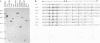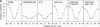Abstract
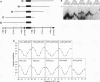
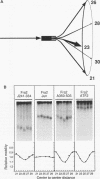
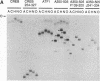
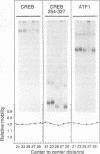
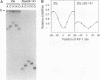
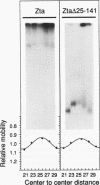
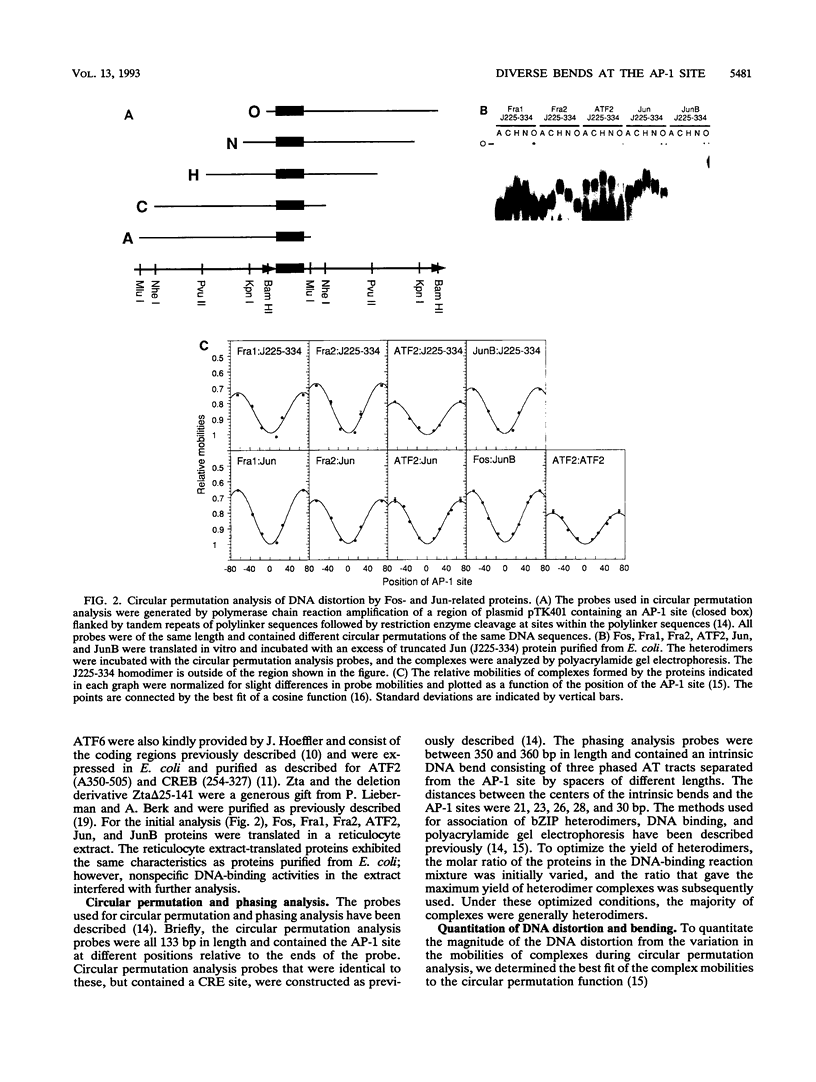
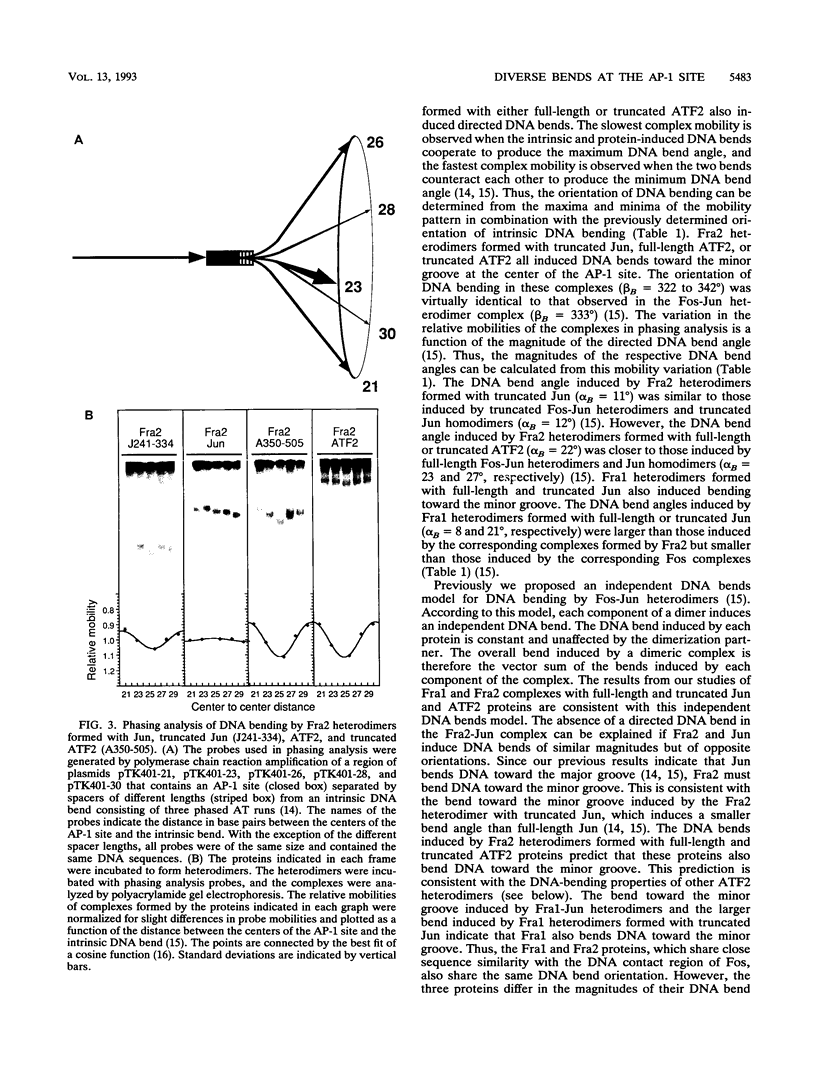
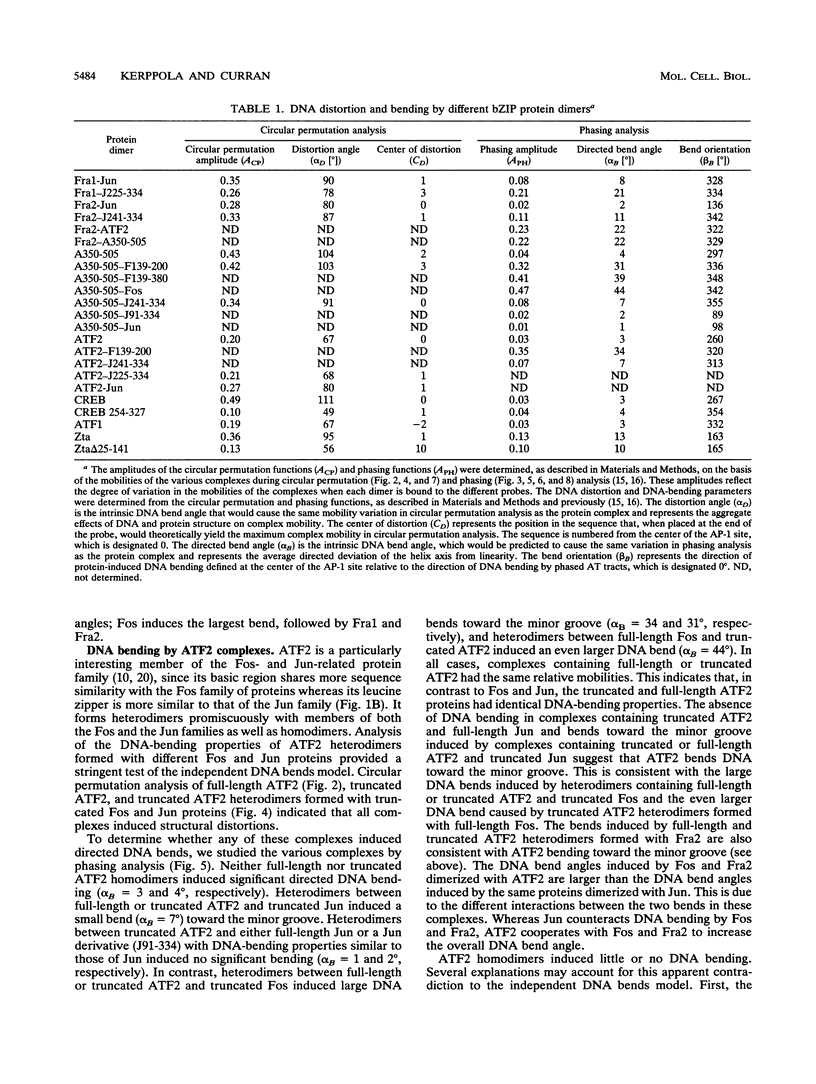
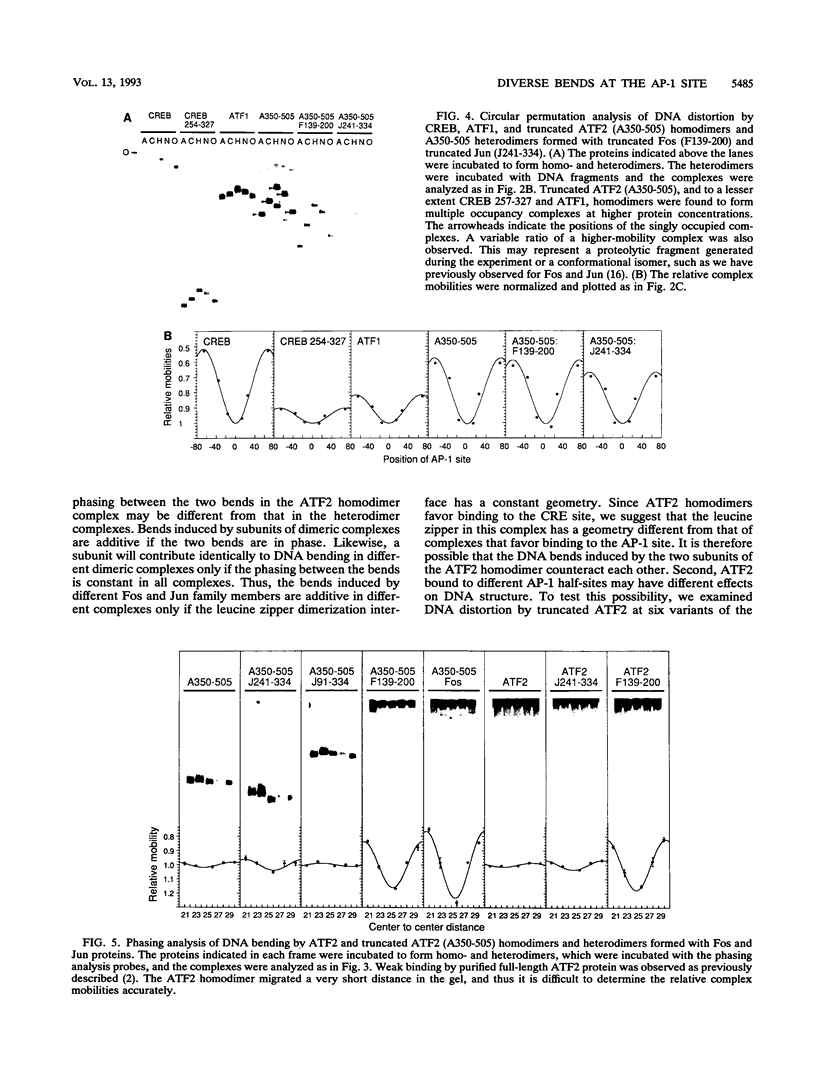
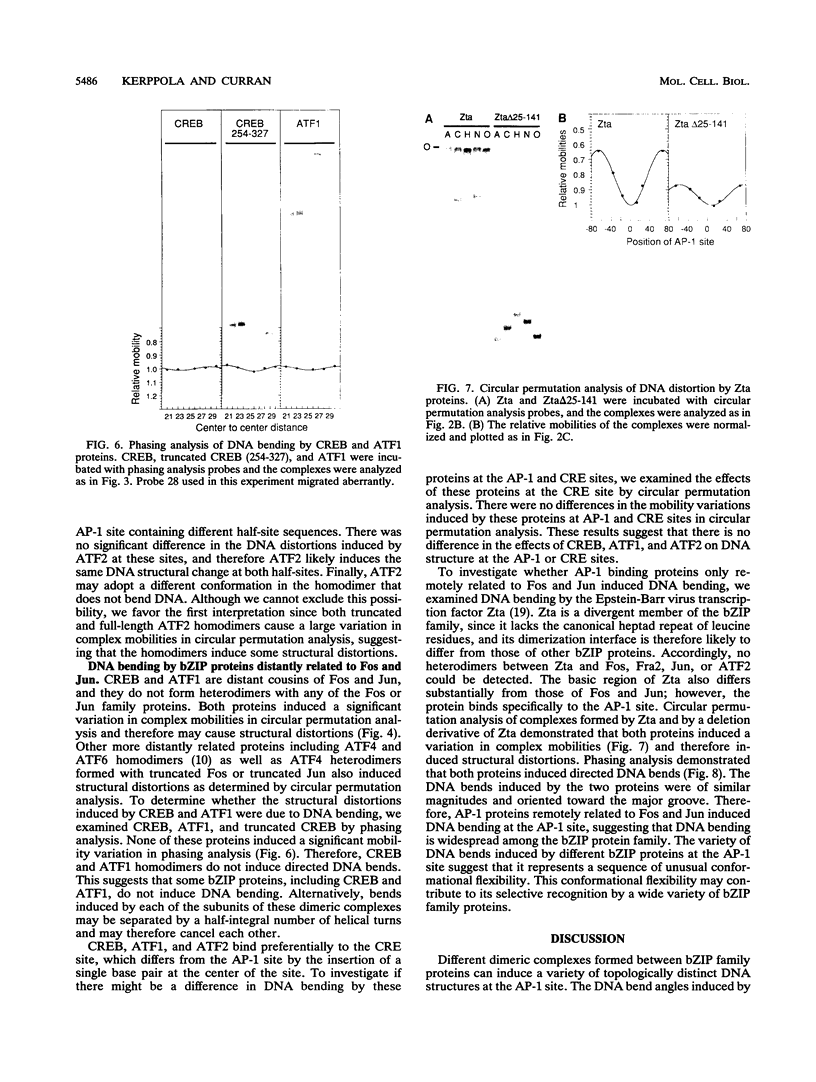
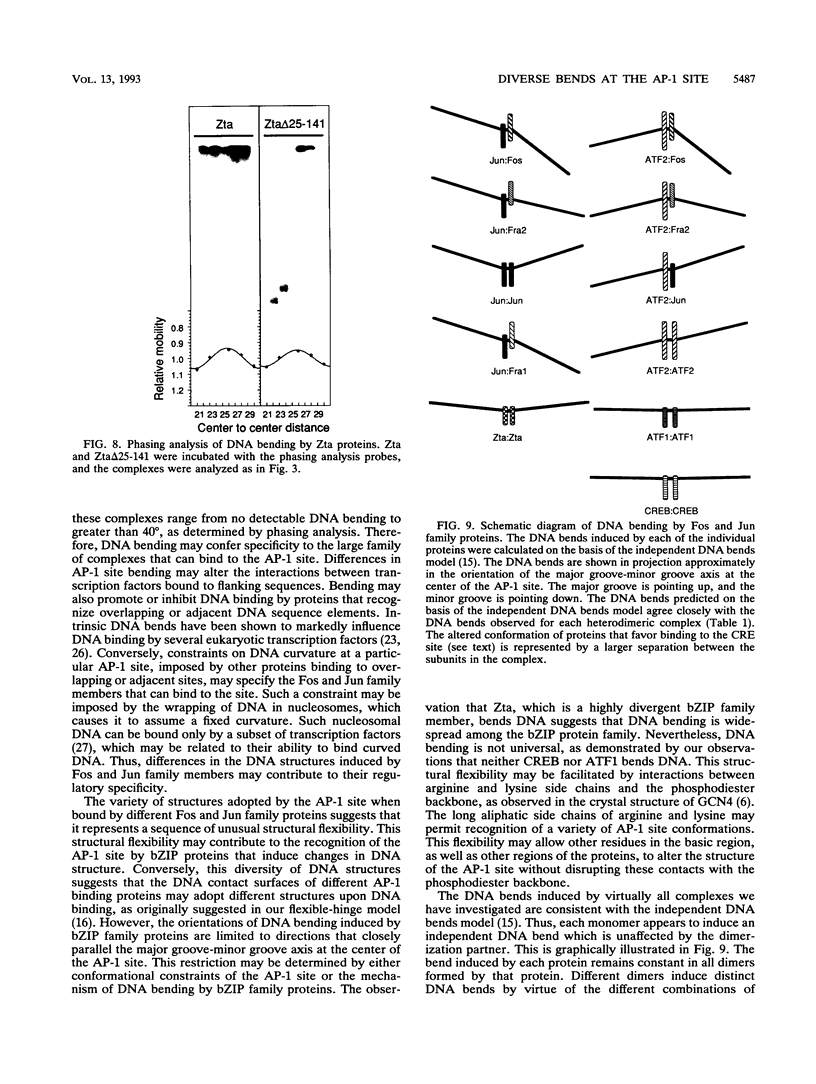
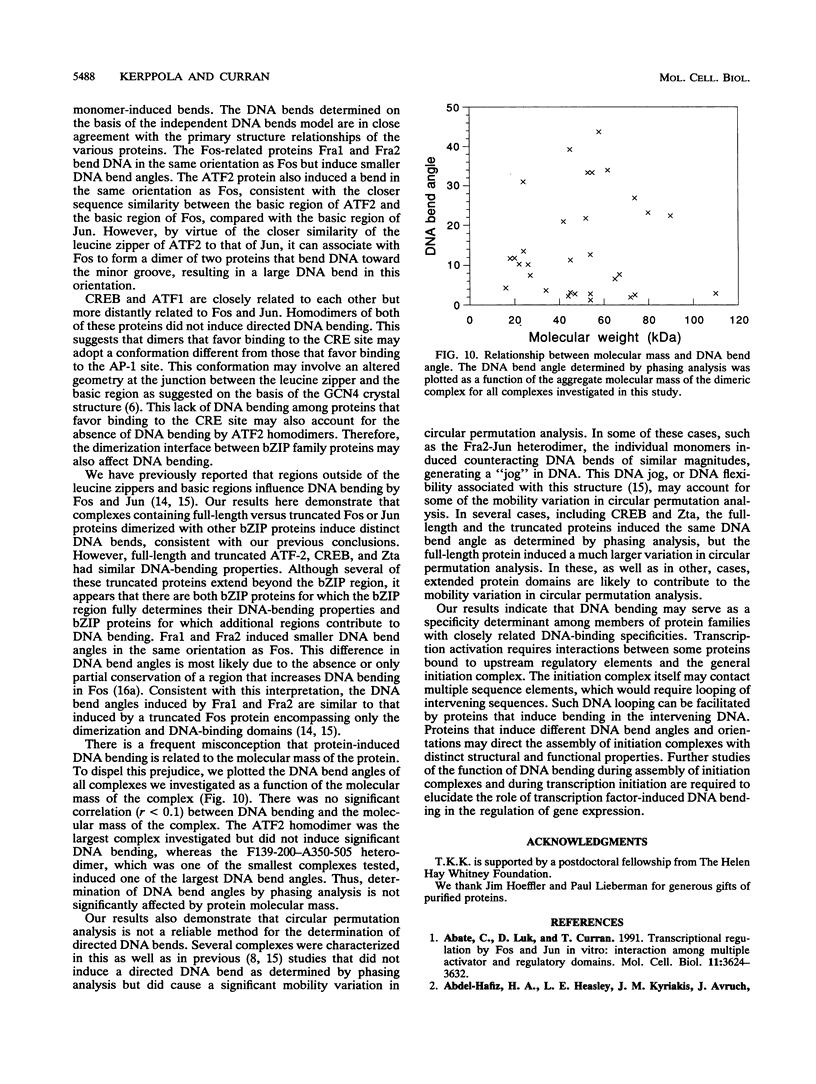
We have investigated DNA bending by bZIP family proteins that can bind to the AP-1 site. DNA bending is widespread, although not universal, among members of this family. Different bZIP protein dimers induced distinct DNA bends. The DNA bend angles ranged from virtually 0 to greater than 40 degrees as measured by phasing analysis and were oriented toward both the major and the minor grooves at the center of the AP-1 site. The DNA bends induced by the various heterodimeric complexes suggested that each component of the complex induced an independent DNA bend as previously shown for Fos and Jun. The Fos-related proteins Fra1 and Fra2 bent DNA in the same orientation as Fos but induced smaller DNA bend angles. ATF2 also bent DNA toward the minor groove in heterodimers formed with Fos, Fra2, and Jun. CREB and ATF1, which favor binding to the CRE site, did not induce significant DNA bending. Zta, which is a divergent member of the bZIP family, bent DNA toward the major groove. A variety of DNA structures can therefore be induced at the AP-1 site through combinatorial interactions between different bZIP family proteins. This diversity of DNA structures may contribute to regulatory specificity among the plethora of proteins that can bind to the AP-1 site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Luk D., Curran T. Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol Cell Biol. 1991 Jul;11(7):3624–3632. doi: 10.1128/mcb.11.7.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hafiz H. A., Heasley L. E., Kyriakis J. M., Avruch J., Kroll D. J., Johnson G. L., Hoeffler J. P. Activating transcription factor-2 DNA-binding activity is stimulated by phosphorylation catalyzed by p42 and p54 microtubule-associated protein kinases. Mol Endocrinol. 1992 Dec;6(12):2079–2089. doi: 10.1210/mend.6.12.1337144. [DOI] [PubMed] [Google Scholar]
- Bengal E., Ransone L., Scharfmann R., Dwarki V. J., Tapscott S. J., Weintraub H., Verma I. M. Functional antagonism between c-Jun and MyoD proteins: a direct physical association. Cell. 1992 Feb 7;68(3):507–519. doi: 10.1016/0092-8674(92)90187-h. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Bessesen D. H., Jackson S. M., Hoeffler J. P. Overexpression and purification of transcriptionally competent CREB from a recombinant baculovirus. Protein Expr Purif. 1991 Oct-Dec;2(5-6):402–411. doi: 10.1016/1046-5928(91)90101-n. [DOI] [PubMed] [Google Scholar]
- Cohen D. R., Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988 May;8(5):2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Parent L. A., Sharp P. A. Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11779–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989 Dec;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler J. P., Lustbader J. W., Chen C. Y. Identification of multiple nuclear factors that interact with cyclic adenosine 3',5'-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol Endocrinol. 1991 Feb;5(2):256–266. doi: 10.1210/mend-5-2-256. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Luk D., Curran T. Fos is a preferential target of glucocorticoid receptor inhibition of AP-1 activity in vitro. Mol Cell Biol. 1993 Jun;13(6):3782–3791. doi: 10.1128/mcb.13.6.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Chambard J. C., Karin M., Olson E. N. Fos and Jun repress transcriptional activation by myogenin and MyoD: the amino terminus of Jun can mediate repression. Genes Dev. 1992 Apr;6(4):676–689. doi: 10.1101/gad.6.4.676. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Sakura H., Kanei-Ishii C., Sudo T., Yoshimura T., Fujisawa J., Yoshida M., Ishii S. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 1989 Jul;8(7):2023–2028. doi: 10.1002/j.1460-2075.1989.tb03610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina H., Sato H., Suzuki T., Sato M., Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci U S A. 1990 May;87(9):3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Ryseck R. P., Bravo R. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene. 1991 Apr;6(4):533–542. [PubMed] [Google Scholar]
- Schüle R., Umesono K., Mangelsdorf D. J., Bolado J., Pike J. W., Evans R. M. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell. 1990 May 4;61(3):497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- Spana C., Corces V. G. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 1990 Sep;4(9):1505–1515. doi: 10.1101/gad.4.9.1505. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman K. S., Northrop J. P., Admon A., Crabtree G. R. Jun family members are controlled by a calcium-regulated, cyclosporin A-sensitive signaling pathway in activated T lymphocytes. Genes Dev. 1993 Feb;7(2):188–196. doi: 10.1101/gad.7.2.188. [DOI] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van Weperen W. W., van der Vliet P. C. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991 Oct;10(10):3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. S., Dang C. V. Opposite orientations of DNA bending by c-Myc and Max. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7635–7639. doi: 10.1073/pnas.89.16.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]