Abstract
Background
Animal models are indispensable to understand the lipid metabolism and lipid metabolic diseases. Over the last decade, the nematode Caenorhabditis elegans has become a popular animal model for exploring the regulation of lipid metabolism, obesity, and obese-related diseases. However, the genomic and functional conservation of lipid metabolism from C. elegans to humans remains unknown. In the present study, we systematically analyzed genes involved in lipid metabolism in the C. elegans genome using comparative genomics.
Results
We built a database containing 471 lipid genes from the C. elegans genome, and then assigned most of lipid genes into 16 different lipid metabolic pathways that were integrated into a network. Over 70% of C. elegans lipid genes have human orthologs, with 237 of 471 C. elegans lipid genes being conserved in humans, mice, rats, and Drosophila, of which 71 genes are specifically related to human metabolic diseases. Moreover, RNA-mediated interference (RNAi) was used to disrupt the expression of 356 of 471 lipid genes with available RNAi clones. We found that 21 genes strongly affect fat storage, development, reproduction, and other visible phenotypes, 6 of which have not previously been implicated in the regulation of fat metabolism and other phenotypes.
Conclusions
This study provides the first systematic genomic insight into lipid metabolism in C. elegans, supporting the use of C. elegans as an increasingly prominent model in the study of metabolic diseases.
Keywords: Caenorhabditis elegans, Lipid metabolism, Comparative genomics, RNAi, Fat storage
Background
Lipids are hydrophobic molecules with thousands of species including fatty acids and fatty acids derivates, e.g., triglycerides, phospholipids, sphingolipids, sterols, and the like. Generally, lipids function as energy storage, components of biological membrane, and signaling molecules, and likewise play a variety of biological roles in almost all aspects of life: growth, development, reproduction, stresses resistance, aging and longevity, etc. [1]. Accordingly, the homeostasis of lipid metabolism must be tightly regulated. Misregulation of lipid metabolism is often associated with many human diseases, particularly metabolic diseases, such as obesity, type 2 diabetes (T2D), cardiovascular diseases (CVD), and non-alcoholic fatty liver (NAFL) [1,2], all of which are increasingly prevalent both in both developed and developing countries, such as China [3].
Previous studies have illustrated that the human systems which control appetite, energy partitioning, and the integration of lipid metabolic processes are highly complex and redundant [4,5]. Therefore, employing animal models is indispensable in further understanding the lipid metabolism and lipid metabolic diseases. In some situations, lower and less complex organisms are particularly useful in providing simpler and clearer views of the fundamental regulation of lipid metabolism as opposed to higher, more complex organisms. Chiefly due to the advantages of rapid generation time over the last decade, easy genetic manipulation and visualization of lipid droplets at whole animal level has made the tiny nematode, C. elegans an emerging model for exploring the biological functions of lipids and the genetic basis of fat storage regulation. To date, research on this organism has uncovered numerous findings in lipids metabolism and the associated fundamental regulatory mechanisms [6-11], thereby prompting further exploration of its utility as an animal model in these types of studies.
As the first multicellular organism to have its genome completely sequenced [12] and further deeply analyzed [13]C. elegans possesses for biosynthesis of polyunsaturated fatty acids [14], mono-methyl branched fatty acids [15], ceramides [16], phospholipids [17-21], and also the encoding of many lipid binding proteins and transporters [22]. This organism, however, lacks some key genes involved in the biosynthesis of sterol, forcing the worm to obtain it from its diet [23,24]. To date, however, how many potential lipid metabolic genes and pathways do exist in the C. elegans genome is not known. Likewise and more importantly for its application as an animal model, the genomic and functional conservation of lipid metabolism from C. elegans to humans is unclear, limiting C. elegans’ utility for in depth exploration of the fundamental mechanisms of lipid metabolism and human metabolic diseases.
In the present study, we analyzed genes involved in lipid metabolism in the C. elegans genome by comparative genomics. We first constructed a database containing 471 lipid metabolic genes from the C. elegans genome, and further assigned most of these into different lipid metabolic pathways. Afterward, we systematically compared these genes in the human, Drosophila, mouse, and rat genomes. Ultimately, we took the advantage of RNA-mediated interference (RNAi) to disrupt the expression of most lipid metabolic genes, allowing us to explore the biological roles these play in C. elegans.
Results
Database construction of C. elegans lipid metabolic genes
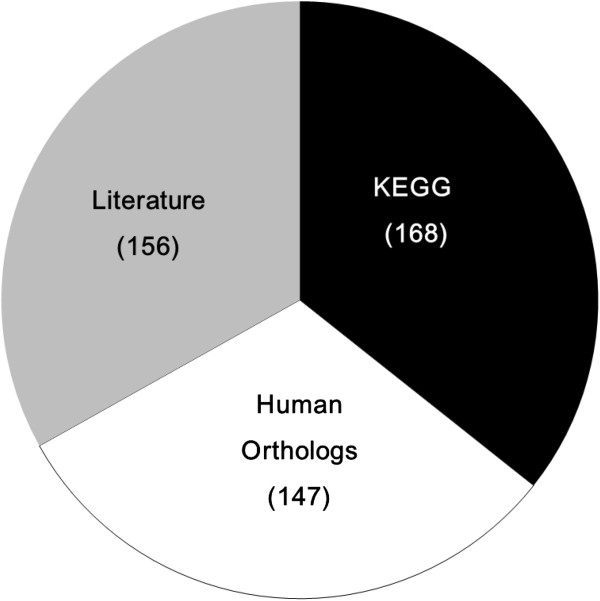
To date, the KEGG database (http://www.genome.jp/kegg/) has collected 168 lipid metabolic genes from the C. elegans genome; this may be far from a complete listing, given the diversity and complexity of lipids in any organism. To garner a full list of lipid metabolic genes from the C. elegans genome, we retrieved lipid metabolic genes by manually searching the WormBase website (http://www.wormbase.org) and published papers collected from NCBI PubMed, as well as the C. elegans orthologs of 407 human lipid metabolic genes and 168 lipid genes from the KEGG database [22,25-28]. Eventually, 471 lipid metabolic genes were retrieved from the C. elegans genome including 147 orthologs of human lipid metabolic genes and 156 genes collected from the literature and WormBase (Figure 1 and Additional file 1: Table S1). In doing so, we constructed the first complete database of lipid metabolic genes of C. elegans, covering most potential lipid metabolic genes that could be found out in C. elegans genome at present (Figure 1 and Additional file 1: Table S1).
Figure 1.
Lipid metabolic genes in C. elegans. The C. elegans genome contains 471 lipid genes, including 168 genes taken from KEGG database colored black, 147 human orthologs colored white, and 156 genes manually picked from WormBase (http://www.wormbase.org) and the published literature colored gray.
The biochemical function of the 471 lipid metabolic genes was further annotated with Ensembl BioMart (http://asia.ensembl.org/biomart/martview/dd89899865f812aff4dcd9ad0dfede49), though the 147 human orthologs and 168 lipid genes from KEGG were already annotated (http://www.genome.jp/kegg/). All 471 genes were also checked again in WormBase (Additional file 1: Table S1). Fuxrthermore, the function annotation obtained from ClueGo [29] showed that 471 lipid genes in C. elegans were not only involved in distinct lipid metabolic processes, but also play multiple roles in response to oxidative stress, aging and longevity, as well as other biological processes (Additional file 2: Figure S1).
Most of the 471 lipid genes in this database are predicted to encode specific enzymes that play roles in defined reactions, though experimental biochemical evidences for this conclusion may be lacking. Some genes predicted to encode the same or similar enzymes were classified into a family. However, they may actually have same, distinct, or no biological conversion/function, as well as may involve into one or more reactions (Table 1). For example, 7 fatty acid desaturase genes were present in the C. elegans genome, but they display completely different preference of substrates except fat-6 and fat-7[14]. Cytochrome P450 enzymes are a superfamily of haem-containing mono-oxygenases with extraordinary diversity in their biochemical function across all kingdoms of life. Within the C. elegans genome, 81 full length cytochrome P450 (CYP) genes have been found, but the exact biological functions of the vast majority of CYPs are largely unknown as of yet [30]. Only 38 of the 81 CYPs may play a role in lipid metabolism, based on our findings from KEGG, WormBase, and the published literature. Another super family is lipase, consisting of 66 genes (Table 1), which may encode hormone-sensitive lipase, phospholipase A2, B1, C, D, triacylglycerol lipase, or unknown lipase (Additional file 1: Table S1).
Table 1.
Lipid metabolic gene families in C. elegans
| Gene | Enzyme | Number in database (all/mapped in pathways) |
|---|---|---|
|
acdh |
acyl-CoA dehydrogenase |
13 (13/13) |
|
ace |
acetylcholinesterase |
4 (4/4) |
|
acl |
acyltransferase |
14 (14/14) |
|
acox |
acyl-CoA oxidase |
8 (8/8) |
|
acs |
acyl-CoA synthetase |
22 (22/22) |
|
alh |
aldehyde dehydrogenase |
13 (13/12) |
|
cpt |
carnitine palmitoyltransferase |
6 (6/6) |
|
cyp |
cytochrome P450s |
38 (81/35) |
|
dhs |
dehydrogenase |
31 (31/9) |
|
dgat |
acyl-CoA:diacylglycerol acyltransferase |
4 (4/4) |
|
ech |
enoyl-CoA hydratase |
11 (11/11) |
|
elo |
elongase |
9 (9/4) |
|
fat |
fatty acid desaturase |
7 (7/7) |
|
lbp |
lipid binding protein |
9 (9/9) |
|
lipl/lips |
lipase |
66 (66/57) |
|
mboa |
lysophospholipid acyltransferase |
7 (7/7) |
|
scrm |
scramblase |
8 (8/0) |
|
stdh |
steroid dehydrogenase |
4 (4/4) |
|
tat |
flippase |
6 (6/0) |
| total | 280 (323/226) |
471 genes potentially involved in 16 lipid metabolic pathways in C. elegans
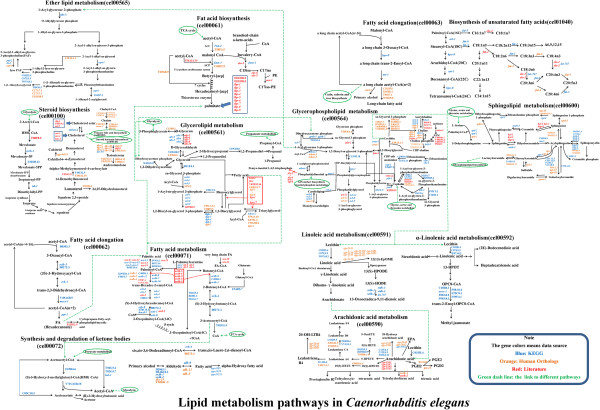
Totally, 168 C. elegans lipid genes from the KEGG database were assigned into 15 lipid metabolic pathways: fatty acid biosynthesis [PATH:cel00061], fatty acid elongation in mitochondria [PATH:cel00062], fatty acid metabolism [PATH:cel00071], synthesis and degradation of ketone bodies [PATH:cel00072], steroid biosynthesis [PATH:cel00100], glycerolipid metabolism [PATH:cel00561], primary bile acid biosynthesis [PATH:cel00120], steroid hormone biosynthesis [PATH:cel00140], glycerophospholipid metabolism [PATH:cel00564], ether lipid metabolism [PATH:cel00565], sphingolipid metabolism [PATH:cel00600], arachidonic acid metabolism [PATH:cel00590], linoleic acid metabolism [PATH:cel00591], alpha-linolenic acid metabolism [PATH:cel00592], and biosynthesis of unsaturated fatty acids [PATH:cel01040]. To clearly distinguish the fatty acid elongation in the cytoplasmic compartment from that in the mitochondria compartment [PATH: cel00062], we renamed this pathway as fatty acid elongation [PATH: cel00063]. Collectively, we built 16 lipid metabolic pathways in C. elegans (Figure 2).
Figure 2.
Lipid metabolism pathways in C. elegans. 16 lipid metabolism pathways are integrated into a lipid metabolism network. Genes were colored based on their data resources (see Figure 1). Black arrows linked different substrates. Two continuous black arrows indicate that there are more than two reactions. The dashed green arrow was used to link different pathways. The Green circle surround linked the pathway name.
To gain a clearer picture of lipid metabolic pathways in C. elegans, all 16 pathways were further integrated into a lipid metabolism network (Figure 2). Additionally, 303 lipid genes including 147 human orthologs and 156 genes reported in the literature were assigned into the 16 pathways based on their annotated or reported biological function (Figure 2). However, due to a lack of both clear evidence of the precise biological function and detailed information, some genes could not be assigned into any pathway, even though their paralogs were assigned in the same pathway. Subsequently, we constructed the first lipid metabolism network with 16 pathways containing 471 lipid genes from C. elegans (Figure 2).
Only 35 of the 81 CYPs could be assigned into different lipid metabolic pathways (Figure 2). Based on KEGG annotation, cyp-33E2 may function in fatty acid metabolism, though it was reported to be involved in the biosynthesis of eicosanoides derived from eicosapentaenoic acid (EPA) [31]. Therefore, we reassigned cyp-33E2 into the arachidonic acid metabolism pathway (Figure 2). Despite cyp-35B1 being shown as a target of insulin/IGF-I-like signaling (IIS) [32], and Cyp-31A2 and Cyp-31A3 being involved in the production of lipids required for eggshell formation [33], their real catalyzing reactions were unclear enough that they could not be assigned into any lipid metabolic pathway.
Genomic comparison of lipid metabolic genes among humans, mice, rats, Drosophila, and C. elegans
Research on animal models has brought numerous powerful and valuable insights to both basic human biology and disease pathology [34-36]. However, under many circumstances, animal models fall short of our expectations, going so far as to result in contradictory or opposite findings. These discrepancies may be due in part to the changes in the conservation of genetically controlled pathways from human to model animals [36-38]. This reality spurred us to inquire about the conservation of lipid metabolism that exists between C. elegans and humans, as well as mice [39,40], rats [41,42], and Drosophila[43-46], all of which are commonly used as models to understand the mechanisms and potential therapies for human metabolic diseases.
All 471 C. elegans lipid genes were used to search for orthologs in the human, mouse, rat and Drosophila genomes using Ensembl BioMart (Methods). Eventually, 581, 585, 563, and 428 lipid genes respectively were obtained in the human, mouse, rat, and Drosophila genomes (Additional file 3: Table S2 and Additional file 4: Figure S2). Comparison of these lipid genes revealed that 237 genes are conserved in all five organisms. Over 70% of C. elegans lipid genes have orthologs in humans, mice, and rats (Table 2). Conversely, 72.12% (419/581) human lipid genes are conserved in C. elegans (Table 2), suggesting a high conservation of lipid metabolism genes between C. elegans and humans, as well as between C. elegans and both mice and rats.
Table 2.
Conservation of lipid metabolic genes in 5 model organisms
| Organisms (lipid genes) | C. elegans (471) Orthologs | Human (581) Orthologs |
|---|---|---|
|
C. elegans (471) |
- |
419(72.12%) |
| Human (581) |
370(78.56%) |
- |
| Mouse (585) |
389(82.59%) |
539(92.77%) |
| Rat (563) |
375(79.62%) |
517(88.98%) |
| Drosophila (428) | 352(74.73%) | 340(58.52%) |
Conservation of C. elegans lipid metabolic genes in human disease
An imbalance of major lipid signaling pathways has previously been implicated in obesity, type 2 diabetes, atherosclerosis, hypertension, non-alcoholic fatty liver disease, etc. [1,47]. To analyze the relationship between lipid genes and human metabolic diseases, human disease genes were retrieved from the OMIM database (http://www.ncbi.nlm.nih.gov/omim/). At present, the OMIM database has collected 19,994 MIM IDs including genes, phenotypes, and locus which we downloaded from the newly updated NCBI FTP. Using Ensembl Biomart ID Mapping, we found 15,181 genes that could be considered human disease genes. Comparison between humans and the other four organisms revealed that mice, rats, C. elegans, and Drosophila have 13259, 12717, 5397, and 5889 respective orthologs, in which 5085 genes are conserved in all five organisms (Figure 3A). Furthermore, 1264 of these 15181 genes may be considered metabolic disease genes that were filtered within the database by keywords ‘Obesity, Diabetes, Metabolic disease, and others major obesity related diseases’. Similarly, 1152, 1051, 638, and 632 orthologs of human metabolic disease genes could be found among mouse, rat, C. elegans, and Drosophila, respectively, in which 346 orthologous genes are conserved in all five organisms (Figure 3B).
Figure 3.
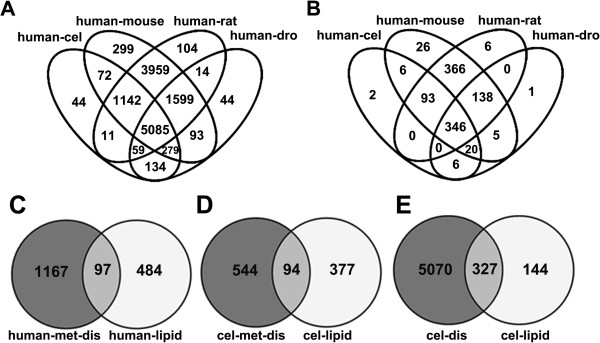
Venn diagram of human disease genes orthologs and the relationship between lipid genes and metabolic disease associated genes. A: Venn diagram of 15181 human disease gene orthologs made with VENNY. Numbers in the overlapping and non-overlapping areas of the diagram indicate the number of genes found by overlapping or unique sets of orthologs in each species; B: Venn diagram of 1264 human metabolic disease genes orthologs; C: Comparison of 581 human lipid genes with human metabolic disease genes (97 of 581 lipid genes overlapped with metabolic disease genes); D: Similarly to the phenomenon in human genes, 94 of 471 C. elegans lipid genes were related to metabolic disease; E: In C. Elegans, 327 lipid genes were associated with human diseases.
In humans, 97 of the 581 lipid genes are overlapped with metabolic disease genes (Figure 3C). Similarly, 94 of the 471 C. elegans lipid genes are orthologs of human metabolic disease genes (Figure 3D). Crucially, all 94 C. elegans lipid genes associated with metabolic diseases are completely overlapped with 97 human metabolic disease genes. In mice, rats, and Drosophila, respectively there are 128, 107, and 97 lipid genes associated with metabolic diseases, with 71 genes conserved across all five species (Additional file 5: Table S3). Moreover, 327 of the 471 lipid metabolism genes in C. elegans are also orthologs of human disease genes (Figure 3E), implying that great number of lipid genes are also involved in other disease processes aside from metabolic syndrome.
Investigated of the biological functions of lipid metabolic genes in C. elegans by RNAi
Among the 471 C. elegans lipid metabolic genes, only a small number have been biochemically and functionally characterized, demonstration that they participate into growth, development, dauer formation, aging and longevity, stress responses, and the like. However, the biological functions of most lipid metabolic genes remain unknown. The current Argilent C. elegans RNAi library covers about 86% of the known and predicted genes, providing a powerful tool to further investigate the biological functions of the lipid genes. We opted to take advantage of RNA-mediated interference (RNAi) to disrupt the expression of each lipid gene and investigate easily visible phenotypes including growth, sterility, and fat storage, indicated by Nile Red staining of fixation [48].
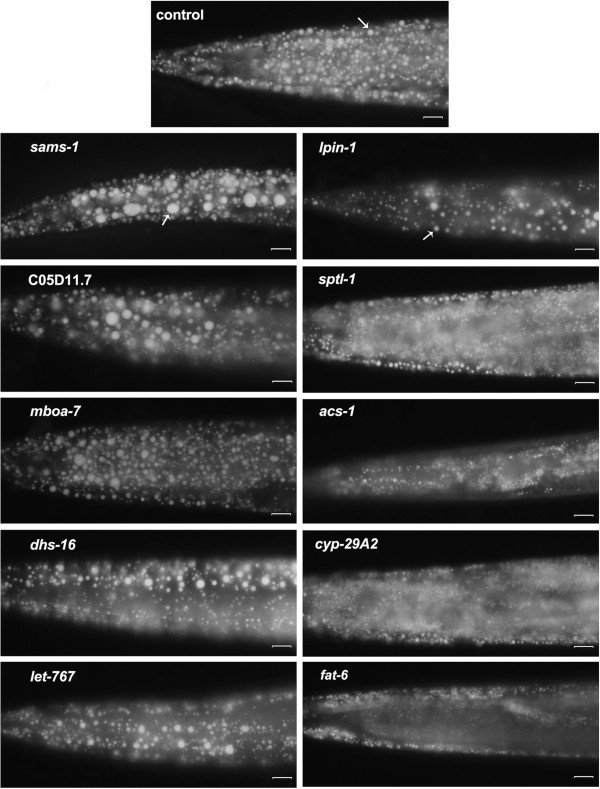
Of the 471 lipid genes in C. elegans, 356 have available RNAi strains. Of those with available strains, RNAi knockdown of 21 genes consistently displayed remarkable phenotypes with significantly altered fat storage, growth defect, lethal, or sterility (Table 3), while10 of these 21 genes significantly affect fat storage. Inactivation of sams-1, mboa-7, and C05D11.7 led to large lipid droplet size (Figure 4). sams-1 encodes S-adenosylmethionine synthetase (SAMS) involved in the biosynthesis of phosphocholine [PATH:Cel00564]. Mutation of sams-1 displayed large size of lipid droplets [20,21]. mboa-7 encodes a member of the membrane-bound O-acyltransferase family, and is required for incorporation of PUFAs into phosphatidylinositol (PI) [PATH:Cel00564] [18]. C05D11.7 is an ortholog of the human patatin-like phospholipase (PNPLA) gene family, catalyzing the hydrolysis of triglycerides in adipose tissue. LET-767 has been reported to be a major 3-ketoacyl-CoA reductase required for the production of monomethyl branched and long chain fatty acids [49]. DHS-16 is a novel 3-hydroxysteroid dehydrogenase regulating the production of dafachronic acids (DAs) [50]. In C. elegans, RNAi knockdown of dsh-16 and let-767 slightly but obviously increased the size of lipid droplets (Figure 4).
Table 3.
Phenotypes of lipid metabolic genes inactivated by RNAi
| Sequence name | Gene name | LDs LD size | Other phenotypes | Pathway | Key references |
|---|---|---|---|---|---|
| C49F5.1 |
sams-1 |
increased |
Growth rate variant, lean, sterile |
Glycerophospholipid metabolism |
[20,21] |
| C05D11.7 |
atgl |
increased |
|
Glycerolipid metabolism |
[51,52] |
| F14F3.3 |
mboa-7 |
increased |
|
Glycerophospholipid metabolism |
[17,18] |
| C10F3.2 |
dhs-16 |
increased |
Egg laying defect, growth rate variant, lethal, slim |
Primary bile acid biosynthesis |
[50] |
| C56G2.6 |
let-767 |
increased |
Growth rate variant, slim, lethal, egg laying defect |
Steroid hormone biosynthesis,Linoleic acid metabolism,Biosynthesis of unsaturated fatty acids |
[49,53] (4) |
| H37A05.1 |
lpin-1 |
decreased |
Growth rate variant, slim, egg laying defect |
Glycerophospholipid metabolism |
[54,55] |
| C23H3.4 |
sptl-1 |
decreased |
Egg laying defect, lethal,pale |
Sphingolipid metabolism |
[16] |
| F46E10.1 |
acs-1 |
decreased |
Growth rate variant, slim, larvae arrest, lethal |
Fatty acid metabolism, Glycerophospholipid metabolism |
[56,57] |
| T19B10.1 |
cyp-29A2 |
decreased |
Egg laying defect, lethal |
Arachidonic acid metabolism |
|
| VZK822L.1 |
fat-6 |
decreased |
Growth rate variant, sterile, pale, slim |
Biosynthesis of unsaturated fatty acids |
[14,58-60] |
| T27F6.6 |
|
|
Egg laying defect |
Sphingolipid metabolism |
|
| ZC416.8 |
cha-1 |
|
Egg laying defect |
Glycerophospholipid metabolism |
[61-64] |
| K12H4.5 |
|
|
Reduced brood size, growth rate variant (l3-l4,72 h normal) |
Glycerophospholipid metabolism |
|
| F25B4.6 |
hmgs-1 |
|
Growth rate variant, larval arrest, larval lethal,sterile |
Synthesis and degradation of ketone bodies |
|
| F56D1.5 |
dhs-5 |
|
Egg laying defect |
Steroid hormone biosynthesis |
|
| T02G5.8 |
kat-1 |
|
Growth rate variant (L3,72 h normal) |
Synthesis and degradation of ketone bodies |
[65,66] |
| F32H2.5 |
fasn-1 |
|
Larval arrest, lethal |
Fatty acid synthesis |
[8,22,67] |
| W09B6.1 |
pod-2 |
|
Larval arrest, lethal |
Fatty acid synthesis |
[22,68,69] |
| F11E6.5 |
elo-2 |
|
Pale, slim |
Fatty acid synthesis |
[14,70] |
| D2024.3 |
elo-3 |
|
Pale, slim, egg laying defect |
unclear |
|
| T01E8.3 | plc-3 | Sterile | Glycerophospholipid metabolism | [71-73] |
Figure 4.
Inactivation of 10 lipid genes by RNAi dramatically changed fat storage in C. elegans. Late L4 worms were fixed with paraformaldehyde and stained with Nile Red [48]. Images were captured using identical settings and exposure time for each image. Inactivation of sams-1, mboa-7, and C05D11.7 led to increased lipid size; RNAi knockdown of dsh-16 and let-767 slightly though noticeably increased the size of lipid droplets in C. elegans; Meanwhile, RNAi inactivation of lpin-1, sptl-1, acs-1, cyp-29A2 and fat-6 resulted in significantly decreased fat storage and comparatively smaller size of lipid droplets. Bar, 10 μm. Arrow indicates the lipid droplet.
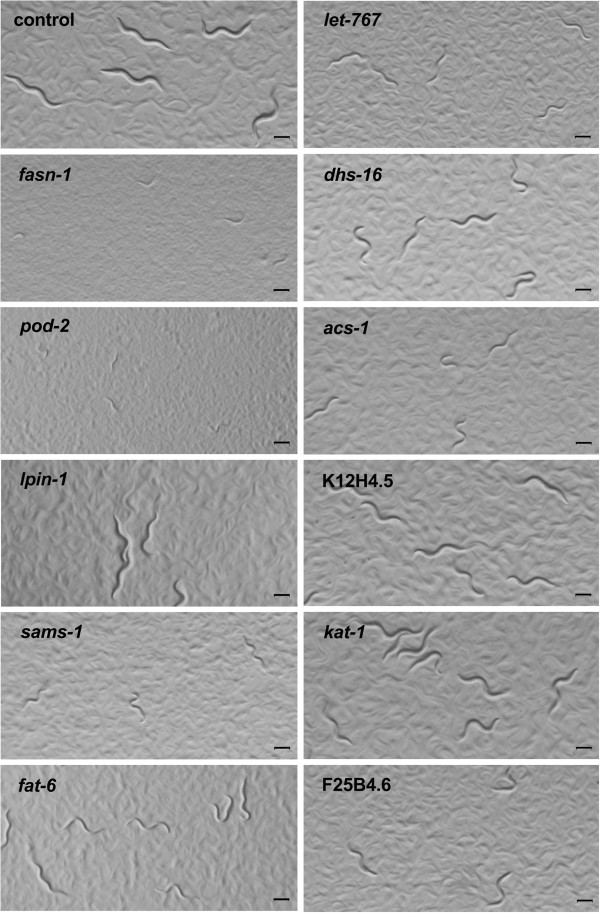
RNAi inactivation of lpin-1, sptl-1, acs-1, cyp-29A2 and fat-6, meanwhile, resulted in significantly decreased fat storage and a smaller size of lipid droplets. Additionally, inactivation of 9 genes led to slower growth (Figure 5). pod-2 and fasn-1 encode acetyl-CoA carboxylase and fatty acid synthase, respectively, and are predicted to catalyze the first and second step in de novo fatty acid biosynthesis [PATH:Cel00061]. Disruption of either of these two key genes by RNAi definitely led to both lethality and larval arrest (Figure 5). Meanwhile, inactivation of 13 other genes resulted in an egg laying variant (Table 3).
Figure 5.
Inactivation of 11 lipid genes by RNAi in C. elegans displayed growth and developmental defects. RNAi of fasn-1 and pod-2 leads to severe larvae arrest. Inactivation of lpin-1, sams-1, fat-6, let-767, dhs-16, acs-1, K12H4.5, F25B4.6 and kat-1 resulted in slower growth. All images were captured using identical settings and exposure time. Worms grown on E. coli HT115 containing an empty vector were used as control. Bar, 200 μm.
Discussion
Understanding the lipid metabolism pathways and the involved genes is critical to further study of the biological functions of lipids and the mechanisms of metabolic diseases. A well-known animal model with a sequenced genome, like C. elegans, can provide a systemic genomic review for the whole lipid metabolism network. Several research groups have bioinformatically or functionally characterized one or several lipid metabolism pathways in C. elegans[14,16,22,26,68,74-76]. In our study, we created a database of 471 C. elegans lipid metabolism genes retrieved from the KEGG data, WormBase, and the published literature (Figure 1 and Additional file 1: Table S1). According to their annotation and real biological functions, these 471 genes could be designed into 16 lipid metabolism pathways integrated into a network (Figure 2). To our knowledge, ours is the first study to systematically analyze the lipid genes, metabolism pathways, and their biological functions in the animal model C. elegans.
Remarkably, aside from the 471 C. elegans lipid genes, we additionally found 581, 585, 563, and 428 respective lipid genes in the human, mouse, rat, and Drosophila genomes. Comparison of these genes revealed that 237 lipid genes are conserved in all five species. As such, this work provides useful information regarding lipid metabolism that will aid research utilizing C. elegans and other organisms as animal models. Among the 471 C. elegans lipid genes, 345 (73.25%) have orthologs in humans. Conversely, 380 of the 581 (65.4%) human lipid genes had orthologs in C. elegans. Notably, of 97 human lipid genes related to metabolic diseases, 94 have orthologs in C. elegans and 71 genes are conserved in the human, mouse, rate, C. elegans and Drosophila genomes, suggesting that lipid genes involved into metabolic diseases are highly conserved among model organisms. Altogether, these results indicate that C. elegans and humans share high conservation in terms of lipid genes and their roles in metabolic diseases, suggesting that C. elegans is likely a useful and insightful model for use not only in research on the fundamental biology of lipid metabolism, but also exploring the mechanisms of human metabolic diseases.
The C. elegans RNAi library provides numerous benefits to investigate the role of a specific gene. We thus systematically analyzed the biological functions of lipid genes with available RNAi strains. Among 471 lipid genes, nearly 40% genes were reported to affect phenotypes (http://www.wormbase.org), e.g. fat content, dauer, lifespan, brood size, growth, etc. In this study, we only focused on the clearly observable phenotypes of growth, sterility, and fat storage. However, due to the RNAi variation and gene redundancy of a family (Table 1), RNAi of many of the lipid genes displayed inconsistent, weak phenotypes that we excluded from this report. Consistently, however, inactivation of 21 lipid genes by RNAi remarkably affects worm fat metabolism, development, and other phenotypes. Of these 21 lipid genes, 6 genes T19B10.1 (cyp-29A2), T27F6.6, K12H4.5, F25B4.6 (hmgs-1), F56D1.5 (dhs-5) and D2024.3 (elo-3) have not been reported in literature (Table 3). Remarkbly, 19 out of 21 lipid genes (all except K12H4.5 and dhs-5; Table 3), are fairly conserved in humans and C. elegans. Moreover, C05D11.7, fat-6, dhs-16, fasn-1, pod-2, elo-3, and lpin-1 have relationships with human metabolic diseases.
C. elegans lpin-1 encodes a putative phosphatidic acid phosphatase orthologous to human LPIN1 (OMIM: 605518), LPIN2 (OMIM: 605519), and LPIN3 (OMIM: 605520) (http://www.wormbase.org). In humans and mice, Lpin-1 is associated with metabolic syndrome and type 2 diabetes [77-80] and mutations in human LPIN2 cause Majeed syndrome [81]. C. elegans lpin-1 has been implicated in affecting fat storage and the breakdown and assembly of the nuclear envelope [54,55]. Consistent with the mammalian Lpin1 mutation, inactivation of C. elegans lpin-1 by RNAi displayed resulted in low fat with small size of lipid droplets, as indicted by Nile Red staining (Figure 4). Human patatin-like phospholipase domain-containing 3 (PNPLA3) is associated with increased liver fat content and liver injury [82]. The C. elegans genome contains at least 5 orthologs of the patatin-like phospholipase (Additional file 1: Table S1). Interestingly, RNAi of C05D11.7, an ortholog of PNPLA3 that was considered to encode adipose triglyceride lipase (ATGL) in C. elegans[51,52], led to higher fat and larger lipid droplets compared with the control in C. elegans (Figure 4), similar to other reports on humans and rodents [83,84].
Although mboa-7 affects PUFA incorporation into PI [18], dhs-16 regulates dafachronic acids metabolism [50], and sptl-1 is involved in the biosynthesis of ceramide [16], they have not been previously implicated in fat storage. RNAi inactivation of mboa-7 and dhs-16 displayed increased lipid droplets size (Figure 4), whereas RNAi inactivation of sptl-1decreased lipid droplet size. The fat content of all three genes should likewise be measured by other methods (e.g., thin-layer chromatography and gas chromatography (TLC/GC)) in future studies. acs-1 affects the incorporation of monomethyl branched chain fatty acid (C17iso) into phospholipids [56] and was reported to increase fat mass [57]. RNAi of acs-1, however, indeed decreased the density and size of lipid droplet (Figure 4). Inactivation of let-767 by RNAi led to slower growth (Figure 5), but once the worms reached the late L4 or early young adult stage, they had increased lipid droplets size (Figure 4). Additionally, RNAi of K12H4.5, kat-1, and F25B4.6 resulted in slower growth (Figure 5), but fat storage looked normal once the worm entered into late L4 or early young adulthood. Several members of cytochrome P450 superfamily were reported to functionally affect lipid metabolism [31,33,85,86]. We only found that RNAi of cyp-29A2 significantly decreased fat storage with small lipid droplets as compared with the control (Figure 4). Consequently, these lipid genes should potentially be considered as a starting point to further investigate their role in the regulation of fat storage.
Conclusions
We created a database of 471 C. elegans lipid metabolism genes that could be designed into 16 lipid metabolism pathways integrated into a network. Additionally, 581, 585, 563, and 428 respective lipid genes in the human, mouse, rat, and Drosophila genomes were also retrieved. Over 70% of C. elegans lipid genes have human orthologs, with 237 of 471 C. elegans lipid genes being conserved in humans, mice, rats, and Drosophila, of which 71 genes are specifically related to human metabolic diseases. Furthermore, RNA-mediated interference (RNAi) of 21 lipid genes affects fat storage, development, and reproduction. This study provides the first genomic view on lipid metabolism in C. elegans, supporting the use of C. elegans as an increasingly prominent model in the study of metabolic diseases.
Methods
Retrieval of C. elegans lipid metabolic genes and database construction
Most of lipid metabolism genes used in this study were initially downloaded from the KEGG pathway database (http://www.genome.ad.jp/kegg/pathway.html), which has to date collected 168, 407, 383, 333, and 170 lipid metabolic genes respectively from the C. elegans, human, mouse, rat, and Drosophila genomes. Of the 407 human lipid genes present in the C. elegans genome, 364 were orthologs. Another 156 lipid metabolism genes in C. elegans were manually taken from WormBase (http://www.wormbase.org) and the published literature, mainly retrieved from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/). Any overlapping genes were excluded with Perl program manually and online using the Venny program (http://bioinfogp.cnb.csic.es/tools/venny/ index.html). In total, 471 C. elegans lipid genes were been retrieved. The BioMart Interface (http://asia.ensembl.org/biomart/martview/53ac7015dbd78f9a5c2f0a3479fa4dc4) was applied to standardize all gene IDs.
Identification of lipid metabolic genes in human, mouse, rat, and Drosophila
We used 471 C. elegans lipid metabolic genes to search for orthologs in the human, mouse, rat and Drosophila genomes by Ensembl BioMart and found 370, 389, 375, and 352 orthologs in human, mouse, rat, and Drosophila, respectively (Table 2). The KEGG database has collected 407 human lipid metabolic genes to date; after combining these and 370 human orthologs of C. elegans lipid genes, the overlapped genes were discarded. Within the human genome, 581 lipid genes have been found, and we used these in reversely to search for orthologs in C. elegans, mouse, rat, and Drosophila genomes (419, 539, 517, and 340 genes, respectively (Table 2)). The KEGG database contains 383, 333, and 170 lipid metabolic genes for the mouse, rat, and Drosophila genomes, respectively. After combining both sets of lipid genes and discarding any overlapped gene in each organism. In total, 585, 563, and 428 lipid genes could be identified in mouse, rat, and Drosophila genomes, respectively, with the additional 581 lipid genes within the human genome (Additional file 3: Table S2 and Additional file 4: Figure S2).
Annotation of lipid metabolic genes in C. elegans
The 168 lipid metabolic genes we retrieved from the KEGG database were already annotated. The remaining 303 lipid genes were annotated via Ensembl BioMart (http://asia.ensembl.org/biomart/martview/53ac7015dbd78f9a5c2f0a3479fa4dc4), Swiss-Prot/TrEMBL (http://www.expasy.org), and Gene Ontology (http://www.geneontology.org/). However, since the annotation databases are incomplete, if a gene could not be annotated from a database, they were annotated by hand using previously published papers (primarily retrieved from PubMed) and WormBase (http://www.wormbase.org/). Following annotation, the biological functions of all 471 genes were manually back checked via WormBase.
The biological processes, KEGG path, gene cellular localization, and molecular function annotation of the 471 genes were processed through ClueGo [29] plug-in of Cytoscape [87].
Retrieval and comparison of disease associated genes in humans, mice, rats, C. elegans, and Drosophila
From the NCBI database (ftp://ftp.ncbi.nih.gov/gene/DATA/), 19994 MIM disease ID genes, phenotypes, and locus were downloaded from NCBI ftp. In total, 15181 genes could be mapped by Ensembl Biomart ID Mapping and were therefore considered human disease genes. All metabolic disease genes were searched from OMIM database (http://www.ncbi.nlm.nih.gov/omim/) using the following keywords: Obesity, Diabetes, Metabolic disease, Sleep apnea, Atherosclerosis, Coronary disease, Hyperlipidemia, and Hypertension. After excluding redundant genes, some further 1264 human metabolic disease genes were identified in our database.
Orthologs information of human gene inferred by Ensembl (release 66) ortholog detection pipeline was obtained from Ensemble’s BioMart interface (http://asia.ensembl.org/biomart/martview/53ac7015dbd78f9a5c2f0a3479fa4dc4). Specifically, we retrieved Ensembl identifiers of orthologs to human genes together with identifiers of their translated protein products. Details of the ortholog detection pipeline are described at http://aug2007.archive.ensembl.org/info/data/compara/homology_method.html. Briefly, gene families were identified from all sequences in the database by WU-Blastp and Smith–Waterman searches, followed by construction of a phylogenetic tree for each gene family to identify orthologous and paralogous relationships between gene pairs.
All orthologs of lipid metabolism genes, human disease genes, and human metabolic genes were then compared by using VENNY, an interactive tool for comparing lists with Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Worm strains, growth condition, and RNA interference
Standard protocols were used to maintain the C. elegans N2 strain growing at 20°C with E. coli OP50 for food, unless specifically noted. RNAi was performed by feeding bacterial strains from the Ahringer C. elegans RNAi library, obtained from Gene Services (Source BioScience) [88,89]. The empty vector L4440 in the Ahringer library E. coli HT115 strain was used as the negative control for RNAi experiments. The current Ahringer C. elegans RNAi library covers about 86% of both known and predicted genes. In our C. elegans lipid metabolism genes database, only 356 genes have available RNAi bacterial strains. Generally, eggs were isolated from gravid adults using hypochlorite treatment and hatched in M9 buffer overnight, and then synchronized L1 worms plated onto NGM plates seeded with E. coli strain HT115 carrying the RNAi clone of a specific lipid gene. Worms were occasionally checked after 48 hr. RNAi of 356 genes was carried out for four biological rounds.
Nile Red staining of fixed nematodes to visualize fat storage
Late L4s or young adult nematodes with one or two eggs within their uterus were washed off growth plates, fixed and then stained with Nile Red as described previously [48,90]. All images were captured using identical settings and exposure time.
Growth rate and sterility analysis
Growth rate analysis was conducted in line with our previous report [90]. All images were captured using identical settings and exposure times. Worms which clearly laid fewer eggs were recorded as having a “egg laying defect”, and those with no eggs were recorded as “sterile”.
Competing interests
All authors declare no conflict of interest.
Authors’ contributions
Conceived and designed the experiments: BL, YR Z. Performed the experiments: YR Z, YH D, XJ Z, HZ W, XY W. Analyzed the data: BL and YR Z. Contributed reagents/materials/analysis tools: BL, YR Z, YH D, XJ Z, HZ W, XY W. Wrote the paper: BL and YR Z. All authors have read and approved the final manuscript.
Supplementary Material
C. elegans lipid metabolic gene database. 471 lipid metabolism genes were retrieved from the C. elegans genome. Different colors were used to highlight the origin of the various lipid genes. The metabolism pathways of each gene mainly come from the KEGG database.
Annotation of biological processes of 471 C. elegans lipid metabolism genes using ClueGO. The chart displays part of significant enrichment analysis of Gene Ontology molecular function in C. elegans lipid metabolic gene database. The x axis stands for the amount molecular function terms in Gene Ontology. One star denotes P < 0.05, while two stars denote P < 0.01.
Lipid metabolism genes in 5 model organisms. All genes IDs were unified as Ensembl gene ID. Respectively, 581, 585, 563, and 428 lipid metabolic genes were present in the human, mouse, rat, and Drosophila genomes.
The list of human, mouse, rat, and Drosophila lipid metabolic genes. 581 (A), 585 (B), 563 (C), and 428 (D) lipid metabolism genes present in human, mouse, rat, and Drosophila genome, respectively. Abbreviation: CEL- C. elegans; DRO- Drosophila.
Lipid metabolism genes related to human metabolic diseases in humans, C. elegans, mice, rats, and Drosophila. Respectively, 97, 94, 128, 107, and 97 lipid genes were overlapped with metabolic diseases in human, C. elegans, mouse, rat, and Drosophila.
Contributor Information
Yuru Zhang, Email: zyuru@mail.kiz.ac.cn.
Xiaoju Zou, Email: xiaoju_zou@yahoo.com.
Yihong Ding, Email: dingyndl@163.com.
Haizhen Wang, Email: whaizhen@mail.kiz.ac.cn.
Xiaoyun Wu, Email: wuxiaoyun79@163.com.
Bin Liang, Email: liangb@mail.kiz.ac.cn.
Acknowledgement
We would like to thank other members of our lab for their comments on this manuscript. This work was supported by Chinese Natural Science Foundation to BL (31171134 and U1202223) and to XJ Zou (31160216), as well as Yunnan Natural Science Foundation to XJ Zou (2011FZ179).
References
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9(2):162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Yang W, Lu J, Weng J, Jia W, Ji L, Xiao J, Shan Z, Liu J, Tian H, Ji Q. Prevalence of diabetes among men and women in China. N Engl J Med. 2010;362(12):1090–1101. doi: 10.1056/NEJMoa0908292. [DOI] [PubMed] [Google Scholar]
- Coll AP, Farooqi IS, O'Rahilly S. The hormonal control of food intake. Cell. 2007;129(2):251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10(7):431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20(2):58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, Faergeman NJ, Van Gilst MR, Ingraham HA, Ashrafi K. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 2010;12(4):398–410. doi: 10.1016/j.cmet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421(6920):268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Mak HY. Lipid droplets as fat storage organelles in Caenorhabditis elegans. J Lipid Res. 2012;53(1):28–33. doi: 10.1194/jlr.R021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BC, Ashrafi K. C. elegans fat storage and metabolic regulation. Biochim Biophys Acta. 2009;1791(6):474–478. doi: 10.1016/j.bbalip.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. C elegans: a model for exploring the genetics of fat storage. Dev Cell. 2003;4(1):131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99(9):5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M, Crawford QT, Seiber M, Wang CY, Han M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004;2(9):E257. doi: 10.1371/journal.pbio.0020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Yin X, Allan R, Lu DD, Maurer CW, Haimovitz-Friedman A, Fuks Z, Shaham S, Kolesnick R. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science. 2008;322(5898):110–115. doi: 10.1126/science.1158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Inoue T, Lee HC, Kono N, Tanaka F, Gengyo-Ando K, Mitani S, Arai H. Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells. 2008;13(8):879–888. doi: 10.1111/j.1365-2443.2008.01212.x. [DOI] [PubMed] [Google Scholar]
- Lee HC, Inoue T, Imae R, Kono N, Shirae S, Matsuda S, Gengyo-Ando K, Mitani S, Arai H. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol Biol Cell. 2008;19(3):1174–1184. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satouchi K, Hirano K, Sakaguchi M, Takehara H, Matsuura F. Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids. 1993;28(9):837–840. doi: 10.1007/BF02536239. [DOI] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147(4):840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Na K, Lee HJ, Lee EY, Paik YK. Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J Biochem. 2011;149(5):529–538. doi: 10.1093/jb/mvr025. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3(2):e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieb WF, Rothstein M. Sterol requirement for reproduction of a free-living nematode. Science. 1968;160(3829):778–780. doi: 10.1126/science.160.3829.778. [DOI] [PubMed] [Google Scholar]
- Merris M, Wadsworth WG, Khamrai U, Bittman R, Chitwood DJ, Lenard J. Sterol effects and sites of sterol accumulation in Caenorhabditis elegans: developmental requirement for 4alpha-methyl sterols. J Lipid Res. 2003;44(1):172–181. doi: 10.1194/jlr.m200323-jlr200. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005;102(38):13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech Ageing Dev. 2006;127(5):458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Chan JP, Hu Z, Sieburth D. Recruitment of sphingosine kinase to presynaptic terminals by a conserved muscarinic signaling pathway promotes neurotransmitter release. Genes Dev. 2012;26(10):1070–1085. doi: 10.1101/gad.188003.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing ST, Ivens A, Butler V, Ravikumar SP, Laing R, Woods DJ, Gilleard JS. The transcriptional response of Caenorhabditis elegans to ivermectin exposure identifies novel genes involved in the response to reduced food intake. PLoS One. 2012;7(2):e31367. doi: 10.1371/journal.pone.0031367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer P, Muller M, Kruger A, Steinberg CE, Menzel R. Cytochrome P450-dependent metabolism of PCB52 in the nematode Caenorhabditis elegans. Arch Biochem Biophys. 2009;488(1):60–68. doi: 10.1016/j.abb.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Kosel M, Wild W, Bell A, Rothe M, Lindschau C, Steinberg CE, Schunck WH, Menzel R. Eicosanoid formation by a cytochrome P450 isoform expressed in the pharynx of Caenorhabditis elegans. Biochem J. 2011;435(3):689–700. doi: 10.1042/BJ20101942. [DOI] [PubMed] [Google Scholar]
- Iser WB, Wilson MA, Wood WH 3rd, Becker K, Wolkow CA. Co-regulation of the DAF-16 target gene, cyp-35B1/dod-13, by HSF-1 in C. elegans dauer larvae and daf-2 insulin pathway mutants. PLoS One. 2011;6(3):e17369. doi: 10.1371/journal.pone.0017369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenati G, Penkov S, Muller-Reichert T, Entchev EV, Kurzchalia TV. Two cytochrome P450s in Caenorhabditis elegans are essential for the organization of eggshell, correct execution of meiosis and the polarization of embryo. Mech Dev. 2009;126(5–6):382–393. doi: 10.1016/j.mod.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Chakraborty C, Hsu CH, Wen ZH, Lin CS, Agoramoorthy G. Zebrafish: a complete animal model for in vivo drug discovery and development. Curr Drug Metab. 2009;10(2):116–124. doi: 10.2174/138920009787522197. [DOI] [PubMed] [Google Scholar]
- Orsucci D, Calsolaro V, Mancuso M, Siciliano G. Neuroprotective effects of tetracyclines: molecular targets, animal models and human disease. CNS Neurol Disord Drug Targets. 2009;8(3):222–231. doi: 10.2174/187152709788680689. [DOI] [PubMed] [Google Scholar]
- Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Ignatchenko A, Whiteley K, Jurisica I, Adamson SL, Rossant J, Kislinger T. Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol Syst Biol. 2009;5:279. doi: 10.1038/msb.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11(6):1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105(19):6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuschke S, Dahm S, Schmidt C, Joost HG, Al-Hasani H. A meta-analysis of quantitative trait loci associated with body weight and adiposity in mice. Int J Obes. 2007;31(5):829–841. doi: 10.1038/sj.ijo.0803473. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Lawson HA, Fawcett GL, Wang B, Pletscher LS, Fox AR, Maxwell TJ, Ehrich TH, Kenney-Hunt JP, Wolf JB. Diet-dependent genetic and genomic imprinting effects on obesity in mice. Obesity. 2010;19(1):160–170. doi: 10.1038/oby.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narishima R, Yamasaki M, Hasegawa S, Fukui T. Genetic obesity affects neural ketone body utilization in the rat brain. Obesity (Silver Spring) 2009;17(3):611–615. doi: 10.1038/oby.2008.566. [DOI] [PubMed] [Google Scholar]
- Tamashiro KL, Terrillion CE, Hyun J, Koenig JI, Moran TH. Prenatal stress or high-fat diet increases susceptibility to diet-induced obesity in rat offspring. Diabetes. 2009;58(5):1116–1125. doi: 10.2337/db08-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Yi NJ, Allison DB, Leips J, Ruden DM. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes Res. 2005;13(9):1596–1605. doi: 10.1038/oby.2005.196. [DOI] [PubMed] [Google Scholar]
- Jiang ZH, Wu XL, Michal JJ, McNamara JP. Pattern profiling and mapping of the fat body transcriptome in Drosophila melanogaster. Obes Res. 2005;13(11):1898–1904. doi: 10.1038/oby.2005.233. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Reiter LT. Recent efforts to model human diseases in vivo in Drosophila. Fly. 2008;2(3):129–132. doi: 10.4161/fly.6332. [DOI] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJF, Nehme NT, Zhang XY, Knauf C, Cani PD, Aumayr K, Todoric J. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140(1):148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Shi Y, Burn P. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat Rev Drug Discov. 2004;3(8):695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS One. 2009;4(10):e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev EV, Schwudke D, Zagoriy V, Matyash V, Bogdanova A, Habermann B, Zhu L, Shevchenko A, Kurzchalia TV. LET-767 is required for the production of branched chain and long chain fatty acids in Caenorhabditis elegans. J Biol Chem. 2008;283(25):17550–17560. doi: 10.1074/jbc.M800965200. [DOI] [PubMed] [Google Scholar]
- Wollam J, Magner DB, Magomedova L, Rass E, Shen Y, Rottiers V, Habermann B, Cummins CL, Antebi A. A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol. 2012;10(4):e1001305. doi: 10.1371/journal.pbio.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107(10):4640–4645. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457(7226):210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- Kuervers LM, Jones CL, O'Neil NJ, Baillie DL. The sterol modifying enzyme LET-767 is essential for growth, reproduction and development in Caenorhabditis elegans. Mol Genet Genomics. 2003;270(2):121–131. doi: 10.1007/s00438-003-0900-9. [DOI] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122(Pt 12):1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Mattaj IW. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci. 2009;122(Pt 12):1963–1969. doi: 10.1242/jcs.044750. [DOI] [PubMed] [Google Scholar]
- Kniazeva M, Shen H, Euler T, Wang C, Han M. Regulation of maternal phospholipid composition and IP(3)-dependent embryonic membrane dynamics by a specific fatty acid metabolic event in C. elegans. Genes Dev. 2012;26(6):554–566. doi: 10.1101/gad.187054.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bakheet R, Parhar RS, Huang CH, Hussain MM, Pan X, Siddiqui SS, Hashmi S. Regulation of fat storage and reproduction by Kruppel-like transcription factor KLF3 and fat-associated genes in Caenorhabditis elegans. J Mol Biol. 2011;411(3):537–553. doi: 10.1016/j.jmb.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;272(1):263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2(7):e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176(2):865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chung SH, Fang-Yen C, Craig C, Kerr RA, Suzuki H, Samuel AD, Mazur E, Schafer WR. A self-regulating feed-forward circuit controlling C. elegans egg-laying behavior. Curr Biol. 2008;18(19):1445–1455. doi: 10.1016/j.cub.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB. Genetic analysis of the cha-1-unc-17 gene complex in Caenorhabditis. Genetics. 1989;122(1):73–80. doi: 10.1093/genetics/122.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Russell RL. Choline acetyltransferase-deficient mutants of the nematode Caenorhabditis elegans. Genetics. 1984;106(2):227–248. doi: 10.1093/genetics/106.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso A, Grundahl K, McManus JR, Asbury JM, Rand JB. Alternative splicing leads to two cholinergic proteins in Caenorhabditis elegans. J Mol Biol. 1994;241(4):627–630. doi: 10.1006/jmbi.1994.1538. [DOI] [PubMed] [Google Scholar]
- Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38(3):363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Nedelcu S, Boulias K, Bishop NA, Guarente L, Horvitz HR. 3-Ketoacyl thiolase delays aging of Caenorhabditis elegans and is required for lifespan extension mediated by sir-2.1. Proc Natl Acad Sci U S A. 2010;107(44):18927–18932. doi: 10.1073/pnas.1013854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Paik YK. A potential role for fatty acid biosynthesis genes during molting and cuticle formation in Caenorhabditis elegans. BMB Rep. 2011;44(4):285–290. doi: 10.5483/BMBRep.2011.44.4.285. [DOI] [PubMed] [Google Scholar]
- Ashrafi K. In: Wormbook. The C. elegans Research Community, editor. Obesity and the regulation of fat metabolism. 10.1895/wormbook.1.130.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa A, Rappleye CA, Aroian RV. Pod-2, along with pod-1, defines a new class of genes required for polarity in the early Caenorhabditis elegans embryo. Dev Biol. 2001;233(2):412–424. doi: 10.1006/dbio.2001.0234. [DOI] [PubMed] [Google Scholar]
- Kniazeva M, Sieber M, McCauley S, Zhang K, Watts JL, Han M. Suppression of the ELO-2 FA elongation activity results in alterations of the fatty acid composition and multiple physiological defects, including abnormal ultradian rhythms, in Caenorhabditis elegans. Genetics. 2003;163(1):159–169. doi: 10.1093/genetics/163.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Gower NJ, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15(8):3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DS, Vazquez-Manrique RP, Gower NJ, Gregory E, Schafer WR, Baylis HA. Inositol 1,4,5-trisphosphate signalling regulates the avoidance response to nose touch in Caenorhabditis elegans. PLoS Genet. 2009;5(9):e1000636. doi: 10.1371/journal.pgen.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Manrique RP, Nagy AI, Legg JC, Bales OA, Ly S, Baylis HA. Phospholipase C-epsilon regulates epidermal morphogenesis in Caenorhabditis elegans. PLoS Genet. 2008;4(3):e1000043. doi: 10.1371/journal.pgen.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8(3):266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Kniazeva M, Euler T, Han M. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 2008;22(15):2102–2110. doi: 10.1101/gad.1692008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A. 2009;106(6):1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27(1):121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Reue K. The role of lipin 1 in adipogenesis and lipid metabolism. Novartis Found Symp. 2007;286:58–68. doi: 10.1002/9780470985571.ch6. discussion 68–71, 162–163, 196–203. [DOI] [PubMed] [Google Scholar]
- Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1(1):73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Reue K, Brindley DN. Thematic Review Series: Glycerolipids. Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J Lipid Res. 2008;49(12):2493–2503. doi: 10.1194/jlr.R800019-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson PJ, Chen S, Tayeh MK, Ochoa L, Leal SM, Pelet A, Munnich A, Lyonnet S, Majeed HA, El-Shanti H. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome) J Med Genet. 2005;42(7):551–557. doi: 10.1136/jmg.2005.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuquoy C, Burnol AF, Moldes M. PNPLA3, a genetic marker of progressive liver disease, still hiding its metabolic function? Clin Res Hepatol Gastroenterol. 2013;37(1):30–35. doi: 10.1016/j.clinre.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50(10):2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulas J, Schmidt C, Rothe M, Schunck WH, Menzel R. Cytochrome P450-dependent metabolism of eicosapentaenoic acid in the nematode Caenorhabditis elegans. Arch Biochem Biophys. 2008;472(1):65–75. doi: 10.1016/j.abb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Aarnio V, Lehtonen M, Storvik M, Callaway JC, Lakso M, Wong G. Caenorhabditis Elegans Mutants Predict Regulation of Fatty Acids and Endocannabinoids by the CYP-35A Gene Family. Front Pharmacol. 2011;2:12. doi: 10.3389/fphar.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Ferguson K, Kadyk L, Watts JL. The role of nuclear receptor NHR-64 in fat storage regulation in Caenorhabditis elegans. PLoS One. 2010;5(3):e9869. doi: 10.1371/journal.pone.0009869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. elegans lipid metabolic gene database. 471 lipid metabolism genes were retrieved from the C. elegans genome. Different colors were used to highlight the origin of the various lipid genes. The metabolism pathways of each gene mainly come from the KEGG database.
Annotation of biological processes of 471 C. elegans lipid metabolism genes using ClueGO. The chart displays part of significant enrichment analysis of Gene Ontology molecular function in C. elegans lipid metabolic gene database. The x axis stands for the amount molecular function terms in Gene Ontology. One star denotes P < 0.05, while two stars denote P < 0.01.
Lipid metabolism genes in 5 model organisms. All genes IDs were unified as Ensembl gene ID. Respectively, 581, 585, 563, and 428 lipid metabolic genes were present in the human, mouse, rat, and Drosophila genomes.
The list of human, mouse, rat, and Drosophila lipid metabolic genes. 581 (A), 585 (B), 563 (C), and 428 (D) lipid metabolism genes present in human, mouse, rat, and Drosophila genome, respectively. Abbreviation: CEL- C. elegans; DRO- Drosophila.
Lipid metabolism genes related to human metabolic diseases in humans, C. elegans, mice, rats, and Drosophila. Respectively, 97, 94, 128, 107, and 97 lipid genes were overlapped with metabolic diseases in human, C. elegans, mouse, rat, and Drosophila.