Abstract
The Total Integrated Archive of short-Read and Array (TIARA; http://tiara.gmi.ac.kr) database stores and integrates human genome data generated from multiple technologies including next-generation sequencing and high-resolution comparative genomic hybridization array. The TIARA genome browser is a powerful tool for the analysis of personal genomic information by exploring genomic variants such as SNPs, indels and structural variants simultaneously. As of September 2012, the TIARA database provides raw data and variant information for 13 sequenced whole genomes, 16 sequenced transcriptomes and 33 high resolution array assays. Sequencing reads are available at a depth of ∼30× for whole genomes and 50× for transcriptomes. Information on genomic variants includes a total of ∼9.56 million SNPs, 23 025 of which are non-synonymous SNPs, and ∼1.19 million indels. In this update, by adding high coverage sequencing of additional human individuals, the TIARA genome database now provides an extensive record of rare variants in humans. Following TIARA’s fundamentally integrative approach, new transcriptome sequencing data are matched with whole-genome sequencing data in the genome browser. Users can here observe, for example, the expression levels of human genes with allele-specific quantification. Improvements to the TIARA genome browser include the intuitive display of new complex and large-scale data sets.
Introduction
Recently, next-generation sequencing technology has been used extensively in biological and clinical research, revealing information on a wide spectrum of human genomic variation, and generating a concomitantly tremendous amount of raw data. This increase in accumulated sequencing data is expected to improve the precision of human genome analysis, and widespread disease-specific and cancer genome sequencing contributes a great effort towards improved diagnosis and therapy. The Cancer Genome Atlas (TCGA) (1–3) and the International Cancer Genome Consortium (ICGC) (4) are performing genomic sequencing of various types of cancers and accumulating their own archiving systems (5). Public databases such as the Sequence Read Archive (SRA) (6, 7), database of Genotypes and Phenotypes (dbGaP) (8), Single Nucleotide Polymorphism Database (dbSNP) (9), Database of Genomic Variants archive (DGVa) (10) and the Catalog Of Somatic Mutations In Cancer (COSMIC) (11) contain both raw sequencing data as well as various types of genomic variants, which can affect human biological function. As the use of genome-wide sequencing increases, so also do the challenges of efficiently managing and retrieving these large-scale data structures. To deal with these challenges for data generated in sequencing projects at Genomic Medicine Institute of Seoul National University (GMI-SNU), we previously developed the Total Integrated Archive of short-Read and Array (TIARA; http://tiara.gmi.ac.kr) database with a focus on integrative browsing of heterogeneous complex data sets through the TIARA genome browser.
The integrative design of TIARA is motivated by several factors. Genomic variants play important roles in bringing about human complex diseases and various cancers. If genomic variants such as Single Nucleotide Polymorphisms (SNPs), short indels and Copy Number Variations (CNVs) can be studied simultaneously, this will help to discover important interactions and more precise etiological factors (12). Moreover, in our previous studies (13–15), we showed that more accurate analyses (i.e. absolute CNV calling) are feasible by using combined analyses, such as massively parallel sequencing with high-resolution comparative genomic hybridization (CGH) array (14, 15). Furthermore, analysis methods based on multiple genomes are essential to properly evaluate the function and meaning of personal genome variants.
In this article, we will set out the basic design of TIARA and introduce several updates to the database. These updates include migration of data to human genome reference NCBI Build 37.3 (hg19), adding functions to the control panel and integrating panels for the viewing of transcriptome sequencing data, including expression levels, variants and aligned reads. Recently, we reported discovery of common and functional rare variants through whole-genome sequencing of 13 human individuals and transcriptome sequencing of 16 at high depth of coverage (16). Investigation of genomic variants between whole-genome sequencing and transcriptome sequencing for matched samples revealed features such as gene-expression levels, allele-specific gene expression and transcriptional base modifications (TBMs) or RNA editing. These data were added to TIARA, allowing browsing of sequencing reads and genomic variants. Table 1 shows the samples that have been deposited in the TIARA database update. The browser facilitates comparison of the genome and transcriptome sequencing results for individual humans, as well as simultaneous and efficient viewing of genomic variants from other high-throughput genome technologies. We believe that this update to TIARA results in a sophisticated database containing complex genomic data structures, presented in a user-friendly browser that will facilitate investigation of ‘omics’ data by researchers worldwide.
Table 1.
The summary of samples deposited in TIARA database
| Legacy from TIARA 2011 | New in TIARA 2013 | |
|---|---|---|
| Whole genome sequencing (12 individuals) | AK1, AK2, AK4, AK6, NA10851 | AK3, AK5, AK7, AK9, AK14, AK20, AK55_Blood, AK55_Cancer* |
| Transcriptome sequencing (16 individuals) | - | AK3, AK4, AK5, AK6, AK7, AK14, AK20, AK_N1, AK_N2, AK_N5, AK_N6, AK_N7, AK_N9, AK_N14, AK_15, AK55_Cancer* |
| High-resolution CGH array (33 individuals) | AK1, AK2, AK4, AK6, AK8, AK10, AK12, AK14, AK16, AK18, AK20, NA18526, NA18537, NA18542, NA18547, NA18552, NA18564, NA18566, NA18570, NA18582, NA18592, NA18942, NA18947, NA18949, NA18951, NA18968, NA18969, NA18972, NA18973, NA18997, NA18999, NA12878, NA19240 | - |
*The sequencing data of AK55 including FASTQ, alignment results and SNPs sare provided only on the anonymous FTP server.
Materials and methods
Whole genome and transcriptome deep sequencing
TIARA contains deposits of sequencing reads for 13 whole genomes and 16 transcriptomes at high depth of coverage from high-throughput sequencing machines including the Illumina Genome Analyzer and AB SOLiD (Supplementary Figure S1). This will provide much more information on rare variants and population characteristics than the five individuals designated AK1, AK2, AK4, AK6 and NA10851, which were previously included in the database (13–19). In this upgrade of the TIARA genome database, the short read (36–151 bp) data originally in FASTQ format, alignment results and genomic variants from the newly included whole genome and transcriptome sequencing have been added. Supplementary Tables S1, S2 and S3 show the summary of sequencing data for individuals stored in TIARA.
Genome variants
The short reads generated by human genome and transcriptome sequencing were previously aligned on human genome reference NCBI Build 36.3 (hg18) using the Genomic Short-read Nucleotide Alignment Program (GSNAP) short-read alignment tool (20), and then human genome variants such as SNPs, short indels and Structural Variations (SVs) were detected and read depths (RDs) were calculated as described in our studies (13–16, 18, 19). We re-aligned those short reads onto human genome reference NCBI Build 37.3 (hg19) and detected genomic variants including SNPs and short indels by the same bioinformatics software pipeline. This allows the TIARA database to retrieve variants called on either hg18 or hg19 as selected by the user.
In addition, CGH array data were previously obtained through experiments using a designed high-resolution CGH array from Agilent Technologies whose probe sequences were based on human genome reference NCBI Build 36.3 (hg18), and CNVs called using the ADM2 algorithm were deposited in the TIARA genome database (14, 17, 21). To improve CNV research, we converted the genomic positions, which were available on human genome reference Build 37.3 (hg19) using a batch coordinate conversion tool provided by UCSC utilities (22) and added the converted positions and log2 ratios to TIARA.
Results
The architecture and development platform of the TIARA system have been retained in this update as described in our original publication (17). TIARA has three types of repositories: (i) a Lucene index file system, which contains genomic variants such as SNPs and short indels, read depths and log2 ratios; (ii) a MySQL database, which contains human reference genome sequences (hg18 and hg19), mapping information of short reads, RefSeq and Ensembl genes (23, 24), gene expression profiles and Asian specific CNV regions (14); and (iii) an anonymous file transfer protocol (FTP) archive, which contains raw files such as FASTQ format read sequences, alignment results and genomic variants in the general feature format. The user-friendly interface of the TIARA genome browser contains eight main components: Control Panel, RefSeq and Ensembl Genes, SNPs, Indels, Integrative Multi-Omics Display Window, Read Depth Display Window, CNV Regions and Log2 Ratio Display Window. The Integrative Multi-Omics Display Window has been implemented in this update to provide improved integrative analysis. Short-read windows now also display transcriptome sequencing data. The arrangement and function of other components are maintained as previously described.
Newly integrated viewing panels
In the new version of the TIARA genome browser, panels are provided to view newly added transcriptome sequencing data. These display windows are fully integrated with other technologies in the browser. The TIARA genome browser displays gene expression levels in Reads Per Kilobase of exon model per Million mapped reads (25), aligned reads supporting SNPs within genes and variants when the user selects RNA-Seq data. Direct comparison of transcriptome and whole genome sequence data for matched individuals allows analysis of allele-specific expression and the impact of variants on expression levels. Interestingly, the user can observe allele-specific expression by comparing the colours of SNPs in the genome and transcriptome sequencing windows (red for heterozygous, blue for homozygous). The TIARA database now contains transcriptome sequencing data for 16 individuals. Furthermore, the addition of whole-genome sequencing data for 10 Asian individuals provides a wealth of rare variants. These can be downloaded via FTP.
Advanced user interface functions
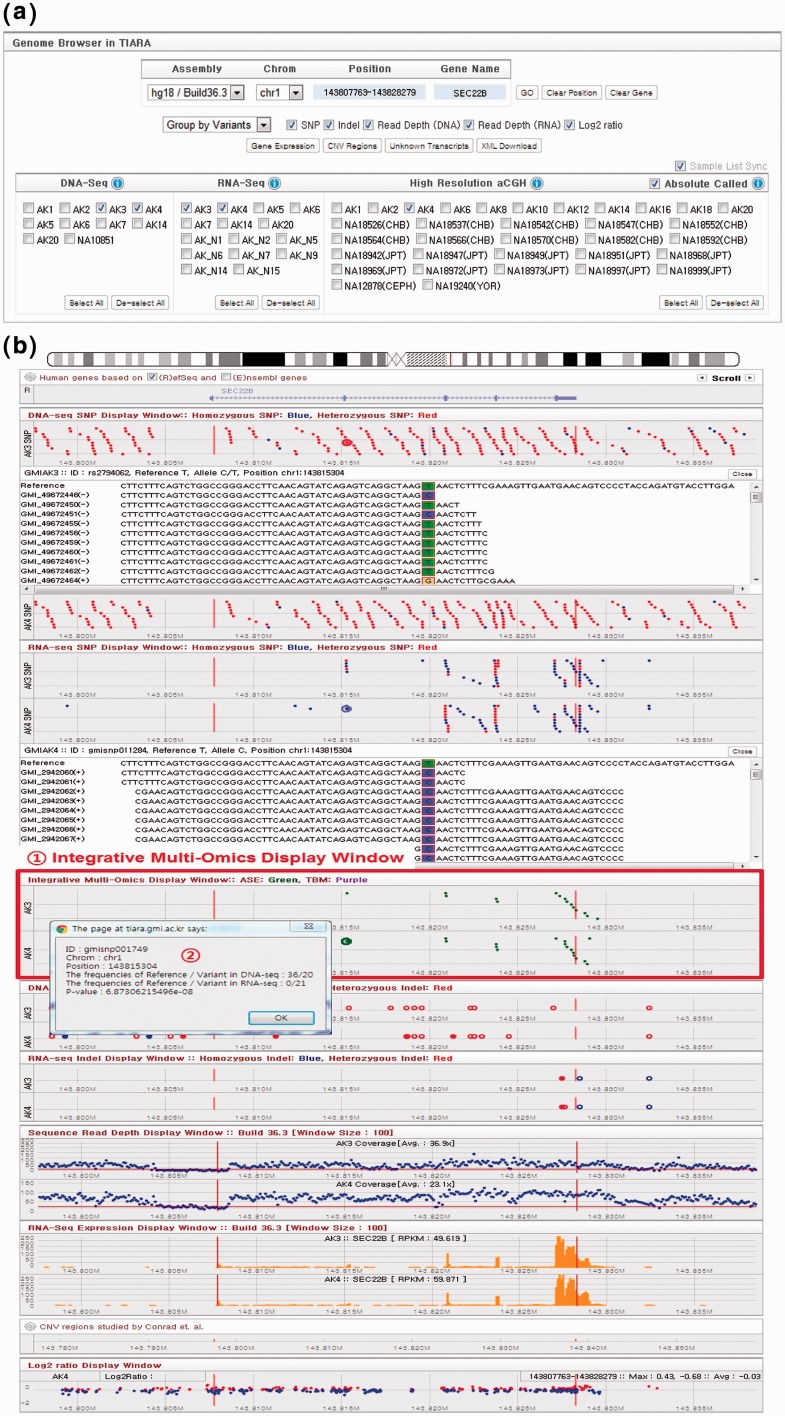
Full details on the Control Panel are provided in Figure 1a, Supplementary Information and the online manual. In particular, to handle the increase in technologies displayed, we have added a new option to group panels by variants or samples (Figure 1b and Supplementary Figure S2). The Integrative Multi-Omics Display Window is shown in (1) of Figure 1b. This window displays instances of allele-specific expression as points coloured green and TBMs as points coloured purple at corresponding genomic positions. For example, the genome browser is directed to the gene SEC22B (chromosome 1 at position 143 815 304 bp) in Figure 1b, where allele-specific expression has been observed. Users may click on one of the green dots representing an instance of allele-specific expression to receive information about the number of reads supporting the reference and variant in whole-genome sequencing and transcriptome sequencing and the statistical significance. This pop-up window is shown in part (2) of Figure 1b (Supplementary Information). This was obtained by clicking on the point shown as an enlarged green dot to the right of the pop-up. Moreover, access has also been provided to gene expression lists, common CNV regions and unknown transcripts, shown in Supplementary Figures S3–S5.
Figure 1.
TIARA genome browser. (a) The control panel of TIARA genome browser. (b) Arrangement of genomic query results according to the types of genomic variants such as SNP, indel, gene expression, allele-specific expression, TBMs, read depth and log2 ratio. The genome browser has been directed to gene SEC22B by entering it into the ‘Gene Name’ text box after selecting samples AK3 and AK4. One SNP from the DNA-Seq SNP display window (single enlarged red dot, second window) has been selected, yielding full read alignment details below, justifying the heterozygous SNP call. Interestingly, allele-specific expression can also be observed for this gene, as indicated by green dots in the Integrative Multi-Omics Display window. The pop-up window, which displays read counts for reference and variant alleles, was obtained by clicking one such point (enlarged green dot).
Discussion
The TIARA database provides access to genomic data from a wide range of technologies, with the fundamental principle of mutual integration and ease of viewing. To show whole-genome sequencing, transcriptome sequencing and CGH array data from the same individual simultaneously, we have upgraded the TIARA genome browser’s display functions. This will facilitate multi-omics and cross-technology analysis of human genome variants. For example, the impact of copy number variation and other genomic variants on the expressed transcriptome is an area that requires simultaneous comparison of multiple data sets. As part of our comprehensive recent studies into the human genome (13–16, 19), we performed sequencing of 13 whole genomes with average coverage over ∼26× and 16 transcriptomes using massively parallel sequencing. We also performed high-resolution CGH array experiments for 33 human samples. The raw data from these experiments have been deposited to the TIARA genome database, as well as variants such as SNPs, short indels and CNVs, detected from the data. At present, the TIARA genome database provides cancer genome sequencing data for one lung cancer patient on anonymous FTP. However, this is an area where a large number of sequencing experiments are being performed worldwide, including cancer genome sequencing of many lung cancer patients at GMI-SNU. As full data sets become available, these will be added to the TIARA database. As well as the familiar bioinformatics challenges of calling somatic mutations, display methods that allow efficient browsing of variants and simultaneous viewing of features such as structural variation and gene expression are important for cancer research. We believe that TIARA will be a useful tool for the human genome research community and will help cancer genome research to realize more precise and effective personalized medicine.
Supplementary Data
Supplementary data are available at Database Online.
Funding
This work was supported by the National Cancer Center Grant (grant # NCC-1210440 to D.H.) and by the Korean Ministry of Knowledge Economy (grant # 10037410 to J.-S.S.). Funding for open access charge: Korean Ministry of Knowledge Economy (10037410).
Conflict of interest. None declared.
Supplementary Material
References
- 1.The Cancer Genome Atlas Research. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodama Y, Shumway M, Leinonen R. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saccone SF, Quan J, Mehta G, et al. New tools and methods for direct programmatic access to the dbSNP relational database. Nucleic Acids Res. 2011;39:D901–D907. doi: 10.1093/nar/gkq1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 11.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 13.Kim JI, Ju YS, Park H, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H, Kim JI, Ju YS, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat. Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju YS, Hong D, Kim S, et al. Reference-unbiased copy number variant analysis using CGH microarrays. Nucleic Acids Res. 2010;38:e190. doi: 10.1093/nar/gkq730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju YS, Kim JI, Kim S, et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat. Genet. 2011;43:745–752. doi: 10.1038/ng.872. [DOI] [PubMed] [Google Scholar]
- 17.Hong D, Park SS, Ju YS, et al. TIARA: a database for accurate analysis of multiple personal genomes based on cross-technology. Nucleic Acids Res. 2011;39:D883–D888. doi: 10.1093/nar/gkq1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong D, Rhie A, Park SS, et al. FX: an RNA-Seq analysis tool on the cloud. Bioinformatics. 2012;28:721–723. doi: 10.1093/bioinformatics/bts023. [DOI] [PubMed] [Google Scholar]
- 19.Ju YS, Lee WC, Shin JY, et al. Fusion of KIF5B and RET transforming gene in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipson D, Aumann Y, Ben-Dor A, et al. Efficient calculation of interval scores for DNA copy number data analysis. J. Comput. Biol. 2006;13:215–228. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- 22.Dreszer TR, Karolchik D, Zweig AS, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu F, Kent WJ, Clawson H, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 24.Flicek P, Amode MR, Barrell D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.