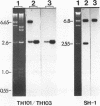
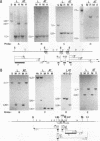
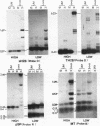
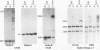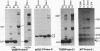Abstract
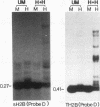
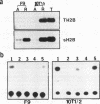
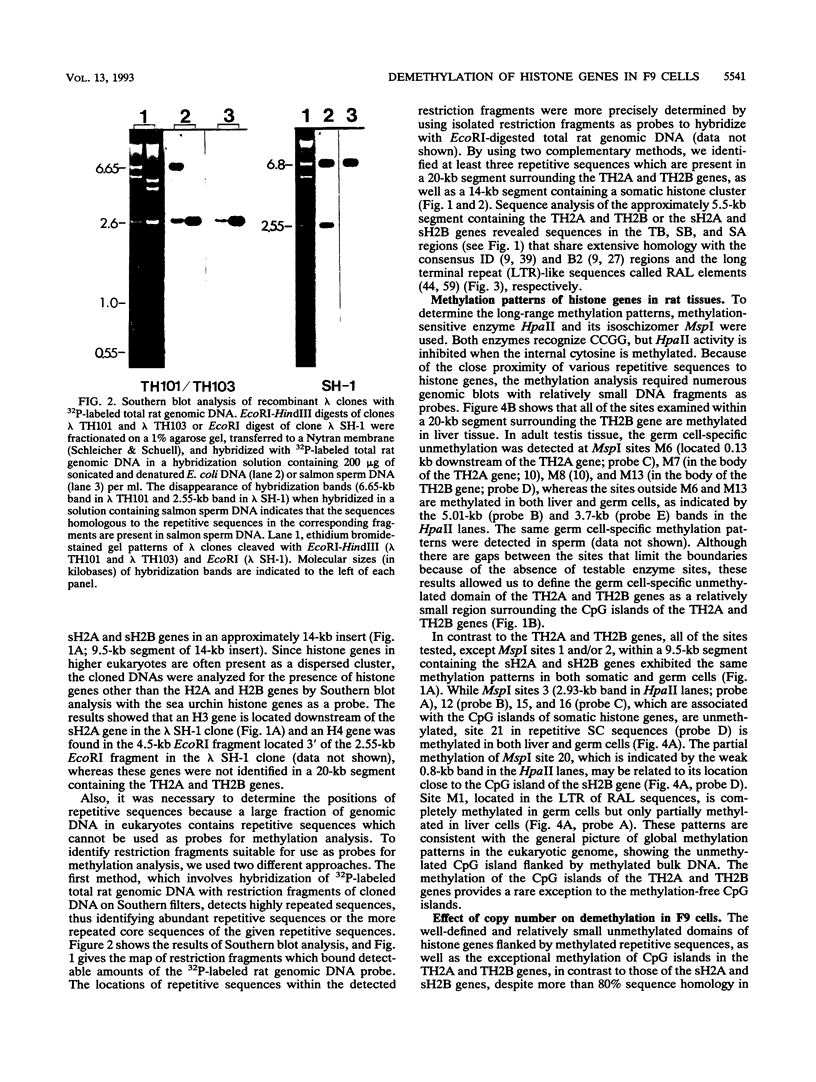
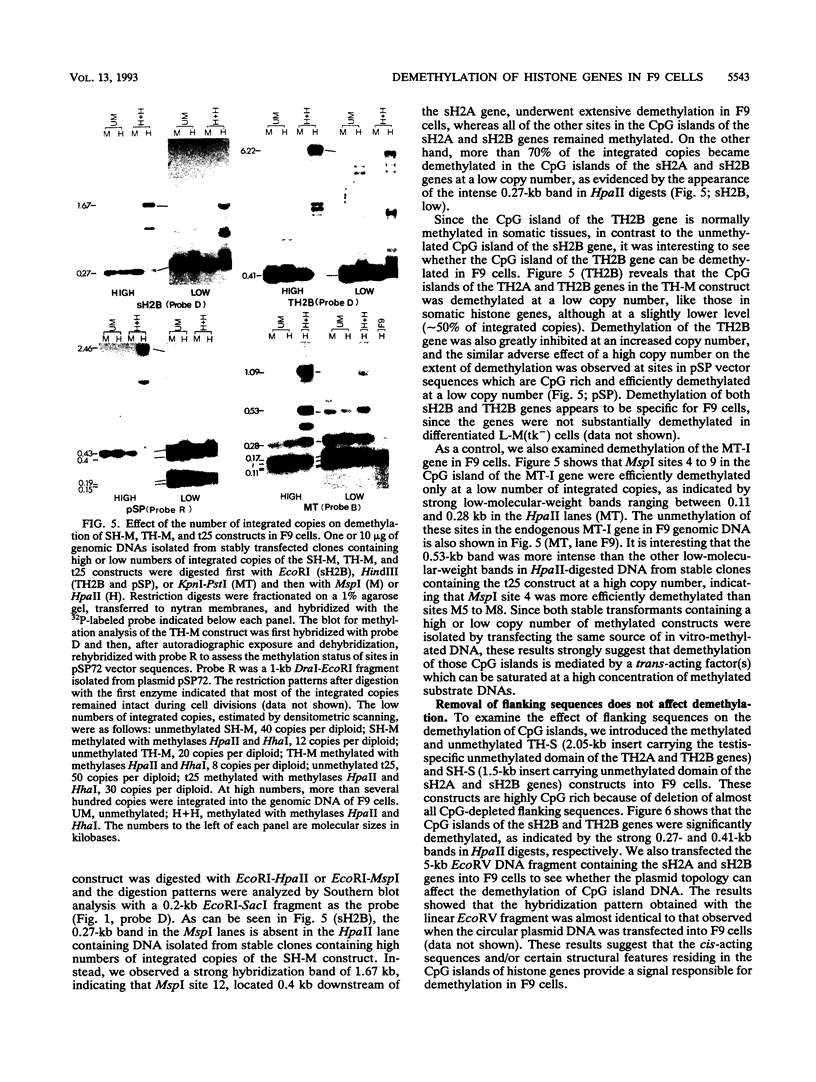
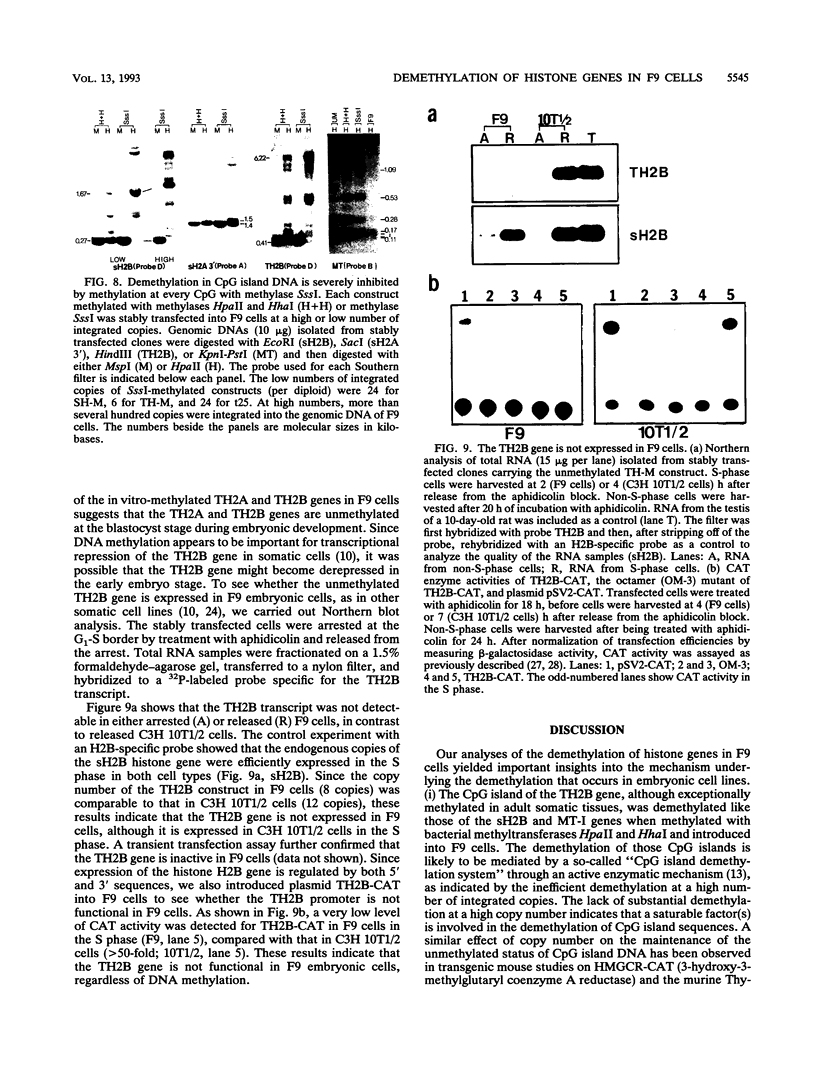
In contrast to many other genes containing a CpG island, the testis-specific H2B (TH2B) histone gene exhibits tissue-specific methylation patterns in correlation with gene activity. Characterization of the methylation patterns within a 20-kb segment containing the TH2A and TH2B genes in comparison with that in a somatic histone cluster revealed that: (i) the germ cell-specific unmethylated domain of the TH2A and TH2B genes is defined as a small region surrounding the CpG islands of the TH2A and TH2B genes and (ii) somatic histone genes are unmethylated in both liver and germ cells, like other genes containing CpG islands, whereas flanking sequences are methylated. Transfection of in vitro-methylated TH2B, somatic H2B, and mouse metallothionein I constructs into F9 embryonal carcinoma cells revealed that the CpG islands of the TH2A and TH2B genes were demethylated like those of the somatic H2A and H2B genes and the metallothionein I gene. The demethylation of those CpG islands became significantly inefficient at a high number of integrated copies and a high density of methylated CpG dinucleotides. In contrast, three sites in the somatic histone cluster, of which two sites are located in the long terminal repeat of an endogenous retrovirus-like sequence, were efficiently demethylated even at a high copy number and a high density of methylated CpG dinucleotides. These results suggest two possible mechanisms for demethylation in F9 cells and methylation of CpG islands of the TH2A and TH2B genes at the postblastula stage during embryogenesis.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Dynan W. S. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol Cell Biol. 1988 May;8(5):2021–2033. doi: 10.1128/mcb.8.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarik D. P., Cook J. A., Pitha P. M. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 1990 Apr;9(4):1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Burbelo P. D., Horikoshi S., Yamada Y. DNA methylation and collagen IV gene expression in F9 teratocarcinoma cells. J Biol Chem. 1990 Mar 25;265(9):4839–4843. [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Cedar H., Stein R., Gruenbaum Y., Naveh-Many T., Sciaky-Gallili N., Razin A. Effect of DNA methylation on gene expression. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):605–609. doi: 10.1101/sqb.1983.047.01.071. [DOI] [PubMed] [Google Scholar]
- Choi Y. C., Chae C. B. DNA hypomethylation and germ cell-specific expression of testis-specific H2B histone gene. J Biol Chem. 1991 Oct 25;266(30):20504–20511. [PubMed] [Google Scholar]
- Falzon M., Kuff E. L. Binding of the transcription factor EBP-80 mediates the methylation response of an intracisternal A-particle long terminal repeat promoter. Mol Cell Biol. 1991 Jan;11(1):117–125. doi: 10.1128/mcb.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra A., Fewell J., Lueders K., Kuff E. In vitro methylation inhibits the promotor activity of a cloned intracisternal A-particle LTR. Nucleic Acids Res. 1986 May 27;14(10):4343–4352. doi: 10.1093/nar/14.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., Keshet I., Shani M., Levine A., Razin A., Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991 May 16;351(6323):239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessmann M., Graessmann A., Wagner H., Werner E., Simon D. Complete DNA methylation does not prevent polyoma and simian virus 40 virus early gene expression. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6470–6474. doi: 10.1073/pnas.80.21.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G., Kolstø A. B., Larsen F., Prydz H. Tissue-specific methylation of a CpG island in transgenic mice. Gene. 1992 Apr 15;113(2):207–214. doi: 10.1016/0378-1119(92)90397-8. [DOI] [PubMed] [Google Scholar]
- Gundersen G., Kolstø A. B., Prydz H. Differential methylation of a CpG-island concatemer in hemi- and homozygous transgenic mice. FEBS Lett. 1991 Dec 16;295(1-3):214–218. doi: 10.1016/0014-5793(91)81421-4. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Walling M. Regulation in vivo of a cloned mammalian gene: cadmium induces the transcription of a mouse metallothionein gene in SV40 vectors. J Mol Appl Genet. 1982;1(4):273–288. [PubMed] [Google Scholar]
- Hasse A., Schulz W. A., Sies H. De novo methylation of transfected CAT gene plasmid constructs in F9 mouse embryonal carcinoma cells. Biochim Biophys Acta. 1992 May 7;1131(1):16–22. doi: 10.1016/0167-4781(92)90092-e. [DOI] [PubMed] [Google Scholar]
- Hoeben R. C., Migchielsen A. A., van der Jagt R. C., van Ormondt H., van der Eb A. J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991 Feb;65(2):904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett S. K., Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991 Sep;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Hwang I. W., Lim K., Chae C. B. Characterization of the S-phase-specific transcription regulatory elements in a DNA replication-independent testis-specific H2B (TH2B) histone gene. Mol Cell Biol. 1990 Feb;10(2):585–592. doi: 10.1128/mcb.10.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Hwang I., Tres L. L., Kierszenbaum A. L., Chae C. B. Molecular cloning and differential expression of somatic and testis-specific H2B histone genes during rat spermatogenesis. Dev Biol. 1987 Nov;124(1):23–34. doi: 10.1016/0012-1606(87)90455-6. [DOI] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruczek I., Doerfler W. Expression of the chloramphenicol acetyltransferase gene in mammalian cells under the control of adenovirus type 12 promoters: effect of promoter methylation on gene expression. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7586–7590. doi: 10.1073/pnas.80.24.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K. D., Weyer U., Doerfler W. Trans effect of the E1 region of adenoviruses on the expression of a prokaryotic gene in mammalian cells: resistance to 5' -CCGG- 3' methylation. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1598–1602. doi: 10.1073/pnas.83.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992 Aug;13(4):1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lock L. F., Takagi N., Martin G. R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987 Jan 16;48(1):39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., Bird A. P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992 Oct 11;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Mehtali M., LeMeur M., Lathe R. The methylation-free status of a housekeeping transgene is lost at high copy number. Gene. 1990 Jul 16;91(2):179–184. doi: 10.1016/0378-1119(90)90086-7. [DOI] [PubMed] [Google Scholar]
- Milner R. J., Bloom F. E., Lai C., Lerner R. A., Sutcliffe J. G. Brain-specific genes have identifier sequences in their introns. Proc Natl Acad Sci U S A. 1984 Feb;81(3):713–717. doi: 10.1073/pnas.81.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Adams R. L., Rinaldi A. Decrease in DNA methylase activity during preimplantation development in the mouse. Development. 1991 May;112(1):189–192. doi: 10.1242/dev.112.1.189. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Monk M. Methylation and the X chromosome. Bioessays. 1986 May;4(5):204–208. doi: 10.1002/bies.950040505. [DOI] [PubMed] [Google Scholar]
- Mummaneni P., Bishop P. L., Turker M. S. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J Biol Chem. 1993 Jan 5;268(1):552–558. [PubMed] [Google Scholar]
- Nakamuta M., Furuich M., Takahashi K., Suzuki N., Endo H., Yamamoto M. Isolation and characterization of a family of rat endogenous retroviral sequences. Virus Genes. 1989 Sep;3(1):69–83. doi: 10.1007/BF00301988. [DOI] [PubMed] [Google Scholar]
- O'Connell C. D., Cohen M. The long terminal repeat sequences of a novel human endogenous retrovirus. Science. 1984 Dec 7;226(4679):1204–1206. doi: 10.1126/science.6505687. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Hansen R. S., Gartler S. M., Riggs A. D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Rupp R. A., Weintraub H. Ubiquitous MyoD transcription at the midblastula transition precedes induction-dependent MyoD expression in presumptive mesoderm of X. laevis. Cell. 1991 Jun 14;65(6):927–937. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- Sanford J. P., Clark H. J., Chapman V. M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987 Dec;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapienza C., Peterson A. C., Rossant J., Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987 Jul 16;328(6127):251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hamada T., Ueda T., Seki R., Higashinakagawa T., Sakaki Y. Inherited type of allelic methylation variations in a mouse chromosome region where an integrated transgene shows methylation imprinting. Development. 1991 Feb;111(2):573–581. doi: 10.1242/dev.111.2.573. [DOI] [PubMed] [Google Scholar]
- Shemer R., Eisenberg S., Breslow J. L., Razin A. Methylation patterns of the human apoA-I/C-III/A-IV gene cluster in adult and embryonic tissues suggest dynamic changes in methylation during development. J Biol Chem. 1991 Dec 15;266(35):23676–23681. [PubMed] [Google Scholar]
- Shemer R., Kafri T., O'Connell A., Eisenberg S., Breslow J. L., Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Inokuchi K., Nienhuis A. W. Site-specific demethylation and normal chromatin structure of the human dihydrofolate reductase gene promoter after transfection into CHO cells. Mol Cell Biol. 1987 Aug;7(8):2830–2837. doi: 10.1128/mcb.7.8.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J., Grant M., LeBon J. M., Okuyama K., Chapman V., Monk M., Riggs A. D. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990 Sep;10(9):4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Stuhlmann H., Jähner D., Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G. W., Searle P. F., Palmiter R. D. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. 1985 Oct 31-Nov 6Nature. 317(6040):828–831. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Fujiyoshi T., Maehara Y., Takahashi K., Yamamoto M., Endo H. A new family of LTR-like sequences abundantly expressed in rat tumors. Nucleic Acids Res. 1986 Dec 9;14(23):9271–9289. doi: 10.1093/nar/14.23.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Tanigawa G., McCarthy P. L., Jr A DNA signal from the Thy-1 gene defines de novo methylation patterns in embryonic stem cells. Mol Cell Biol. 1990 Aug;10(8):4396–4400. doi: 10.1128/mcb.10.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Langner K. D., Jüttermann R., Müller U., Zock C., Klimkait T., Doerfler W. Reactivation of the methylation-inactivated late E2A promoter of adenovirus type 2 by E1A (13 S) functions. J Mol Biol. 1988 Jul 20;202(2):255–270. doi: 10.1016/0022-2836(88)90456-1. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988 Sep 23;241(4873):1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]
- Yisraeli J., Adelstein R. S., Melloul D., Nudel U., Yaffe D., Cedar H. Muscle-specific activation of a methylated chimeric actin gene. Cell. 1986 Aug 1;46(3):409–416. doi: 10.1016/0092-8674(86)90661-6. [DOI] [PubMed] [Google Scholar]