Abstract
Myosin V is biomolecular motor with two actin-binding domains (heads) that take multiple steps along actin by a hand-over-hand mechanism. We used high-speed polarized total internal reflection fluorescence (polTIRF) microscopy to study the structural dynamics of single myosin V molecules that had been labeled with bifunctional rhodamine linked to one of the calmodulins along the lever arm. With the use of time-correlated single-photon counting technology, the temporal resolution of the polTIRF microscope was improved ∼50-fold relative to earlier studies, and a maximum-likelihood, multitrace change-point algorithm was used to objectively determine the times when structural changes occurred. Short-lived substeps that displayed an abrupt increase in rotational mobility were detected during stepping, likely corresponding to random thermal fluctuations of the stepping head while it searched for its next actin-binding site. Thus, myosin V harnesses its fluctuating environment to extend its reach. Additional, less frequent angle changes, probably not directly associated with steps, were detected in both leading and trailing heads. The high-speed polTIRF method and change-point analysis may be applicable to single-molecule studies of other biological systems.
Introduction
Myosin V is a molecular motor that translocates along actin filaments in many cell types, transporting cargos toward the barbed end of actin or tethering them at their destinations (1–10). Defects in myosin V are associated with human pigmentation, immunological, and neurological disorders (1). Myosin V consists of two N-terminal globular ATP- and actin-binding domains (heads), a lever arm comprising α-helical light-chain-binding motifs and six tightly bound calmodulins (CaMs) or CaM-like light chains per head, a coiled-coil dimerization domain, and finally a C-terminal cargo-binding tail that is usually absent in studies using recombinant proteins (2).
In eukaryotic cells, single molecules or very small groups of myosin V molecules can transport cargos by producing ∼50 mechanical steps without dissociating upon each diffusional encounter with actin, a property termed processivity (4,6,8). Many ensemble and single-molecule techniques have been applied toward determining the chemomechanical cycle of myosin V (4–8,10–13), leading to a consensus view of the mechanism in which a state with both heads bound to actin, at a 36 nm separation, occupies much of the ATPase cycle. The biochemical and structural states of the two heads are coordinated, probably via intermolecular strain, which minimizes backward stepping and dissociation of the entire molecule (2). A 36 nm step is taken when the trailing head dissociates, swings 72 nm forward (toward the barbed end), and becomes the leading head. A rapid forward-directed working stroke rotates the lever arm of the attached head, accounting for most (20–24 nm) of the step (7). The remaining distance is covered by thermal fluctuations of the free head until it binds to an actin subunit ∼36 nm ahead of the bound head. Evidence for this postulated thermal-search period was obtained by high-speed atomic force microscopy (AFM) (10) and by tracking gold particles (14), microtubules (15), or actin (16) bound to the lever arm. All of these experiments required investigators to attach a relatively large probe to the lever arm/or slow down its motion by a nearby adherent surface.
Smaller motions and rotations of the lever arm have been associated with ADP release from the trailing head, possibly accounting for ∼5 nm of stepping (7). Additional small steps and lever arm rotations were detected when the leading-head lever arm stroked before (17) or after (18) the trailing head detached, and also apparently were independent of productive stepping (10,19). The notion that the orientation of the lever arm can change prior to stepping is supported by cryoelectron microscopy (20,21), whereas combined single-molecule fluorescence localization and polarization experiments support a straight-legged model for stepping (19). The roles of these substeps (if any) in facilitating productive translocation or in coordinating the two heads are not clear.
Single-molecule fluorescence techniques (8,22–24) are powerful tools for detecting the position and orientation changes of macromolecules with the use of relatively small organic fluorophore or quantum dot reporter probes. Polarized total internal reflection fluorescence (polTIRF) microscopy measures the orientation and rotational motions of dipolar fluorophores by exploiting their preference for absorbing light and emitting fluorescence polarized along their dipole axis. When the fluorophore is bound to the macromolecule in a known position and direction, as is the case with bifunctional rhodamine (BR) linked to a pair of nearby cysteine residues engineered into CaM (25), molecular rotations can be inferred from the probe angles changes (12,23,26,27).
In addition to the three-dimensional (3D) orientation of the fluorophore and its linked protein domain, one can determine rotational mobility, including microsecond wobbling motions that are not generally accessible in ensemble fluorescence anisotropy experiments (22,28,29). Microsecond-timescale wobbling of a protein domain might be functionally important, such as in detecting the thermal search period during myosin V stepping. Until now, the capability of polTIRF to measure wobble has not been utilized for dynamic measurements, except to verify stable probe attachment. This is partly because the observation time required to cycle through the range of input polarizations that are necessary to measure the 3D orientation and wobble (40–80 ms) is longer than the expected lifetime of many structural states, including myosin V’s postulated thermal search.
In this work, we studied recombinant myosin V with BR linked to one of the lever-arm CaM subunits to determine, at high time resolution, the events that occurred while it was taking translocation steps. We improved the time resolution of the polTIRF microscope ∼50-fold by incorporating a single-photon counting device and switching the polarization of the fluorescence excitation between the requisite directions every 100 μs instead of every 5–10 ms as in most of our previous studies (12,23,26–28,30). Even when the 532 nm laser excitation was made stronger, the signals were noisy due to the limited number of photons collected during each 100 μs polarization time window. To help interpret the signals, we used a maximum-likelihood, multitrace change-point algorithm (31) that objectively identifies changes in intensity (change points), indicating structural changes with single-photon precision and no binning of the raw data. We determined the orientation of the BR and the extent of wobble during the intervals between intensity changes, and found that the changes often corresponded to structural motions of the molecule. Short-lived substeps that displayed abrupt increases in probe wobble were detected with the pattern expected for hand-over-hand stepping, likely corresponding to the random thermal fluctuations of the detached head during its search for the next actin-binding site. In molecules showing these angular fluctuations, they typically occurred when the trailing head stepped, as expected. Additional, less frequent angle changes of the lever arm were detected prior to stepping in both leading and trailing positions. To our knowledge, this work represents the first use of single-molecule polTIRF microscopy to report microsecond-timescale motions relevant to the function of a biophysical macromolecule. This method may be applicable to single-molecule studies of other biological systems.
Materials and Methods
Processive motility assay
Recombinant chicken myosin V with its full-length lever arm (amino acids 1–1099) and a FLAG affinity tag at its C-terminus was coexpressed with CaM in SF9 cells (32). Chicken CaM with residues Pro-66 and Ala-73 mutated to cysteine was expressed in Escherichia coli (33), purified, and labeled with BR (29,34) (a generous gift from Dr. J.E.T. Corrie). Myosin V was labeled by exchanging endogenous wild-type chicken CaM (WT-CaM) with exogenous BR-CaM at low stoichiometry (12,29). G-actin was obtained from rabbit skeletal muscle (35). Alexa 647-labeled F-actin was prepared from G-actin and Alexa-647 actin (Molecular Probes, Carlsbad, CA) at a 5:1 ratio and 1 μM total actin subunit concentration and stabilized with 1.1 μM phalloidin (Molecular Probes). In some experiments, 0.05 μM biotin-actin (Cytoskeleton, Denver, CO) was also incorporated during polymerization.
PolTIRF experiments were performed as described previously (29,36). Briefly, all reagents were made in myosin buffer (M5; pH = 7.6) containing 25 mM KCl, 20 mM HEPES, 5 mM MgCl2, and 1 mM EGTA, except for the motility buffer (M5+) used for single-molecule myosin V motility assays, which consisted of M5 buffer with 1–40 μM ATP, 100 mM dithiothreitol (DTT), 100 μg/ml WT-CaM, and ∼100 pM of BR-CaM-labeled myosin V. For some experiments, 2,3-butanedione monoxime (BDM; B0753; Sigma, St. Louis, MO), prepared fresh from powder, was included in the final motility buffer (final concentration 50–100 mM) and the pH was readjusted as needed with KOH. Sometimes the M5+ buffer included 10 mM phosphocreatine (P-7936; Sigma) and 0.3 mg/ml creatine phosphokinase (prepared daily from powder; C3755; Sigma), but no influence on myosin velocity or stepping kinetics was detected. To avoid BR blinking, 100 mM DTT was used as a reducing agent with no deoxygenating system (23).
Myosin V processivity experiments were performed in a flow cell consisting of a plasma-cleaned quartz slide (high purity fused silica grade quartz, 212000-001; Quartz Scientific, Fairport Harbor, OH), spin coated with 2 mg/ml poly(methyl methacrylate) (secondary standard grade, 37-003-7; Sigma-Aldrich, St. Louis, MO), and a glass coverslip (No. 1; Fisher Scientific, Hampton, NH) held together by pieces of double-sided tape (Cat. No. 665; Scotch). In most experiments, 60 nM N-ethylmaleimide (NEM)-treated myosin II (37) was flowed into the lane and incubated for 5 min, followed by a rapid flow of a 200 nM Alexa 647-actin filaments. The surface was passivated by a 5 min incubation with 1 mg/ml unlabeled bovine serum albumin (BSA; A0281; Sigma Aldrich). Finally, the motility buffer containing the myosin and ATP was flowed into the chamber. Slight variations in the protocol were also implemented, including substituting the NEM myosin and Alexa-647-labeled actin with 1 mg/ml biotinylated BSA (A8549; Sigma Aldrich), and incubation with 0.5 mg/ml streptavidin (S-4762; Sigma Aldrich) and biotin-Alexa 647-actin filaments, but no significant differences were detected.
High-speed polTIRF setup
We improved the time resolution of the polTIRF microscope (28,29,36) by decreasing the time each laser polarization illuminated the sample from the earlier 5 ms periods to 0.1 ms, and by storing the arrival time and the polarization state of the illumination for each detected photon. Two alternating beams from a 532 nm Nd:YAG laser (Fig. S1 in the Supporting Material) were projected through a coupling prism at a glancing incident angle, producing an evanescent field at the quartz slide/aqueous interface. Switching between the two beams (termed path 1 and path 2) was achieved by computer-controlled voltages applied to a Pockels cell (PC0) and a polarizing beam-splitting (PBS0) cube. Similarly, the polarization of the laser in each beam was switched between four different linear polarizations (s, 90°; p, 0°; L, +45°; and R, −45° relative to the plane defined by the incident and reflected beams; Fig. S1) with an additional Pockels cell and a Berek compensator in each beam (PC1/BC1 and PC2/BC2). Each polarization illuminated the sample for 0.1 ms in the sequence s1, p1, p2, s2, R1, L1, L2, and R2 (letter indicates incident polarization, number indicates path).
Fluorescence emission was collected by a Leica (Heerbrugg, Switzerland) 100× 1.2 NA water immersion lens, passed through a long-pass blocking filter, and either imaged onto an intensified charge-coupled device (CCD) camera (Cascade II; Photometrics, Tucson, AZ) or directed through a polarizing beam splitter (PBS1) that resolved its x and y components between two avalanche photodiodes (APDx and APDy). A time-correlated single-photon counting PC adaptor board (TCSPC, SPC-130; Becker and Hickl, Berlin, Germany) operating in FIFO mode, and a pulse router (HRT-82; Becker and Hickl) were modified to record the excitation polarization state according to a suggestion by Dr. Wolfgang Becker of Becker and Hickl (details available upon request). Photon pulses during the first 3 μs of each polarization interval, while the Pockel cell voltage was settling, were ignored. A 10 MHz pulse train from a digital delay generator (DG645; Stanford Research Systems, Sunnyvale, CA) triggered the timing of the TCSPC circuit and a 10 kHz pulse train, generated by same DG645 unit, triggered digital counters that sequenced the high-voltage amplifiers driving the three Pockels cells. For visualization purposes, single-photon counts from each of the 16 combinations of excitation path and polarization and emission polarization were converted to and plotted as polarized fluorescence intensities (PFIs) by counting the number of photons detected during successive constant time bins (here 0.1 ms). An analytical model of the expected PFIs was fitted by maximum likelihood to the 16 PFIs, or averages of the 16 PFIs over intervals between sudden change points (see below) to determine the orientation of the probe absorption and emission dipole (see Fig. 3 B, inset) (26,28,29); θ is the axial probe angle relative to the microscope optical axis, and ϕ is its azimuth around the optical axis. Using the orientation of the actin, known from the initial fluorescence video snapshot, θ and ϕ are converted to β and α, the axial and azimuthal probe angles relative to the actin (26,28,29). The probe wobbles due to protein motions and to faster local motions relative to the CaM. On timescales slower than the fluorescence lifetime (4 ns) but faster than the measurement interval (0.1 ms), the extent of probe motion is described as a cone of half-angle δ that is centered on the orientation (θ, ϕ). A control experiment in which the microscope stage was moved stepwise successively in 100 nm increments, and with individual BR-CaMs firmly adhered to the slide, showed that the measured probe angles and wobble were not deflected as the slide moved across the stage area (∼1.5 μm × 1.5 μm) from which fluorescence was captured by the photodiodes.
Figure 3.
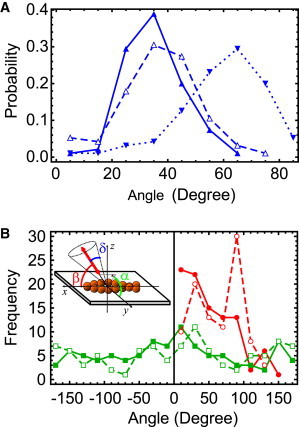
Distributions of probe angle and wobble before and after steps including a detectable high-wobble state (98 events in 73 molecules). (A) The distributions of δ (blue) during the intervals immediately before (▲ with solid line) and after (▵ with dashed line) steps in which a high wobble state was detected peak at lower values (∼30°) compared with the distribution of δ during the high-wobble state (▾ with dotted line), which peaks at large values (∼70–80°). (B) Distributions of β (red) and α (green) during the intervals immediately before (● and ▪ with solid lines) and after (○ and □ with dashed lines) steps with detectable high-wobble states. Steps with a high-wobble state allow trailing and leading lever arm states to be identified unambiguously as the intervals before and after the step, respectively. The inset illustrates the angle conventions for β, α, and δ relative to the actin filament.
Immediately before each polarization recording, two images of the field of candidate fluorophores were recorded with the CCD camera, superimposed on an image of the Alexa-647-labeled actin filaments and displayed on a monitor. These images were used to estimate the position and direction of the BR-CaM-labeled myosin V. A moving molecule was selected for polarization analysis and identified by a mouse click. A custom-built LabView program centered the molecule above the objective via a piezoelectric stage, inserted a removable mirror that directed the fluorescence emission onto the APDs, and sent a trigger signal that reset the Pockels cell drivers to the initial polarization setting and started the TCSPC. Photon counts were recorded for 5 s, corresponding to 6250 cycles of the 16 polarization channels. During recording of polarized fluorescence, spatial information from the fluorophore was not available.
Multiple-channel change-point algorithm
In a previous study (31) we developed a multiple-channel change-point (MCCP) algorithm tailored to single-photon polTIRF experiments. In this algorithm, a likelihood function is used to compare two hypotheses: 1), the hypothesis that there is a single, abrupt intensity change at the ith photon in an interval containing N photons in time T; and 2), the null hypothesis that there is no intensity change at the ith photon. Assuming that the fluorescence emission is random, the only user-defined parameter is the probability of a false-positive change point, here chosen to be 5%, which defines a threshold that the likelihood function must exceed for significance. The likelihood function, Li, is calculated for each photon as follows:
where ni,j is an N × 16 dimensional matrix of the accumulated number of photons i for each of the j PFIs (Fig. S7), ti indicates the arrival time of the ith photon, and Nj is the total number of photons in the jth PFI. A nonuniform distribution of false positives across the interval is mitigated by applying correction terms to the likelihood function and also including a small exclusion region that prevents change points from being detected in the first or last 2.5% of the photons in the interval (31,38).
The algorithm is applied iteratively to the data until no additional change points are detected. The likelihood surfaces for the change points at 0.46 s and 0.49 s (points 5 and 6 correspond to the magenta and red lines in Fig. S3 C) exceed the threshold (black line) for 95% confidence by over 10 log units and correspond to abrupt changes in the PFIs (four of 16 are shown in Fig. S3 A) and the kink in the accumulated photon trace (Fig. S3 B). The likelihood surface of a third change point at ∼0.4 s (point 4, blue line) is broader and less pronounced with only a small change in the PFIs, but it still exceeds the threshold for significance by ∼8 log units. The peaks of the likelihood functions determine the most likely location of the change points (vertical black dashed lines), and the relative sharpness of the likelihood peaks determines the 95% confidence interval (CI; gray-shaded regions), which for change points 4, 5, and 6 are 1000, 226, and 248 photons or 15, 3.6, and 4.5 ms, respectively.
Ambiguities in the dipole model for orientation and wobble
The 16 combinations of eight time-multiplexed input polarizations and two simultaneously measured emission polarizations used here, including ±45° linear polarized excitations, enable unambiguous determination of the probe angle within a hemisphere of orientation (26,29,39). In addition to allowing a greater range of angles to be discerned, the ±45° laser polarizations also resolve a more subtle ambiguity associated with δ. When the probe is completely free to rotate on the microsecond timescale, δ should be 90° (θ and ϕ are not defined at high δ); however, when only s and p (i.e., 0° and 90°) input laser polarizations are used (12) and the probe has high wobble, an orientation in the laboratory frame of reference of (θ, ϕ) = (54.7°, 45°) and δ ≈ 50° is usually reported by the dipole model because the corresponding eight PFIs are the same as in the case when δ = 90°. In previous work, this artifact was not noticed because periods of large probe wobble were not explicitly investigated.
Choice of hemisphere
For any given measurement of dipole orientation, either one of two vectors ((θ, ϕ) or (180° − θ, ϕ ± 180°)) may apply due to the twofold symmetry of the probe dipole. The BR probe bound to the protein is not symmetrical, however, due to its specific attachments to the two different Cys residues, so it is meaningful to consider which of the dipole ends is being reported by the angle and, importantly, to maintain the same end of the probe in successive measurements on a given molecule. If the true rotational motions of the molecule are smaller than 180° and within 90° of an average angle, then all of the orientations for one end of the dipole will fall into one hemisphere. A hemisphere that fulfills this requirement is centered on the director axis determined from the measured orientations during a processive run (see Supporting Material). Which end of the director axis to align with the analysis hemisphere is still to be chosen, but for relative motions during a run, it is irrelevant. The opposite hemisphere corresponds to the molecule walking on the opposite side of the actin filament (27), an experimental detail that is not known for any individual molecule. The end closest to the initial direction of motion of the molecule, as measured from the CCD images recorded prior to the polarization analysis, is chosen as the hemisphere director. When (θ, ϕ) is rotated into the actin frame of reference (β, α), the hemisphere does not change.
Results
Rotational dynamics using high-speed polTIRF microscopy and change-point analysis
We improved the maximum time resolution of standard polarized fluorescence microscopy (5 ms recording gates, 20–40 ms cycle time (28,29)) by switching the excitation laser polarizations every 0.1 ms and cataloging the arrival time of each detected photon (Fig. S1). To obtain 20–50 photons within the 0.1 ms gate time, we increased the laser power from our standard 20 mW up to ∼50 mW in a ∼2000 μm2 area at the sample. In many recordings the BR probe photobleached before a myosin step occurred. The recordings used for analysis represented molecules that photobleached after 1–2 s of high-intensity illumination, on average, and after zero to six myosin stepping events (see Materials and Methods). Fig. S2 shows the photon counts recorded from the 16 combinations of input and detector polarization binned at 0.1 ms intervals. As the molecule walked processively along actin in the presence of 1–40 μM Mg.ATP, tilting of the BR (and myosin lever arm) back and forth between two fairly well defined angles was clearly evident from abrupt and simultaneous changes in many of the polarized photon count rates, as previously reported (12). All of the channels photobleached to the background in a single step, consistent with signals from a single fluorophore.
To convert noisy polTIRF photon counting data into interpretable intensity and angular time courses, a specialized change point analysis (38) was developed (31). Instead of binning the photons into intensity traces, the raw time-stamped data are fitted directly by a likelihood function that compares the probability of a change in the photon collection rate with that of the constant rate. By scanning this likelihood function over all of the photons in an interval from all of the 16 polarization channels, we can identify the location and uncertainty of the most likely intensity change. By iteratively applying this algorithm through the whole trace, we can identify angle changes, bleaching events, and any other large-magnitude changes in the individual channels (dashed lines in Fig. 1) that are clearly seen by eye. Smaller-magnitude but statistically robust changes are also detected.
Figure 1.
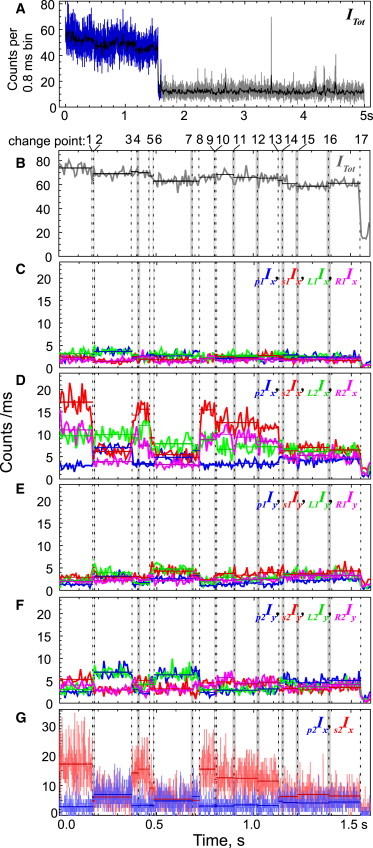
High-time-resolution PFIs. (A) Sixteen PFIs are summed into one total intensity with photons binned within each 0.8 ms cycle (blue and gray) or averaged over 10 cycles (black) that is relatively constant before photobleaching to background intensity at ∼1.5 s. (B and C–F) The total intensity (B) and the 16 PFIs (C–F) comprising the interval before photobleaching (blue region in panel A) are shown expanded, with intensities averaged over 10 cycles. The individual measured intensities depend on the relative orientation between the fluorophore dipole axis and the direction of the input laser polarizations p (blue), s (red), L (green), and R (magenta), and the polarized fluorescence detected on APDx (C and D) and APDy (E and F) for beams 1 and 2, respectively (see Fig. S1 for details on the experimental setup). (G) To illustrate the high time resolution of each PFI, the photons collected during each 0.8 ms cycle of the laser polarizations are shown for p2Ix (blue) and s2Ix (red) binned at 0.8 ms. The MCCP algorithm gives the times of intensity change points (vertical black dashed lines) along with a 95% CI (gray area). The average intensity between neighboring change points (horizontal lines) is also shown in B–G.
The fluorescence intensity for each channel during the interval is the average determined by summing the photons and dividing by the duration between adjacent change points. Each set of 16 intensities is used as input to a maximum-likelihood fitting algorithm that estimates the dipole angles, (θ, ϕ); wobble, δ; and brightness, κ, of the probe during each interval between change points (horizontal lines in Fig. 2, B–D). To interpret the angles in the biological frame of reference, the orientation relative to the microscope optical axis (θ, ϕ) is reexpressed in terms of a frame referred to the long axis of the actin filament, (β, α), where β is the polar angle with respect to the barbed (+) end of the actin filament and α is the azimuthal angle around the actin filament (12,29) (inset in Fig. 3 B). Due to the dipole character of probe excitation and emission, any one orientation = (β, α) cannot be distinguished from an equivalent orientation pointing in the opposite direction = (180°-β, α ± 180°). Instead of imposing a fixed hemisphere for reporting the angles of all molecules (26,39,40), we use an objective method to determine a favorable hemisphere for each molecule, based on the initial direction of motility and the average direction of the molecule’s probe orientations (i.e., the orientation of the director; see Materials and Methods). To determine the CIs for the measured angles, we calculate four additional estimates of each interval’s intensity based on the statistical uncertainty of localizing the adjacent change points (see Materials and Methods and Fig. S3 B).
Figure 2.
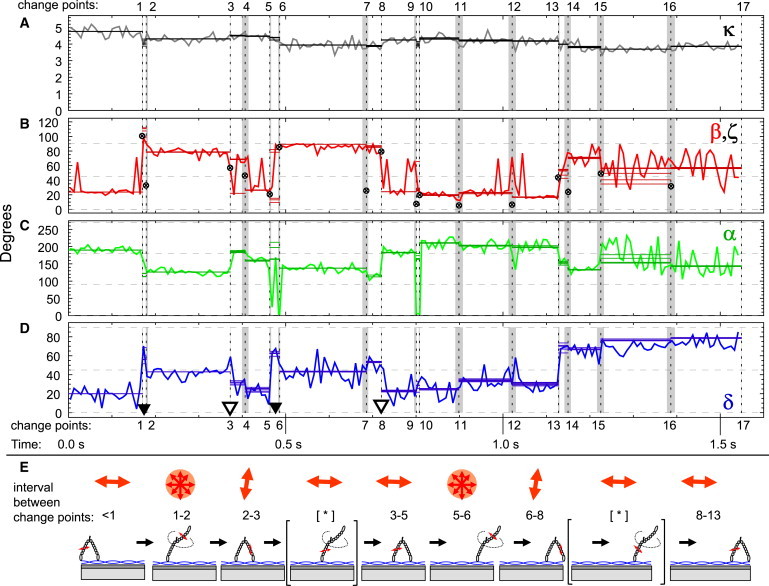
Results of the dipole model for the 8.0 ms binned intensities (jagged lines) and the change-point intensities corresponding to the CIs (five horizontal lines), which are often very close to each other because the uncertainty in localizing the change points is small. (A) The fitted intensity scale factor κ (gray) is similar to the total intensity (Fig. 1B) and is relatively constant despite large changes in the underlying PFIs, the hallmark of tilts. (B) The polar angle β (red) shows four large angle changes (denoted by ▼ and ▿ in panel D) that correspond to myosin stepping events at change points 1, 3, 5, and 8, where β alternates between a low value of 22° and a high value of 81°. The magnitude of the total angular displacement ζ between change-point intervals is represented by small crosses (⊗). (C) The azimuthal angle α (green) also changes with each step, alternating between 190° and 140°. (D) The extent of rapid probe motion during the measurement (wobble) is represented by a cone with half-angle δ (blue) and shows relatively low values during a dwell and increases during steps from the low to high β states (between change points 1-2 and 5-6), but not after steps from the high to low β states (i.e., change points 3-4 and 7-8). Wobble at the end of the trace is also elevated, possibly indicating a laser-induced breakage of one of the bifunctional bonds on the probe, resulting in a monofunctional attachment. (E) Cartoon depicting the BR-CaM-labeled myosin lever arm translocating along an actin filament (blue helix) that is attached to the quartz slide (light gray) by NEM myosin II (dark gray). The probe on the lever arm is indicated by double-headed red arrows. Increases in probe wobble (red disks) occur during every other step, consistent with the hand-over-hand mechanism and thermal search.
Large changes in β and α (Fig. 2, B and C) correspond to changes in the polarized count rates and indicate tilting between two stable states with orientation (β, α) ≈ (22°, 190°) and (81°, 140°) (Fig. S4). In Fig. 2, four major stepping events (▼ and ▿) are observed at 0.17, 0.37, 0.46, and 0.72 s as described previously (12). The angle changes in β and α can be combined into the angle subtended by the probe vector in the two orientations, ζi = cos−1 ≈ 75° (where − and + indicate the state before and after stepping, respectively).
Importantly, δ (microsecond wobble) transiently increased during the recording on alternate steps when β changed from low (trailing position) to high (leading position) values, consistent with a labeled trailing head detaching from actin (▼ in Fig. 2 D), undergoing rapid rotational motions and then rebinding to actin as the new lead head with β ≈ 90° (Fig. 2 E). The wobble parameter remains below 50° during the steps when β changes from high to low values (▿). This is most easily interpreted as the nonstepping head undergoing a working stroke while it is strongly bound to actin, as depicted in Fig. 2 E (a cartoon showing the sequence of events with the expected alternating periods of high wobble). A gallery of angular time courses from additional molecules (Fig. S5) demonstrates the same stepping pattern with periods of high wobble when the probe switches from a low to a high β angle, corresponding to the labeled head detaching and wobbling while searching for its next actin-binding site.
Binned data
For visualization purposes, the photon arrival events were accumulated in bins to produce PFIs with time resolution traded against photon shot noise (0.8 ms bins, gray/blue curves in Fig. 1, A and G, and 8 ms bins in Fig. 1, B–F). Despite the high laser power, intensities binned at 8 ms are relatively noisy, resulting in large fluctuations in the individual PFIs and in the estimated orientations. The variability of (β, α) during an otherwise constant dwell is larger than might be expected from the variance in the intensity traces, due to the highly nonlinear relationship between the magnitude of the PFIs and the corresponding dipole orientation. The wobble parameter δ is also noisy (Fig. 2 D) but can be seen to increase in magnitude prior to the first and third tilting events when β switches from low to high values (▼ in Fig. 2). During the final 1/3 of the recording, the probe is in a state of prolonged high-wobble (δ>75°) immediately prior to photobleaching, a characteristic that was observed in many other molecules (Fig. S6).
Dynamics of periods of increased probe wobble during steps
After manually screening for molecules with robust tilting (i.e., strong anticorrelated changes in the PFIs), we detected 1224 tilting events in 306 molecules, 69 of which contained a total of 84 substeps exhibiting increased wobble (inverted solid triangles in Fig. 3 A) relative to the period just before (solid triangles) and after (open triangles) each step. The fraction of molecules with at least one such high-wobble substep was ∼23%, but the fraction of steps with a high-wobble intermediate was only ∼7%. One likely cause of the low detection of these substeps is the limited number of photons collected during short-duration events. Consistent with this view, molecules recorded at higher laser power contained more high-wobble substeps (∼10%) than those recorded at low power (∼4%). The substep durations are distributed exponentially with an average duration of ∼13 ms (solid black dots in Fig. 4 A). A fraction of missing events is expected at or below the detection limit from the exponential distribution (open black dots). Nevertheless, some molecules with a very high signal/background ratio and multiple myosin steps with strong polarization changes did not exhibit detectable high-wobble substeps (Fig. S6 C).
Figure 4.
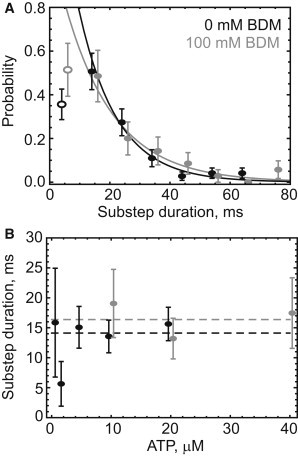
(A) Histogram of the duration of the high-wobble interval between large orientation changes that likely represent detached heads. Single exponential fits, excluding the first bin (open symbol) in each distribution, result in average lifetimes of 13 ± 2 ms (n = 99) and 16 ± 4 ms (n = 53) in the absence (black) and presence (gray) of 50–100 mM BDM. (B) The duration of the high-wobble state for molecules in the absence (black) and presence (gray) of BDM over a range of Mg.ATP concentrations agrees within uncertainty (SE) with the mean duration (horizontal dashed line), indicating that the kinetics of this state are independent of Mg.ATP concentration. Experiments with BDM in the solution were performed at a higher ATP concentration to compensate for the reduction in velocity (18). The outlier at 2 μM Mg.ATP represents only six events detected at this concentration.
Another way to test the origin of the substeps is to collect the angles before and after the high-wobble period (solid red symbols and open red symbols in Fig. 3 B, respectively). Considering all high-wobble substeps (98 events in 73 molecules), there is a strong tendency for low β angles to precede the high wobble state (red points with solid line) and high β angles to follow it. This provides strong support for the sequence of events drawn in Fig. 2, allowing the preceding orientation (low β) to be identified with the trailing lever arm, the state following the step (high β) with the leading lever arm and the high-wobble period with the search. The distributions of α before (solid green points in Fig. 3 A) and after (open green points) stepping are uniform and the same before and after stepping.
Previous work has shown that the myosin inhibitor butanedione monoxime (BDM) slows the stepping rate of myosin V by inhibiting the release of ADP (18) and also slows the rebinding rate of the detached head (14). Therefore, we performed motility experiments in the presence of 50 or 100 mM BDM and ∼2-fold higher MgATP concentration to obtain comparable rates of stepping. On the molecules with the most robust measurements of stepping, we detected 43 substeps in 39 out of 123 molecules containing a total of 380 steps (11% of steps or 32% of molecules contained high-wobble substeps). The detection efficiencies again depended on laser power, indicating detection limited by photon counts (16% of steps and 34% of molecules contained substeps at high laser powers, compared with 6% of steps and 26% of molecules at low laser power). Fitting single exponentials to histograms of the substep durations in the absence (black) or presence (gray, Fig. 4 A) of BDM led to estimates of the average substep durations of 12.7 ± 2 ms and 16.4 ± 4 ms respectively (±95% CI). Varying the ATP concentration from 1 to 20 μM did not systematically affect the duration of the high-wobble substep in the absence (black) or presence (gray, Fig. 4 B) of BDM. These effects of BDM are smaller than expected from results obtained by Dunn and Spudich (14) in gold-particle-labeled myosin V, possibly because our molecules contained only CaM in the lever arm, whereas the molecules used in the previous study also included essential light chains, which possibly affected the detached head rebinding rate.
Minor angle changes
In some molecules (52 events in 42 molecules), we detected transient increases in wobble without corresponding maintained tilting motions of the myosin. Instead, the orientations before and after the high-wobble substep were approximately the same (Fig. S6 D), consistent with the labeled head detaching from actin and rebinding actin without stepping (also known as a foot-stomp (10,19)). The average duration of these events was 33 ms (n = 22 events in 16 molecules). Nine of these molecules showed this type of nonproductive high-wobble period in the presumptive trailing head (β < 50°) and seven showed it in the leading head (β > 50°). Neither the ATP concentration nor the presence of BDM appeared to influence the occurrence of these nonstep increases in wobble.
Smaller-magnitude angle changes also occurred between the major steps. For the molecule in Fig. 2, the lever arm appeared to undergo additional minor tilting at change point 7 on the leading head and at points 4 and 9–12 on the trailing head. The angular displacements (ζ) during these motions were usually <10° but could be up to 45°. Change points 14–17 in Fig. 2 during the high-wobble period before photobleaching represent statistically significant small changes in intensity, but they lack well-defined, quantifiable orientations.
The numbers of minor events detected by the change-point algorithm before and after high-wobble substeps were similar for the trailing and leading heads (solid and open symbols in Fig. 5 A). For both the leading and trailing heads, the majority of steps occurred with no additional angle changes and the overall number of events averaged ∼1.1 small change points per major step. Neither the magnitude (Fig. 5 B) nor the kinetics (data not shown) of these nonstep angle changes differed appreciably between the leading- and trailing-head states. By manually reviewing these recordings, as well as those of molecules in which no high-wobble states were detected during stepping (and thus the leading and trailing states were less clear), we confirmed that few molecules exhibited small, lasting angle changes preceding their steps. Thus, at the ATP concentrations used here (1–20 μM), the lever orientation is predominantly constant prior to major rotations of the lever arm, consistent with mainly straight-leg stepping (19).
Figure 5.
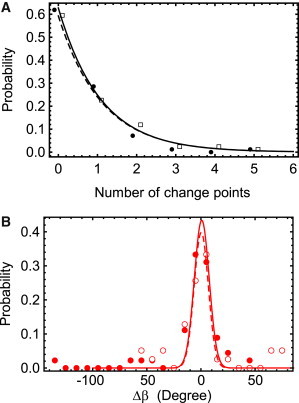
Distributions of nonstep angle changes (n = 73) obtained using only molecules with steps exhibiting a high wobble state so that the dwells could be unambiguously assigned to the trailing and leading heads. (A) The distributions of the number of additional angle changes on the trailing (n = 59, solid line) and leading (n = 69, dashed line) heads are similar. (B) Histograms of the change in β angle during the dwell before (solid symbols) and after (open symbols) large wobble events. Similar histograms for Δα are included in Fig. S8.
Photophysical effects
Approximately 30% of the recordings contained an extended period of high probe wobble immediately before photobleaching (for example, the last ∼0.5 s of Fig. 2 D and Fig. S6). Previous studies reported occasional increases in fluorescence intensity or spectral shifting prior to photobleaching (30,41). We also observed an increase in fluorescence before photobleaching in some traces; however, the increase in probe wobble was much more common. Photobleaching times in this population were not different from the normal traces, and the likelihood of obtaining a period of high wobble before photobleaching did not depend on laser intensity. The phenomenon is probably due to a photophysical artifact of laser light damage to the probe or probe-labeled lever arm, based on several observations. Processes that adversely affect actin binding or ATPase activity would also be expected to produce a population of recordings with a prolonged terminal state of constant angle, but molecules without such sustained high-wobble episodes tilted back and forth until they photobleached. Consequently, we speculate that one of the bifunctional linkers in the probe may break under the high laser illumination, resulting in a singly attached, highly mobile probe that does not discriminate between attached actomyosin states and detached ones.
Short bursts of multiple, very rapid polarization change points were also detected infrequently in a subset of molecules (Fig. S6). The timescale of these motions (∼18 ms) was much faster than myosin stepping and had no bias toward the leading or trailing head. There were more of these rapid events at higher laser powers, suggesting that they may be another photophysical effect.
Discussion
High-speed polTIRF detects periods of brief detachment during myosin V stepping
In previous polTIRF studies of myosin V (26,27,39), the time resolution was limited to 20 ms and the range of discernible angles was often restricted to 1/8 of a sphere due to symmetries that resulted from confining the input laser polarizations to three orthogonal planes (12,28). Here, laser polarizations aligned in four directions (−45°, 0°, 45°, and 90°) relative to the plane defined by the input and reflected laser beams (the scattering plane) in both beams decreased the ambiguity fourfold, resulting in a hemisphere of unambiguously distinguishable orientations (26,27,29) (Fig. S1). This scope of angles is the maximum limit imposed by the intrinsic dipole symmetry of the probe.
Statistically valid change points in the photon collection rates identified times of structural changes, such as tilting and increased wobble. An important assumption of the change-point approach is that the molecule resides in discrete structural states and switches abruptly between these states, resulting in sudden changes in the polarized fluorescence photon count rates. This feature implies that the rates are constant between change points, and that the total number of counts divided by the interval between change points is a good estimate of the fluorescence intensity. The 16 intensities were fitted numerically by a maximum-likelihood model of fluorescence that depended on the dipole orientation (β, α), and wobble, δ (12,19,27–29). Wobble quantifies the depolarization of the fluorescence signal on timescales slower than the fluorescent lifetime (∼4 ns) but faster than the measurement timescale (∼0.1 ms). The expected microsecond rotational diffusion timescale of a free myosin V head is thus quantified, for the first time to our knowledge, by this δ parameter.
Increases in δ during a switch in lever-arm orientation were measured in many molecules (Fig. S5). The timing of the structural changes derives purely from the fluorescence intensities independently of any binning of data or of any model predicting the relative intensities. The brief increases in δ are consistent with a highly mobile state of the trailing head after it detaches from actin during the thermal search for its next actin-binding site. This assignment is supported by several of our results. First, the results of fitting the dipole model to the 8 ms binned data confirm that the high-wobble substeps occur between steps (Fig. 3 A) and are characterized by randomly distributed β and α, consistent with microsecond tumbling of the detached head. Second, in >90% of recordings where more than one high-wobble substate was detected, they were observed on alternating steps, as expected (for example, Fig. 2 E and Fig. S5). Third, the average time for rebinding was independent of ATP concentration (Fig. 4 B). Lastly, the ∼13 ms duration of these short intervals (Fig. 4 A) agrees well with the period of high positional fluctuations of gold-nanoparticle-labeled myosin V during steps along actin (14).
Experimental evidence for the single-head-attached myosin intermediate was obtained by optical trapping (17), fluorescence microscopy (15,16), and high-speed AFM (10), but measurement of the rebinding rate in these studies was complicated by a bead, a large reporter molecule, or surface interactions. The mean waiting time for rebinding of the free head to actin was theoretically estimated to be 0.1–1 ms from first passage time calculations (7), the processive run length (42), and Brownian dynamics simulations (43). Both BR-CaM-labeled myosin V in the current work (Fig. 4) and single-particle tracking of 40 nm gold-particle-labeled CaM (14) indicated rebinding times of 10–15 ms, even though the 40 nm gold particle probably slowed diffusion of the head (the presence of a single rhodamine fluorophore (∼ 600 Da) is not expected to have a significant influence on the dynamics). The slower binding compared with the expectation from diffusion-limited models strongly suggests that a structural or biochemical step, such as reversal of the working stroke (44,45), weak-to-strong actomyosin binding (11), or Pi release (13), and not Brownian dynamics, limits the rebinding rate. The increase in probe wobble during a step supports models of hand-over-hand motion (8) with high rotational mobility of the head as it steps from the trailing to the leading position. Models in which the head remains in close proximity to the actin filament and skips or slides along the filament while reaching for the next strongly bound actin subunit (47) are thus rendered less likely.
The polarization switching devices (Pockels cells), photon detectors, and single-photon counting circuit are all fast electronic elements with approximately microsecond or faster response times. Consequently, the factor that limits the time resolution is the number of detected photons from the single probe (here ∼50 photons/ms). The periods of increased wobble during stepping of the labeled head last ∼10–20 ms, leading to ∼500–1000 total photons detected on average from these intervals. These values are just above the threshold required to detect change points with 95% confidence, which probably accounts for fewer high-wobble substeps being detected (14%, compared with the expected 50% of all steps). The excitation intensity is not near saturation of the absorption dipole, so higher excitation laser powers would increase the numbers of detected photons per time interval, but at the expense of shorter recording times before photobleaching. Of the ∼1000 total molecules recorded at the laser intensities used, ∼40% photobleached before a step occurred, which limited the practical range of input intensities.
Other factors may result in undercounting the expected number of substeps. First, short-lived events that contain enough photons to trigger detection of a change point but not enough to be reliably fit by the dipole model can be missed because the fitting typically underestimates the wobble of low-photon states. Also, the number of stepping events during a recording might be overestimated because at low ATP and ADP concentrations, some large rotational motions are not associated with stepping (10,19).
Distributions of orientation and wobble during myosin V stepping
The distributions of probe orientations with each step were characterized by two peaks in β at ∼20° and ∼85° (Fig. 2 A and Fig. S4), which result in lever-arm orientations of 60° and 120° for the leading and trailing configurations when the local orientation of the BR probe on the lever arm is taken into account (12,27). In previous experiments in which both position and orientation were measured (48), the smaller magnitude peak in βprobe was determined to be the trailing head, in agreement with the relative orientation between the probe axis and the myosin lever arm (27,49). In the high-speed polTIRF experiments described here, this assignment of angles to the two positions is additionally supported by the observation that the high-wobble search period is observed when βprobe switches from smaller to larger angle (Figs. 2 and 3 A, and Fig. S5), thereby confirming the trailing to leading positions, respectively, as suggested by the earlier studies.
The fairly uniform α distributions (Fig. 2 B and Fig. S4 B) are consistent with myosins binding at random azimuthal angles around the actin filament. The magnitude of the power stroke is estimated from angle changes of the stepping head with Δβ, Δα, and ζ equal to −68° ± 7°, −2.4° ± 4°, 83° ± 4°, respectively (±SE). The small magnitude of Δα, leading to similar magnitudes of Δβ and ζ, confirms that the power stroke is aligned closely along the actin filament axis.
Minor angle changes
Previous single-molecule work on myosin V using optical traps (7,17,18), electron micrographs (20,21), fluorescence (19), and atomic force microscopy (10) suggested that in addition to the high-wobble search period during an active step, there may be mechanistically important substeps before or after a step. The base of the lever arm in the leading head might tilt forward while the remainder of the lever is restrained by its attachment to the rear lever arm, causing a kink to form the so-called telemark configuration (19–21,50). In our data, nonstep change points were sometimes detected during the intervals between steps, but there was no bias toward leading or trailing heads in either the number of events (Fig. 5 A) or their magnitude (Fig. 5 B). The occasional larger-magnitude angle changes that were detected may be consistent with an alternate reaction pathway (51). In our experiments, the most common configuration observed while the molecule waited for ATP binding was one in which both heads were bound to actin and both lever arms were relatively straight. Upon ATP binding, the stepping motion normally was rapid and proceeded without a preceding tilting of either lever arm. These results apply to the specific conditions optimized for detection of the high-wobble state, i.e., a relatively narrow range of ATP concentrations, no added ADP, and a relatively brief recording interval limited by photobleaching. High-speed AFM images of myosin V also showed mainly straight lever arms at micromolar ATP in the absence of ADP, but the leading lever arms became kinked more often at higher ADP concentrations (10). polTIRF measurements over a broader range of nucleotide concentrations might identify a role for such auxiliary rotations in the myosin molecule.
Conclusions
An upgraded polTIRF microscope was used to investigate myosin V motility with high time resolution. Brief ∼10–15 ms substeps, characterized by highly disordered lever-arm orientations, were detected during stepping events and likely correspond to the detached head searching by random fluctuations for its next actin-binding site. The majority of myosin V molecules contained straight lever arms in both leading and trailing positions (i.e., not bent or kinked), and additional rotations of the lever arm were uncommon on both the leading and trailing heads. Since biological macromolecules generally function in an environment characterized by significant thermal fluctuations, other important enzymatic steps and short-lived intermediates might be detected by rotational flexibility that can be measured at the single-molecule level using high-speed polTIRF microscopy.
Acknowledgments
We thank Drs. Yujie Sun and Martin Pring for helpful discussions, Xiaonan Cui for help in labeling BR-CaM, and H. Lee Sweeney for providing the myosin V.
This work was supported by National Institutes of Health grant R01 GM086352 (Y.E.G), and National Science Foundation grants DGE-02-21664 (J.F.B.), EF-0928048 (P.C.N.), and DMR08-32802 (P.C.N. and Y.E.G.). The authors thank the Aspen Center for Physics, National Science Foundation Grant No. 1066293, for hospitality during various phases of this work.
Supporting Material
References
- 1.Reck-Peterson S.L., Provance D.W., Jr., Mercer J.A. Class V myosins. Biochim. Biophys. Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- 2.Sweeney H.L., Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y., Goldman Y.E. Lever-arm mechanics of processive myosins. Biophys. J. 2011;101:1–11. doi: 10.1016/j.bpj.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta A.D., Rock R.S., Cheney R.E. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 5.Rief M., Rock R.S., Spudich J.A. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakamoto T., Amitani I., Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 7.Veigel C., Wang F., Molloy J.E. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 8.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder H.W., 3rd, Mitchell C., Goldman Y.E. Motor number controls cargo switching at actin-microtubule intersections in vitro. Curr. Biol. 2010;20:687–696. doi: 10.1016/j.cub.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kodera N., Yamamoto D., Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 11.De La Cruz E.M., Wells A.L., Sweeney H.L. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forkey J.N., Quinlan M.E., Goldman Y.E. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld S.S., Sweeney H.L. A model of myosin V processivity. J. Biol. Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 14.Dunn A.R., Spudich J.A. Dynamics of the unbound head during myosin V processive translocation. Nat. Struct. Mol. Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 15.Shiroguchi K., Kinosita K., Jr. Myosin V walks by lever action and Brownian motion. Science. 2007;316:1208–1212. doi: 10.1126/science.1140468. [DOI] [PubMed] [Google Scholar]
- 16.Komori Y., Iwane A.H., Yanagida T. Myosin-V makes two Brownian 90 degrees rotations per 36-nm step. Nat. Struct. Mol. Biol. 2007;14:968–973. doi: 10.1038/nsmb1298. [DOI] [PubMed] [Google Scholar]
- 17.Cappello G., Pierobon P., Prost J. Myosin V stepping mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:15328–15333. doi: 10.1073/pnas.0706653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uemura S., Higuchi H., Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 19.Syed S., Snyder G.E., Goldman Y.E. Adaptability of myosin V studied by simultaneous detection of position and orientation. EMBO J. 2006;25:1795–1803. doi: 10.1038/sj.emboj.7601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker M.L., Burgess S.A., Knight P.J. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 21.Oke O.A., Burgess S.A., Trinick J. Influence of lever structure on myosin 5a walking. Proc. Natl. Acad. Sci. USA. 2010;107:2509–2514. doi: 10.1073/pnas.0906907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forkey J.N., Quinlan M.E., Goldman Y.E. Protein structural dynamics by single-molecule fluorescence polarization. Prog. Biophys. Mol. Biol. 2000;74:1–35. doi: 10.1016/s0079-6107(00)00015-8. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg S.A., Quinlan M.E., Goldman Y.E. Rotational motions of macro-molecules by single-molecule fluorescence microscopy. Acc. Chem. Res. 2005;38:583–593. doi: 10.1021/ar040137k. [DOI] [PubMed] [Google Scholar]
- 24.Joo C., Balci H., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 25.Irving M., St Claire Allen T., Goldman Y.E. Tilting of the light-chain region of myosin during step length changes and active force generation in skeletal muscle. Nature. 1995;375:688–691. doi: 10.1038/375688a0. [DOI] [PubMed] [Google Scholar]
- 26.Beausang J.F., Schroeder H.W., 3rd, Goldman Y.E. Twirling of actin by myosins II and V observed via polarized TIRF in a modified gliding assay. Biophys. J. 2008;95:5820–5831. doi: 10.1529/biophysj.108.140319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis J.H., Beausang J.F., Goldman Y.E. The azimuthal path of myosin V and its dependence on lever-arm length. J. Gen. Physiol. 2012;139:101–120. doi: 10.1085/jgp.201110715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forkey J.N., Quinlan M.E., Goldman Y.E. Measurement of single macromolecule orientation by total internal reflection fluorescence polarization microscopy. Biophys. J. 2005;89:1261–1271. doi: 10.1529/biophysj.104.053470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beausang J.F., Sun Y., Goldman Y.E. Orientation and rotational motions of single molecules by polarized total internal reflection fluorescence microscopy. In: Selvin P.R., Ha T., editors. Single Molecule Techniques. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2008. pp. 121–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan M.E., Forkey J.N., Goldman Y.E. Orientation of the myosin light chain region by single molecule total internal reflection fluorescence polarization microscopy. Biophys. J. 2005;89:1132–1142. doi: 10.1529/biophysj.104.053496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beausang J.F., Goldman Y.E., Nelson P.C. Changepoint analysis for single-molecule polarized total internal reflection fluorescence microscopy experiments. Methods Enzymol. 2011;487:431–463. doi: 10.1016/B978-0-12-381270-4.00015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell T.J., Morris C., Sweeney H.L. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. USA. 2002;99:14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putkey J.A., Slaughter G.R., Means A.R. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. J. Biol. Chem. 1985;260:4704–4712. [PubMed] [Google Scholar]
- 34.Corrie J.E., Craik J.S., Munasinghe V.R. A homobifunctional rhodamine for labeling proteins with defined orientations of a fluorophore. Bioconjug. Chem. 1998;9:160–167. doi: 10.1021/bc970174e. [DOI] [PubMed] [Google Scholar]
- 35.Pardee J.D., Spudich J.A. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 36.Beausang, J. F. 2010. Single molecule investigations of DNA looping using the tethered particle method and translocation by acto-myosin using polarized total internal reflection fluorescence microscopy. PhD dissertation, University of Pennsylvania, Philadelphia, PA.
- 37.Veigel C., Bartoo M.L., Molloy J.E. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys. J. 1998;75:1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watkins L.P., Yang H. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y., Schroeder H.W., 3rd, Goldman Y.E. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol. Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reifenberger J.G., Toprak E., Selvin P.R. Myosin VI undergoes a 180 degrees power stroke implying an uncoupling of the front lever arm. Proc. Natl. Acad. Sci. USA. 2009;106:18255–18260. doi: 10.1073/pnas.0900005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wazawa T., Ishii Y., Yanagida T. Spectral fluctuation of a single fluorophore conjugated to a protein molecule. Biophys. J. 2000;78:1561–1569. doi: 10.1016/S0006-3495(00)76708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith D.A. How processive is the myosin-V motor? J. Muscle Res. Cell Motil. 2004;25:215–217. doi: 10.1023/b:jure.0000038171.22680.2e. [DOI] [PubMed] [Google Scholar]
- 43.Craig E.M., Linke H. Mechanochemical model for myosin V. Proc. Natl. Acad. Sci. USA. 2009;106:18261–18266. doi: 10.1073/pnas.0908192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinosita K., Jr., Shiroguchi K., Itoh H. On the walking mechanism of linear molecular motors. Adv. Exp. Med. Biol. 2007;592:369–384. doi: 10.1007/978-4-431-38453-3_31. [DOI] [PubMed] [Google Scholar]
- 45.Shiroguchi K., Chin H.F., Kinosita K., Jr. Direct observation of the myosin Va recovery stroke that contributes to unidirectional stepping along actin. PLoS Biol. 2011;9:e1001031. doi: 10.1371/journal.pbio.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reference deleted in proof.
- 47.Okada T., Tanaka H., Yanagida T. The diffusive search mechanism of processive myosin class-V motor involves directional steps along actin subunits. Biochem. Biophys. Res. Commun. 2007;354:379–384. doi: 10.1016/j.bbrc.2006.12.200. [DOI] [PubMed] [Google Scholar]
- 48.Toprak E., Enderlein J., Selvin P.R. Defocused orientation and position imaging (DOPI) of myosin V. Proc. Natl. Acad. Sci. USA. 2006;103:6495–6499. doi: 10.1073/pnas.0507134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker D., Bryant Z., Delp S.L. Coarse-grained structural modeling of molecular motors using multibody dynamics. Cell Mol. Bioeng. 2009;2:366–374. doi: 10.1007/s12195-009-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder G.E., Sakamoto T., Selvin P.R. Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand-over-hand via telemark configuration. Biophys. J. 2004;87:1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kad N.M., Trybus K.M., Warshaw D.M. Load and Pi control flux through the branched kinetic cycle of myosin V. J. Biol. Chem. 2008;283:17477–17484. doi: 10.1074/jbc.M800539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


