The defects point to a severe interference with the architecture of the drug binding sites that could not be rescued by those drugs.
W158 and W799 are situated in the coupling helices that embed into a groove on the NBDs. These are distant from the drug binding sites but may be important in stabilizing the hairpin structure of the coupling helices for proper connection. A phenylalanine is found at the homologous positions of the related bacterial ABC transporters MsbA and Sav1866 [46,47]. However, phenylalanine substitutions of W158 and W799 in Pgp led to severely impaired mating efficiencies. It is currently not well understood how coupling signals are transmitted between NBDs and TMDs and whether they differ between ABC proteins transporting different substrates. The severe defects seen in W158F and W799F point to the importance of properly coupled interdomain communication in Pgp.
Finally, it is appropriate to comment on the tryptophan-deficient Pgp protein that was previously constructed by replacing all eleven endogenous Trps with phenylalanine (Mdr3F1–11) [31]. That protein exhibited altered drug resistance patterns in mammalian cells. It also did not confer FK506 resistance nor could it functionally complemented Ste6, perhaps due to severely compromised protein expression in yeast, and has not been deployed in Trp fluorescence studies. Chimeric constructs between Wt and the Mdr3F1–11 mutant suggested that reduced activity was not caused by loss of any uniquely important Trp but rather a cumulative loss of several Trps. Interestingly, several studies on gradient-driven transporters showed that those proteins are much more tolerant to the straight-forward strategy of replacing Trp by phenylalanine. Successful examples are the outer membrane protein A (five Trps replaced) [60], the lac permease (six Trps replaced) [30], the Na+/H+ antiporter NhaA from E. coli (six Trps replaced) [24], and the human sodium/glucose cotransporter SGLT1 (13 replaced and one was essential for function) [29]. In contrast, for Pgp the current study highlights the importance of thoroughly testing individual Trp substitutions before attempting to combine mutations into a low-tryptophan Pgp protein that is functional. This was exemplified in the N-triple mutant, a combination of W132Y, W158F and W311Y substitutions that individually were all highly resistant to FK506, but each mutant showed significantly reduced resistance to one of the other drugs and W132Y and W158F also showed impaired mating efficiencies; this N-triple combination completely lost function (Figure 4). In stark contrast, combining W228F, W311Y, W694L, and W1104Y, which each maintained high activity in FK506 resistance and mating assays resulted in a fully active protein that could be purified in sufficient quantities and biochemically characterized (Figures 5 and 6). The purified pair (W228F/W311Y and W694L/W1104Y) and Quad (W228F/W311Y/W694L/W1104Y) mutants exhibited levels of drug stimulated ATPase activity similar to Wt Pgp with multiple drugs, confirming that as a whole polyspecificity was maintained in these mutants.
The intrinsic fluorescence of the Quad mutant, although reduced by ~1/3 compared to Wt Pgp, remains too high for site-specific drug binding studies. This was exemplified in Trp fluorescence quenching studies using the fluorescent transport substrate rhodamine 123. Rhodamine, like many Pgp substrates, absorbs light between 300 to 400 nm and can serve as FRET acceptor from Trps in close proximity (calculated R0 at which 50% transfer occurs is 17Å). Wt Pgp contains eight Trps in the TMDs that are within 15 to 25 Å from the QZ59-SSS binding sites; except W44 that is ~40 Å away (Figure 1). The exact location of the rhodamine binding site(s) in Pgp is currently unknown. The similar quenching characteristics of Wt and the quad mutant, together with a Hill coefficient greater than one (1.5 and 1.9, respectively) suggest two binding site for rhodamine that are likely within FRET range of most of the eight transmembrane Trps in Wt Pgp (five in the quad mutant) and contribute to the quenching either by FRET or due to perturbation of the Trp probe’s immediate environment.
We conclude that Trp removal is a challenging task especially for complex systems such as mammalian ABC transporters containing multiple TMDs and large cytoplasmic domains [53]. Many individual substitutions of Trps at the membrane surface significantly reduced Pgp drug resistance and/or yeast cell mating efficiency suggesting that they are important in maintaining polyspecificity and functional integrity of the drug binding sites. However, four mutants, W228F, W311Y, W694L, and W1104Y (three of which are located in the membrane) and their combinations retained high drug resistance and yeast cell mating in vivo as well as drug-stimulated ATPase activities similar to WT Pgp. Removal of those four Trp residues reduced the intrinsic fluorescence of Pgp, but not enough for its use in site-specific drug binding studies. Together, these results are a first step toward understanding the role of Trps in Pgp and towards the development of a tryptophan-less Pgp, that will be vital for studying the drug binding and transport mechanism of Pgp.
Supplementary Material
Highlights.
Substitution of the eleven native Trps in the P-glycoprotein multidrug transporter.
Several Trps at the membrane interface are critical for substrate polyspecificity.
Trps in the coupling helices are important for interdomain communication.
Single Trp mutants can be combined while maintaining drug polyspecificity.
Multi-Trp mutant protein is fully active with reduced intrinsic fluorescence.
ACKNOWLEDGMENTS
We thank Guillermo Altenberg for careful reading of the manuscript, Roger B. Sutton for help with modeling a lipid POPC bilayer in the Pgp structure (shown in the Graphical abstract) using the program Orientation of Proteins in Membranes (OPM), and Ping Bai and Sri Karan Botta for excellent technical assistance. This work was supported by the Cancer Prevention and Research Institute of Texas RP101073, the National Institute of Health RGM102928 and U54-GM94610, and the South Plains Foundation.
ABBREVIATIONS
- Pgp
P-glycoprotein
- ABC
ATP Binding Cassette
- TMD
transmembrane domain
- NBD
nucleotide binding domain
- Trp
tryptophan
- DDM
n-Dodecyl-β-D-maltopyranoside
- NATA
N-acetyl-L-tryptophanamide
- PMPC
1-palmitoyl-2-myristoyl-sn-glycero-3-phosphocholine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Eckford PD, Sharom FJ. ABC efflux pump-based resistance to chemotherapy drugs. Chemical Reviews. 2009;109:2989–3011. doi: 10.1021/cr9000226. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro AB, Ling V. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur J Biochem. 1997;250:122–129. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- 3.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, Cellular, and Pharmacological Aspects of the Multidrug Transporter. Annual Review of Pharmacology and Toxicology. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 5.J.K. Karaszi E, Homolya L, Szakacs G, Hollo Z, Telek B, Kiss A, Rejto L, Nahajevszky S, Sarkadi B. Kappelmayer J Calcein assay for multidrug resistance reliably predicts therapy response and survival rate in acute myeloid leukaemia. British Journal of Haematology. 2001;112:308–314. doi: 10.1046/j.1365-2141.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 6.Leith CP, Kopecky KJ, Chen IM, Eijdems L, Slovak ML, McConnell TS, Head DR, Weick J, Grever MR, Appelbaum FR, Willman CL. Frequency and clinical significance of the expression of the multidrug resistance proteins MDR1/P-glycoprotein, MRP1, and LRP in acute myeloid leukemia: a Southwest Oncology Group Study. Blood. 1999;94:1086–1099. [PubMed] [Google Scholar]
- 7.Cordon-Cardo C, O'Brien JP, Boccia J, Casals D, Bertino JR, Melamed MR. Expression of the multidrug resistance gene product (P-glycoprotein) in human normal and tumor tissues. J Histochem Cytochem. 1990;38:1277–1287. doi: 10.1177/38.9.1974900. [DOI] [PubMed] [Google Scholar]
- 8.Schinkel AH. P-Glycoprotein, a gatekeeper in the blood-brain barrier. Adv Drug Deliv Rev. 1999;36:179–194. doi: 10.1016/s0169-409x(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 9.Watkins PB. The barrier function of CYP3A4 and P-glycoprotein in the small bowel. Advanced Drug Delivery Reviews. 1997;27:161–170. doi: 10.1016/s0169-409x(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 10.Pariante CM. The role of multi-drug resistance p-glycoprotein in glucocorticoid function: studies in animals and relevance in humans. Eur J Pharmacol. 2008;583:263–271. doi: 10.1016/j.ejphar.2007.11.067. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 12.Tamaki A, Ierano C, Szakacs G, Robey RW, Bates SE. The controversial role of ABC transporters in clinical oncology. Essays in Biochemistry. 2011;50:209–232. doi: 10.1042/bse0500209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Rzhetsky A, Allikmets R. The Human ATP-Binding Cassette (ABC) Transporter Superfamily. Genome Research. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 14.Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal Duplication and Homology with Bacterial Transport Proteins in the Mdr1 (P-Glycoprotein) Gene from Multidrug-Resistant Human-Cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 15.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-Glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo TW, Bartlett MC, Clarke DM. Simultaneous Binding of Two Different Drugs in the Binding Pocket of the Human Multidrug Resistance P-glycoprotein. J. Biol. Chem. 2003;278:39706–39710. doi: 10.1074/jbc.M308559200. [DOI] [PubMed] [Google Scholar]
- 17.Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- 18.Gutmann DAP, Ward A, Urbatsch IL, Chang G, van Veen HW. Understanding polyspecificity of multidrug ABC transporters: closing in on the gaps in ABCB1. Trends in Biochemical Sciences. 2010;35:36–42. doi: 10.1016/j.tibs.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo TW, Bartlett MC, Clarke DM. The "LSGGQ" motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J Biol Chem. 2002;277:41303–41306. doi: 10.1074/jbc.C200484200. [DOI] [PubMed] [Google Scholar]
- 20.Urbatsch IL, Sankaran B, Bhagat S, Senior AE. Both P-glycoprotein nucleotide-binding sites are catalytically active. J Biol Chem. 1995;270:26956–26961. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- 21.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 22.Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PM, O'Mara ML, George AM. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends in Biochemical Sciences. 2009;34:520–531. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Kozachkov L, Padan E. Site-directed tryptophan fluorescence reveals two essential conformational changes in the Na+/H+ antiporter NhaA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15769–15774. doi: 10.1073/pnas.1109256108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirnova I, Kasho V, Sugihara J, Kaback HR. Probing of the rates of alternating access in LacY with Trp fluorescence. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21561–21566. doi: 10.1073/pnas.0911434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber J, Senior AE. Features of F-1-ATPase catalytic and noncatalytic sites revealed by fluorescence lifetimes and acrylamide quenching of specifically inserted tryptophan residues. Biochemistry. 2000;39:5287–5294. doi: 10.1021/bi992730t. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Siemiarczuk A, Sharom FJ. Intrinsic fluorescence of the P-glycoprotein multidrug transporter: sensitivity of tryptophan residues to binding of drugs and nucleotides. Biochemistry. 2000;39:14927–14938. doi: 10.1021/bi0018786. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Tyagi NK, Goyal P, Pandey D, Siess W, Kinne RKH. Sodium-independent low-affinity D-glucose transport by human sodium/D-glucose cotransporter 1: Critical role of tryptophan 561. Biochemistry. 2007;46:2758–2766. doi: 10.1021/bi061696x. [DOI] [PubMed] [Google Scholar]
- 30.Menezes ME, Roepe PD, Kaback HR. Design of a membrane transport protein for fluorescence spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1638–1642. doi: 10.1073/pnas.87.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwan T, Loughrey H, Brault M, Gruenheid S, Urbatsch IL, Senior AE, Gros P. Functional analysis of a tryptophan-less P-glycoprotein: a tool for tryptophan insertion and fluorescence spectroscopy. Mol Pharmacol. 2000;58:37–47. doi: 10.1124/mol.58.1.37. [DOI] [PubMed] [Google Scholar]
- 32.Vernet T, Dignard D, Thomas DY. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52:225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- 33.Raymond M, Gros P, Whiteway M, Thomas DY. Functional complementation of yeast ste6 by a mammalian multidrug resistance mdr gene. Science. 1992;256:232–234. doi: 10.1126/science.1348873. [DOI] [PubMed] [Google Scholar]
- 34.Beaudet L, Gros P. Functional dissection of P-glycoprotein nucleotide-binding domains in chimeric and mutant proteins. Modulation of drug resistance profiles. J Biol Chem. 1995;270:17159–17170. doi: 10.1074/jbc.270.29.17159. [DOI] [PubMed] [Google Scholar]
- 35.Bai JP, Swartz DJ, Protasevich, Brouillette CG, Harrell PM, Hildebrandt E, Gasser B, Mattanovich D, Ward A, Chang G, Urbatsch IL. A Gene Optimization Strategy that Enhances Production of Fully Functional P-Glycoprotein in Pichia pastoris. Plos One. 2011;6 doi: 10.1371/journal.pone.0022577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE. Large Scale Purification of Detergent-soluble P-glycoprotein from Pichia pastoris Cells and Characterization of Nucleotide Binding Properties of Wild-type, Walker A, and Walker B Mutant Proteins. J. Biol. Chem. 1999;274:34711–34718. doi: 10.1074/jbc.274.49.34711. [DOI] [PubMed] [Google Scholar]
- 37.Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Figler RA, Omote H, Nakamoto RK, Al-Shawi MK. Use of chemical chaperones in the yeast Saccharomyces cerevisiae to enhance heterologous membrane protein expression: high-yield expression and purification of human P-glycoprotein. Arch Biochem Biophys. 2000;376:34–46. doi: 10.1006/abbi.2000.1712. [DOI] [PubMed] [Google Scholar]
- 39.Van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Analytical Biochemistry. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- 40.Cogan EB, Birrell GB, Griffith OH. A robotics-based automated assay for inorganic and organic phosphates. Analytical Biochemistry. 1999;271:29–35. doi: 10.1006/abio.1999.4100. [DOI] [PubMed] [Google Scholar]
- 41.Eftink MR. Fluorescence techniques for studying protein structure. Methods of Biochemical Analysis. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- 42.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GRS, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fern ndez-Suarez XM, Harrow J, Herrero J, Hubbard TJP, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SMJ. Ensembl 2012. Nucleic Acids Research. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamberlain AK, Lee Y, Kim S, Bowie JU. Snorkeling preferences foster an amino acid composition bias in transmembrane helices. Journal of Molecular Biology. 2004;339:471–479. doi: 10.1016/j.jmb.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 44.Granseth E, von Heijne G, Elofsson A. A study of the membrane-water interface region of membrane proteins. Journal of Molecular Biology. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 45.Liang J, Adamian L, Jackups R. The membrane-water interface region of membrane proteins: structural bias and the anti-snorkeling effect. Trends in Biochemical Sciences. 2005;30:355–357. doi: 10.1016/j.tibs.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 47.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderegg RJ, Betz R, Carr SA, Crabb JW, Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 1988;263:18236–18240. [PubMed] [Google Scholar]
- 49.Lugo MR, Sharom FJ. Interaction of LDS-751 with the drug-binding site of P-glycoprotein: a Trp fluorescence steady-state and lifetime study. Arch Biochem Biophys. 2009;492:17–28. doi: 10.1016/j.abb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Urbatsch IL, al-Shawi MK, Senior AE. Characterization of the ATPase activity of purified Chinese hamster P-glycoprotein. Biochemistry. 1994;33:7069–7076. doi: 10.1021/bi00189a008. [DOI] [PubMed] [Google Scholar]
- 51.Muino PL, Callis PR. Solvent Effects on the Fluorescence Quenching of Tryptophan by Amides via Electron Transfer. Experimental and Computational Studies. Journal of Physical Chemistry B. 2009;113:2572–2577. doi: 10.1021/jp711513b. [DOI] [PubMed] [Google Scholar]
- 52.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 53.Koike K, Oleschuk CJ, Haimeur A, Olsen SL, Deeley RG, Cole SP. Multiple membrane-associated tryptophan residues contribute to the transport activity and substrate specificity of the human multidrug resistance protein, MRP1. J Biol Chem. 2002;277:49495–49503. doi: 10.1074/jbc.M206896200. [DOI] [PubMed] [Google Scholar]
- 54.Loo TW, Clarke DM. Correction of Defective Protein Kinesis of Human P-glycoprotein Mutants by Substrates and Modulators. Journal of Biological Chemistry. 1997;272:709–712. doi: 10.1074/jbc.272.2.709. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Loo TW, Bartlett MC, Clarke DM. Modulating the folding of P-glycoprotein and cystic fibrosis transmembrane conductance regulator truncation mutants with pharmacological chaperones. Molecular Pharmacology. 2007;71:751–758. doi: 10.1124/mol.106.029926. [DOI] [PubMed] [Google Scholar]
- 56.Gautherot J, Durand-Schneider AM, Delautier D, Delaunay JL, Rada A, Gabillet J, Housset C, Maurice M, Ait-Slimane T. Effects of Cellular, Chemical, and Pharmacological Chaperones on the Rescue of a Trafficking-defective Mutant of the ATP-binding Cassette Transporter Proteins ABCB1/ABCB4. Journal of Biological Chemistry. 2012;287:5070–5078. doi: 10.1074/jbc.M111.275438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito K, Olsen SL, Qiu W, Deeley RG, Cole SP. Mutation of a single conserved tryptophan in multidrug resistance protein 1 (MRP1/ABCC1) results in loss of drug resistance and selective loss of organic anion transport. J Biol Chem. 2001;276:15616–15624. doi: 10.1074/jbc.M011246200. [DOI] [PubMed] [Google Scholar]
- 58.Ito K, Oleschuk CJ, Westlake C, Vasa MZ, Deeley RG, Cole SPC. Mutation of Trp(1254) in the multispecific organic anion transporter, multidrug resistance protein 2 (MRP2) (ABCC2), alters substrate specificity and results in loss of methotrexate transport activity. Journal of Biological Chemistry. 2001;276:38108–38114. doi: 10.1074/jbc.M105160200. [DOI] [PubMed] [Google Scholar]
- 59.Oleschuk CJ, Deeley RG, Cole SPC. Substitution of Trp(1242) of TM17 alters substrate specificity of human multidrug resistance protein 3. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2003;284:G280–G289. doi: 10.1152/ajpgi.00331.2002. [DOI] [PubMed] [Google Scholar]
- 60.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.