Abstract
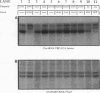
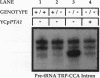
We have identified an essential Saccharomyces cerevisiae gene, PTA1, that affects pre-tRNA processing. PTA1 was initially defined by a UV-induced mutation, pta1-1, that causes the accumulation of all 10 end-trimmed, intron-containing pre-tRNAs and temperature-sensitive but osmotic-remedial growth. pta1-1 does not appear to be an allele of any other known gene affecting pre-tRNA processing. Extracts prepared from pta1-1 strains had normal pre-tRNA splicing endonuclease activity. pta1-1 was suppressed by the ochre suppressor tRNA gene SUP11, indicating that the pta1-1 mutation creates a termination codon within a protein reading frame. The PTA1 gene was isolated from a genomic library by complementation of the pta1-1 growth defect. Episome-borne PTA1 directs recombination to the pta1-1 locus. PTA1 has been mapped to the left arm of chromosome I near CDC24; the gene was sequenced and could encode a protein of 785 amino acids with a molecular weight of 88,417. No other protein sequences similar to that of the predicted PTA1 gene product have been identified within the EMBL or GenBank data base. Disruption of PTA1 near the carboxy terminus of the putative open reading frame was lethal. Possible functions of the PTA1 gene product are discussed.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Kirchner G., Chen J. Y., Vijayraghavan U., Jacobson A., Martin N. C., Abelson J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990 Sep 25;265(27):16216–16220. [PubMed] [Google Scholar]
- Atkinson N. S., Dunst R. W., Hopper A. K. Characterization of an essential Saccharomyces cerevisiae gene related to RNA processing: cloning of RNA1 and generation of a new allele with a novel phenotype. Mol Cell Biol. 1985 May;5(5):907–915. doi: 10.1128/mcb.5.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N. S., Hopper A. K. Chromosome specificity of polysomy promotion by disruptions of the Saccharomyces cerevisiae RNA1 gene. Genetics. 1987 Jul;116(3):371–375. doi: 10.1093/genetics/116.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Cameron G. N. The EMBL data library. Nucleic Acids Res. 1988 Mar 11;16(5):1865–1867. doi: 10.1093/nar/16.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Y., Kirchner G., Aebi M., Martin N. C. Purification and properties of yeast ATP (CTP):tRNA nucleotidyltransferase from wild type and overproducing cells. J Biol Chem. 1990 Sep 25;265(27):16221–16224. [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987 Oct;105(4):1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Winey M. Split tRNA genes and their products: a paradigm for the study of cell function and evolution. Yeast. 1989 Nov-Dec;5(6):405–427. doi: 10.1002/yea.320050602. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Olson M. V. Transcription and processing of cloned yeast tyrosine tRNA genes microinjected into frog oocytes. Nature. 1979 Mar 8;278(5700):137–143. doi: 10.1038/278137a0. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Winey M., Ursic D., Webb F., Culbertson M. R. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992 May;12(5):2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Ribonucleases, tRNA nucleotidyltransferase, and the 3' processing of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl B. E., Pringle J. R. Molecular analysis of Saccharomyces cerevisiae chromosome I: identification of additional transcribed regions and demonstration that some encode essential functions. Genetics. 1991 Feb;127(2):287–298. doi: 10.1093/genetics/127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983 Feb;32(2):537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Homyak M., Edenfield J., Eisenberg D. Profile scanning for three-dimensional structural patterns in protein sequences. Comput Appl Biosci. 1988 Mar;4(1):61–66. doi: 10.1093/bioinformatics/4.1.61. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Pringle J. R. Genetic analysis of Saccharomyces cerevisiae chromosome I: on the role of mutagen specificity in delimiting the set of genes identifiable using temperature-sensitive-lethal mutations. Genetics. 1991 Feb;127(2):279–285. doi: 10.1093/genetics/127.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Rauhut R., Vijayraghavan U., Abelson J. Accumulation of pre-tRNA splicing '2/3' intermediates in a Saccharomyces cerevisiae mutant. EMBO J. 1990 Apr;9(4):1245–1252. doi: 10.1002/j.1460-2075.1990.tb08232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Banks F. A yeast mutant which accumulates precursor tRNAs. Cell. 1978 Jun;14(2):211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Hopper A. K. Genetic methods for study of trans-acting genes involved in processing of precursors to yeast cytoplasmic transfer RNAs. Methods Enzymol. 1990;181:400–421. doi: 10.1016/0076-6879(90)81139-l. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Schultz L. D., Shapiro R. A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980 Mar;19(3):741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Hurt D. J., Wang S. S., Lin Y. H., Hopper A. K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987 Mar;7(3):1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Oeller P. W., Yde Steensma H., Hirschman J., Ruezinsky D., Coleman K. G., Pringle J. R. Temperature-sensitive lethal mutations on yeast chromosome I appear to define only a small number of genes. Genetics. 1984 Sep;108(1):67–90. doi: 10.1093/genetics/108.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Knapp G., Ogden R. C., Peebles C. L., Abelson J. Splicing of yeast tRNA precursors: structure of the reaction intermediates. Cell. 1979 Sep;18(1):37–45. doi: 10.1016/0092-8674(79)90351-9. [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Rohlman C. E., Molony L. A., Engelke D. R. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991 Feb;11(2):721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Borts R. H., Haber J. E. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987 Feb;115(2):233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S. M., Phizicky E. M. An enzyme from Saccharomyces cerevisiae uses NAD+ to transfer the splice junction 2'-phosphate from ligated tRNA to an acceptor molecule. J Biol Chem. 1991 Jun 25;266(18):11986–11992. [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Ohya Y., Ohsumi Y., Anraku Y. Nucleotide sequence of the CLS4 (CDC24) gene of Saccharomyces cerevisiae. Gene. 1987;54(1):125–132. doi: 10.1016/0378-1119(87)90354-4. [DOI] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981 Jan 15;145(2):405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987 Jun 5;49(5):613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983 Feb;32(2):525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Phizicky E. M., Schwartz R. C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986 Feb 25;261(6):2978–2986. [PubMed] [Google Scholar]
- Rauhut R., Green P. R., Abelson J. Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J Biol Chem. 1990 Oct 25;265(30):18180–18184. [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. A synthetic substrate for tRNA splicing. Anal Biochem. 1987 Oct;166(1):90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- Rose M. D. Isolation of genes by complementation in yeast. Methods Enzymol. 1987;152:481–504. doi: 10.1016/0076-6879(87)52056-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Russell R. R. Temperature-sensitive osmotic remedial mutants of Escherichia coli. J Bacteriol. 1972 Nov;112(2):661–665. doi: 10.1128/jb.112.2.661-665.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. Nonsense suppressors of yeast cause osmotic-sensitive growth. Proc Natl Acad Sci U S A. 1977 Jan;74(1):305–309. doi: 10.1073/pnas.74.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U., Company M., Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989 Aug;3(8):1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Hopper A. K. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol Cell Biol. 1988 Dec;8(12):5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M. R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988 Apr;118(4):609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko G. S., Park F. J., Jones E. W. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics. 1982 Dec;102(4):679–690. doi: 10.1093/genetics/102.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W. H., Wills N., Broach J. R. A general screen for mutant of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics. 1989 Sep;123(1):55–68. doi: 10.1093/genetics/123.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]