Abstract
Mitochondria must grow with the growing cell to ensure proper cellular physiology and inheritance upon division. We measured the physical size of mitochondrial networks in budding yeast and found that mitochondrial network size increased with increasing cell size and that this scaling relation occurred primarily in the bud. The mitochondria to cell size ratio continually decreased in aging mothers over successive generations. However, regardless of mother age or mitochondrial content, all buds attained the same average ratio. Thus, yeast populations achieve a stable scaling relation between mitochondrial content and cell size despite asymmetry in inheritance.
Main Text
The amount of mitochondria in the cell varies in response to metabolic demands (1), requiring active size regulation mechanisms. The mitochondrial to cell size ratio is relatively constant in mammalian cells (2) and two yeast species (3, 4). We used the budding yeast, Saccharomyces cerevisiae to study how the relationship between mitochondrial and cell size is achieved in cells growing and dividing asymmetrically (fig. S1). Yeast mitochondria are three-dimensional networks of dynamic membrane bound tubules localized at the cell periphery (5). To study mitochondrial size scaling, we developed a method to quantify the 3D skeletons of mitochondrial networks (Figs. 1A, S2) using spinning disk confocal Z-stacks of live yeast cells expressing mitochondrial matrix-targeted GFP (Green Fluorescent Protein). We converted network length to mitochondrial volume (μm3) assuming a constant tubule diameter. Absolute network length accuracy was within 85% of manual measurements, with an average reproducibility of 96% (6).
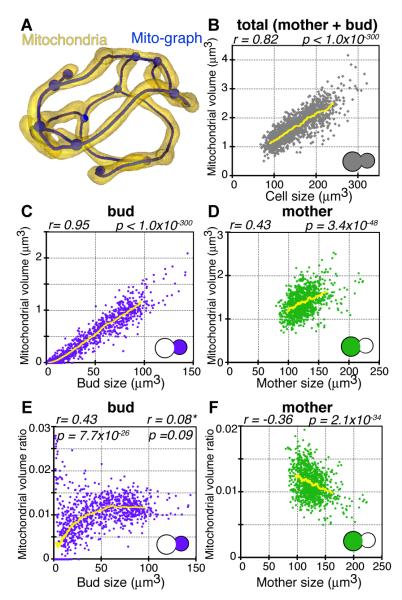
Fig. 1.
Mitochondrial-cell size scaling is strongest in the bud. (A) Example of a mitochondrial network graph (blue), generated from 3D confocal images of a mitochondrion (yellow), imaged using matrix-targeted GFP in a live yeast cell. (B) Mitochondrial volume vs. cell size for all cells analyzed in the timecourse population (n = 1430), including budding and non-budding cells. The population maintained a consistent average mitochondrial to cell volume ratio. (C, D) Mitochondrial volume in the bud or mother and (E, F) Mitochondrial volume ratio in the bud or mother vs. bud or mother size, respectively for all budding cells (n = 1053). (C, D) Buds displayed a much stronger correlation between mitochondrial and cell volume than mothers. Pearson’s correlation coefficient, r, and significance value, p, are shown. Values for r and p on left and right sides in (E) are for the bud population greater/less than 40μm3. Yellow lines indicate rolling average.
We imaged timecourses of live yeast cells growing for one to two generations and measured the total cell and mitochondrial volumes for mother and bud compartments. We found a strong correlation between total mitochondrial and cell volumes (Fig. 1B). However, when we compared mother and bud compartments separately, the constant mitochondrial volume ratio found in the population as a whole was dominated by the scaling in the bud (Fig. 1C, D). Mitochondrial volume ratio in the buds increased and then leveled off when buds were about half their final size (Fig. 1E). Bud size reflects the progression of budding, suggesting buds accumulate mitochondria until they reach a set-point of mitochondrial volume relative to bud size. In contrast, in mothers the mitochondrial volume ratio decreased with increasing cell size (Fig. 1F). Mother size reflects generational age (7), suggesting older mothers don’t preserve the mitochondrial volume ratio set-point they inherited when they were buds.
Analysis of the dynamics of cell growth and mitochondrial accumulation during budding timecourses revealed dramatic asymmetry in mitochondrial accumulation between mother and bud (Fig. 2A). During budding, mothers experienced an overall loss of both mitochondrial volume and the resultant volume ratio, while at the same time mitochondrial content increased in the bud proportional to bud growth (Figs. 2, S3, S4). The mitochondrial accumulation rate in mothers was most negative at the moment that it was greatest in the bud (fig. S3B), suggesting the bud might gain mitochondria at the expense of its mother. Although first and 2+ generation mothers differed significantly in cell size and mitochondrial content at the start of budding, the average kinetics of both bud growth and mitochondrial accumulation in their buds was indistinguishable (Figs. 2C, S3), resulting in the same average volume ratios at division (Fig. 2D). Thus, regardless of the initial size or mitochondrial content of mothers, they generate, on average, identical buds and do so with the same dynamics throughout budding.
Fig. 2.
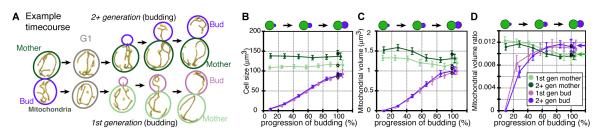
Mitochondrial accumulation dynamics during progression of budding. (A) Overview of timecourse experiment. First generation mothers were considered separately from older, 2+, generation mothers. Yellow and blue represent mitochondria and network graphs. Colors for first generation (1st gen) and older generation (2+ gen) mothers and buds correspond to the colors used in all other figures. (B) Average cell size, (C) mitochondrial volume, and (D) mitochondrial volume ratio for mothers (green) and buds (purple) averaged throughout progression of budding, in percent units where 100% is average time until division for that generation. Pre-division data points, outlined in black, are included at t=100%. Error bars: +/− 95% confidence intervals and numbers of cells in (6). (B) As expected, mothers grew very little (7), while the buds grew rapidly, with a peak growth rate halfway through budding (fig. S3A). (C) Mothers lost, while their buds gained, mitochondrial volume. (D) The resultant mitochondrial volume ratio in the bud increased and then plateaued, (similar to Fig. 1E). At division, mothers had a significantly lower mitochondrial volume ratio than their buds (arrows in D depict mean values; p-val = 0.005 by rank sum, n=25).
Yeast cells displayed “mitochondrial content asymmetry” with a lower mitochondrial volume ratio in the mother than in the bud upon division (Fig. 2D). We asked if mitochondrial content is replenished during G1. The mitochondrial volume ratio tended to increase for cells with lower, and decrease for cells with higher initial volume ratios (Figs. 3A, S5), suggesting a partial homeostatic restoration towards a unique mitochondrial volume ratio set-point. We found no evidence that cells delayed G1 exit in response to a decreased volume ratio (fig. S6), but instead that cells may respond to deviations in the appropriate volume ratio by adjusting their rate of mitochondrial biogenesis (Figs. 3B, S7). Analysis of mitochondrial content versus generational age (Fig. 3C) showed that sequential loss during budding and partial regain during G1 resulted in continual loss of mitochondrial volume ratio in aging mother cells (Figs. 3D, S8, 9). However, these much older mothers still generated buds with the same average volume ratio at division. By generating buds with identical mitochondrial content, the population can renew and maintain a narrow distribution of mitochondrial volume ratios despite asymmetry in the inheritance of mitochondrial content between mother and bud (fig. S10).
Fig. 3.
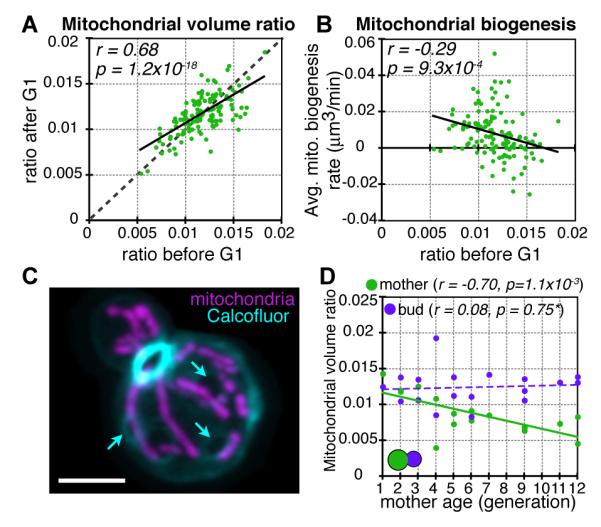
Homeostatic control of mitochondrial content during cell aging. (A) In individual cells, mitochondrial volume ratio is strongly correlated between the start and end of G1 (n = 130). Linear regression trendline (black) and line of equal volume ratio at the start and end of G1 (dotted) are shown. (B) Average mitochondrial biogenesis rate during G1 is negatively correlated with mitochondrial volume ratio at start of G1 (n = 129). (C) Example of bud scar staining with Calcofluor (cyan) and mitochondria (magenta) for a mother of generational age 10. Images are maximum intensity projections of a z-stack. Scale bar is 3μm. Arrows point to three example bud scars. (D) Pre-division mitochondrial volume ratio vs. mother age for Calcofluor-labeled mothers (green) and buds (purple; 6). Volume ratio is maintained in buds but decreases in mothers as they age. Solid lines in (A), (B), and (D) represent statistically significant linear regression trendlines and Pearson’s correlation coefficient, r, and significance value, p, are shown. Dotted line in (D) represents the best-fit trendline.
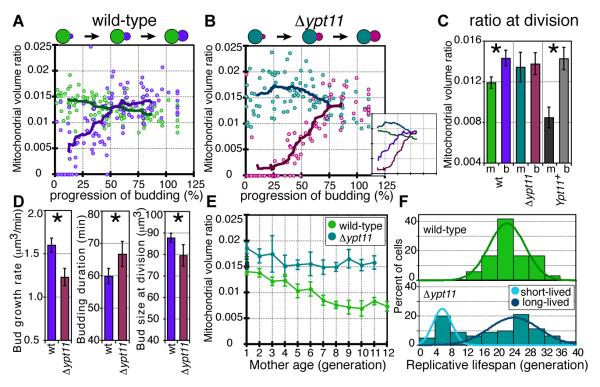
There are two possibilities for how proper mitochondrial volume ratio is achieved in the bud: a passive mechanism not modulated as a function of mitochondrial content, where scaling would arise inherently, e.g. allometric growth (8) or an active mechanism capable of sensing and feedback. In the passive case, a delay in mitochondrial inheritance would lead to less mitochondria entering the bud and a decreased volume ratio in the newborn mothers. In the active case cells could sense and compensate for the delay to achieve the target volume ratio in the bud upon division. In Δypt11 mutants, which exhibit delayed appearance of mitochondria in the bud (6, 9-10), we found that while buds were delayed in mitochondrial inheritance (figs. S11, 12), their volume ratio increased rapidly such that volume ratios in Δypt11 and wild-type buds were indistinguishable at division (Fig. 4A, B), consistent with an active mechanism. Meanwhile, mother mitochondrial content increased transiently, presumably because mitochondria could not be redistributed into the buds. Mother-daughter mitochondrial content asymmetry was thus eliminated in the absence, and was enhanced with overexpression, of Ypt11p (Fig. 4C), suggesting that Ypt11p contributes to this asymmetry. However in all three cases, the proper volume ratio was achieved in the bud at division. We thus propose that yeast cells can somehow sense mitochondrial accumulation kinetics in the bud and respond to ensure that the target volume ratio is achieved in time for division. The compensatory behavior observed in Δypt11 mutants likely involves one or both of the remaining mitochondrial inheritance pathways (6,10), which themselves may also contribute to mother-daughter mitochondrial content asymmetry.
Fig. 4.
Mitochondrial accumulation dynamics and aging in Δypt11 mutant. (A) Wild-type and (B) Δypt11 buds reach the same mitochondrial volume ratio at division despite different accumulation dynamics during budding. Mitochondrial volume ratio of (A) wild-type and (B) Δypt11 mothers (green and turquoise, respectively) and buds (purple and magenta, respectively) as a function of progression of budding. Average timecourse behavior was obtained by converting bud sizes from single timepoint data into progression of budding (6, fig. S16). Thick and thin lines indicate the rolling average and 95% confidence interval, respectively. (C) Ypt11p is involved in generating mother-daughter mitochondrial content asymmetry. The average mitochondrial volume ratio at division for wild-type (n=39), Δypt11 (n=28), and Ypt11p-overexpressing (Ypt11+; n=36; 12) mothers (m) and buds (b). Asterisks: p-values of 1.9×10−5 and 5.6 ×10−9 by rank sum. (D) Average bud growth rate, duration of budding (n = 56 and 24 for wild-type and Δypt11 buds, respectively), and bud size at division (n = 106 and 53, respectively) for wild-type (purple) and Δypt11 (magenta) buds during budding. Asterisks: p-values of 5.3×10−6, <1×10−14, and 4.6 ×10−4 by rank sum. (E) Age-dependent loss of mitochondrial volume ratio from mothers. The average mitochondrial volume ratio for wild-type (green) and Δypt11 (turquoise) mothers of increasing generational age. Darker 1st generation data-points represent average values of mothers at their “birth” (6). Error bars: +/− 95% confidence intervals and numbers of cells in (6). (F) Replicative lifespan histogram for wild-type (green, n=36) and Δypt11 (turquoise, n=80) cells calculated using live-cell imaging in a microfluidic device (6). The Δypt11 distribution was found to be bimodal, consisting of two Gaussian-like populations (light and dark turquoise curves) and the maximal lifespan (cells living beyond 30 generations, fig. S15) was found to be significantly higher for Δypt11 than wild-type cells (statistics details in 6).
Δypt11 buds could potentially reach the proper mitochondrial volume ratio by slowing their growth and “waiting” for the delayed mitochondrial content to catch up. We analyzed timecourses of Δypt11 cells and found that buds indeed grew at slower rates and for a longer time prior to division (Fig. 4D). Mutants with the longest budding durations exhibited greatly decreased mitochondrial volume ratios at the time when normal Δypt11cells would divide (fig. S13). However, Δypt11 cells divided with buds of a smaller size than wild-type buds, suggesting, surprisingly, that cell size may be affected by mitochondrial inheritance kinetics. Yeast cell size is known to respond to changes in metabolism brought about by nutritional environment (11), suggesting perhaps a delay in mitochondrial inheritance somehow altered the cell’s metabolic state even though its environment remained unchanged.
We tested whether simply delaying redistribution of mitochondria into the bud could alter the age-dependent loss of volume ratio observed in wild-type mothers (Fig. 3D). Aging Δypt11 mothers lost mitochondrial volume ratio much more slowly than wild-type mothers (Figs. 4E, S14). Replicative lifespan distributions of Δypt11 mutants (6) were similar to Δmmr1 mitochondrial inheritance mutants (12). The Δypt11 distribution was statistically bimodal (6): consisting of a shorter-lived population, not seen in wild type, and a longer-lived population with both a slightly increased average and significantly increased maximum lifespan compared to wild type (Figs. 4F, S15, 6). Because of their decreased lifespans, short-lived Δypt11 cells represent a decreasing proportion of the aging Δypt11 population (e.g. 19% and 5% of cells at generations 6 and 10). Thus, the Δypt11 cells with increased mitochondrial volume ratios at later generations consisted almost entirely of the second population with a longer lifespan than wild-type cells. Thus, in addition to the quality of mitochondria retained in the mother (12), their quantity may also contribute to their replicative lifespans.
The failure of mother cells to maintain mitochondrial volume ratio suggests that they either cannot sense reduced content or are unable to compensate for the loss. Failing to preserve her own mitochondrial volume ratio could be the cost incurred for generating the healthiest, fittest offspring upon division.
Supplementary Material
Acknowledgments
We thank J. Nunnari, J. Shaw, and A. Lewandowska for plasmids, strains, and discussions, G. Pesce for sharing unpublished results and discussions, and A. Caudy, E. Hong, KC Huang, A. Rosebrock, and the Marshall Lab for reading the manuscript. Microscopy was performed at the UCSF Nikon Imaging Center. This work was supported by Sandler and Boyer Postdoctoral Fellowships (SMR); the Herbert Boyer Junior Faculty Endowed Chair Award, and NIH grant R01GM097017 (WFM); an NIH NRSA fellowship (YHMC); FAPESP (05/00587-5, 07/50882-9) and CNPq (301303/06-1) grants (LdFC and MPV); NIH grants R01GM070808, R01GM026259, P50Gm081879, and a Packard Fellowship (YZ and HL); a China Scholarship Council (CSC) scholarship (YZ); and NIH grant 5RO1GM097213-02 (PY and JCF). Image analysis algorithms are described in detail in methods (6). The data are included in the main and supplementary figures.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS
References and Notes
- 1.Devin A, Rigoulet M. Regulation of mitochondrial biogenesis in eukaryotic cells. Toxicol. Mech. Methods. 2004;14:271. doi: 10.1080/15376520490479620. [DOI] [PubMed] [Google Scholar]
- 2.Posakony JW, England JM, Attardi G. Mitochondrial growth and division during the cell cycle in HeLa cells. J. Cell Biol. 1977;74:468. doi: 10.1083/jcb.74.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka K, Kanbe T, Kuroiwa T. Three-dimensional behaviour of mitochondria during cell division and germ tube formation in the dimorphic yeast Candida albicans. J. Cell Sci. 1985;73:207. doi: 10.1242/jcs.73.1.207. [DOI] [PubMed] [Google Scholar]
- 4.Uchida M, et al. Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast. 2011;28:227. doi: 10.1002/yea.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunnari J, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materials and methods are available as supporting material on Science Online.
- 7.Woldringh CL, Huls PG, Vischer NO. Volume growth of daughter and parent cells during the cell cycle of Saccharomyces cerevisiae a/alpha as determined by image cytometry. J. Bacteriol. 1993;175:3174. doi: 10.1128/jb.175.10.3174-3181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huxley JS, Tessier G. Terminology of relative growth. Nature. 1936;137:780–781. [Google Scholar]
- 9.Itoh T, Watabe A, Toh EA, Matsui Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:7744. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frederick RL, Okamoto K, Shaw JM. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner JJ, Ewald JC, Skotheim JM. Cell Size Control in Yeast. Curr. Biol. 2012;22:R350. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFaline-Figueroa JR, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- 14.Karren MA, Conrood EM, Anderson TK, Shaw JM. The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J Cell Biol. 2005;171:291. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. 2010;14:20.1. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3370. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Z, et al. Molecular phenotyping of aging in single cells using a novel microfluidic device. Aging Cell. 2012;11:599. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao G, Wan W, Zhang S, Redden DT, Allison DB. Testing for differences in distribution tails to test for differences in ‘maximum’ lifespan. BMC Med. Res. Methodol. 2008;8:49. doi: 10.1186/1471-2288-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder WJ, Avila LS, Hoffman W. Visualizing with VTK: A tutorial. IEEE Computer Graphics and Applications. 2000;20:20. [Google Scholar]
- 20.Cornea ND, Silver D, Yuan XS, Balasubramanian R. Computing hierarchical curve-skeletons of 3D objects. Visual Computer. 2005;21:945. [Google Scholar]
- 21.Lam L, Lee SW, Suen CY. Thinning Methodologies - a Comprehensive Survey. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1992;14:869. [Google Scholar]
- 22.Chen H, Clyborne W, Sedat JW, Agard DA. PRIISM: an integrated system for display and analysis of three-dimensional microscope images. SPIE-the International Society for Optical Engineering. 1992;1660:784. [Google Scholar]
- 23.Sprague BL, et al. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 2003;84:3529. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996;84:389. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]
- 25.Chan YHM, Marshall WF. Scaling properties of cell and organelle size. Organogenesis. 2009;6:88. doi: 10.4161/org.6.2.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen P, et al. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 2007;18:3523. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann FR, Nurse P. Nuclear size control in fission yeast. J. Cell Biol. 2007;179:593. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 1977;105:79. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 29.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2001;2:339. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 30.Kuo TC, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 2011;13:1214. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boldogh IR, Ramcharan SL, Yang HC, Pon LA. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol Biol Cell. 2004;15:3994. doi: 10.1091/mbc.E04-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortsch J, Hummel E, Krist M, Westermann B. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J Cell Biol. 2011;194:473. doi: 10.1083/jcb.201012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.