Abstract
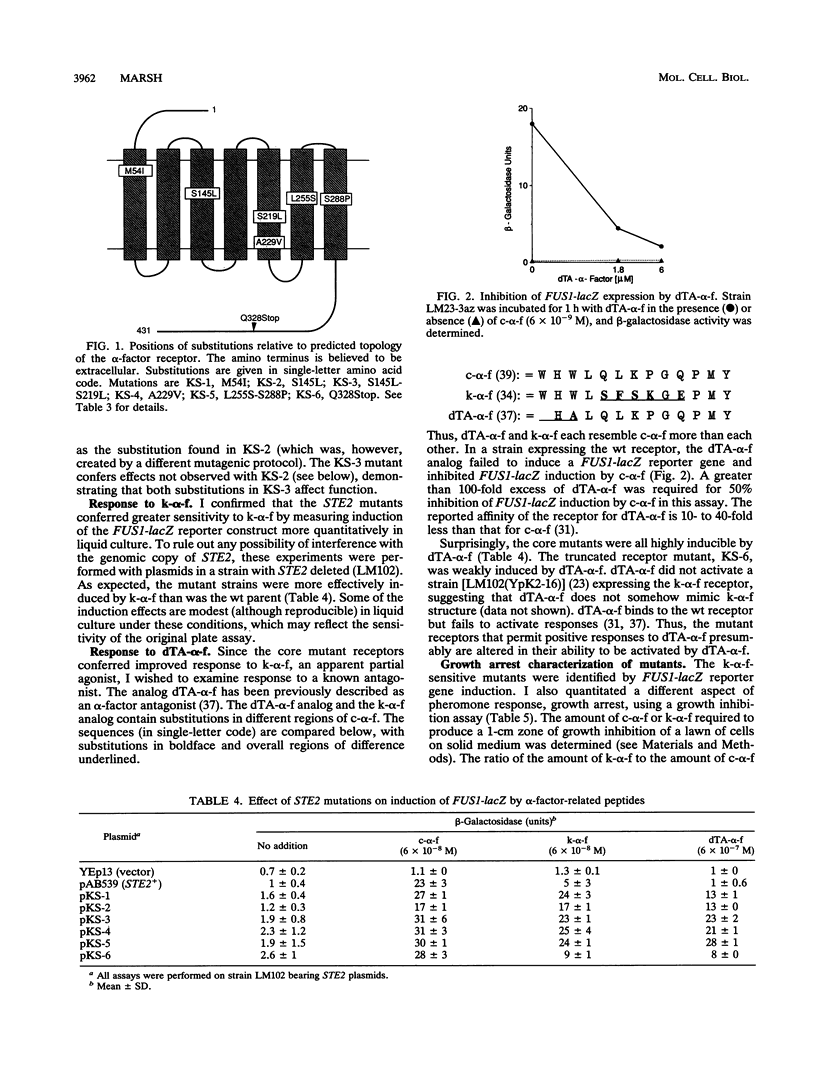
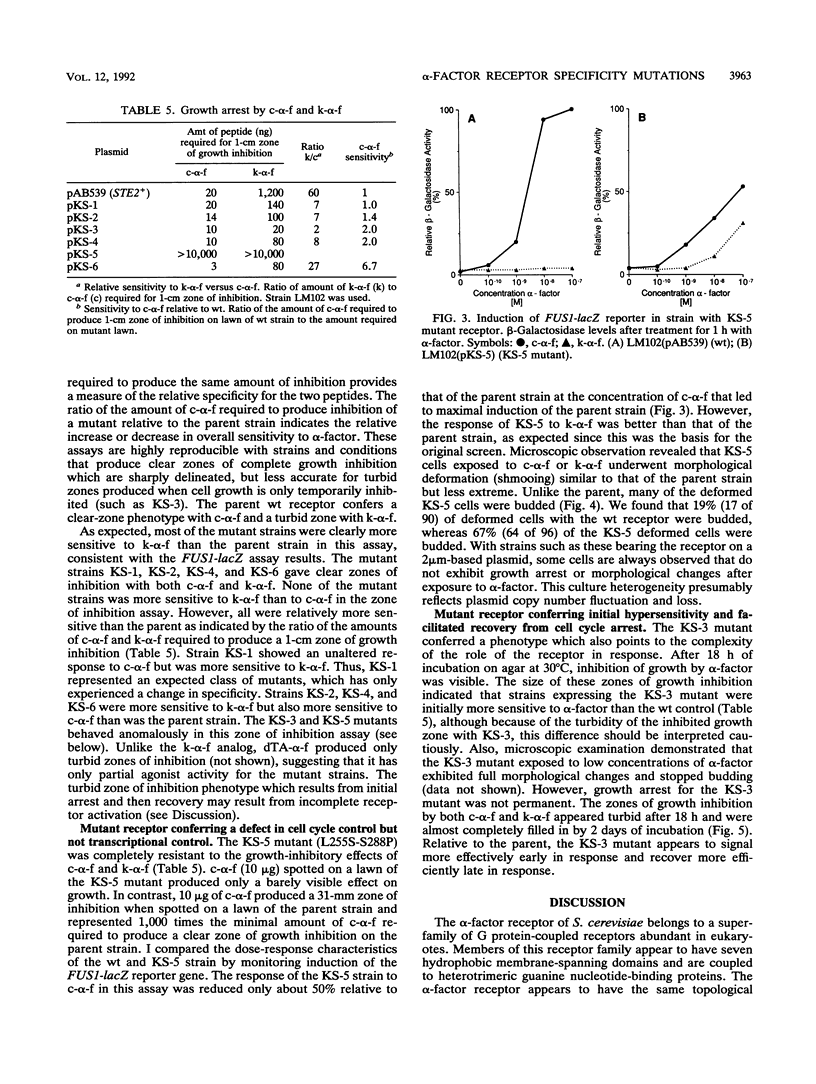
Mutations in the Saccharomyces cerevisiae alpha-factor receptor that lead to improved response to Saccharomyces kluyveri alpha-factor were identified and sequenced. Mutants were isolated from cells bearing randomly mutagenized receptor gene (STE2) plasmids by an in vivo screen. Five mutations lead to substitutions in hydrophobic segments in the core of the receptor (M54I, S145L, S145L-S219L, A229V, L255S-S288P). Remarkably, strains expressing these mutant receptors exhibited positive pheromone responses to desTrp1,Ala3-alpha-factor, an analog that normally blocks these responses. The M54I mutation appeared to affect only ligand specificity. The other mutations conferred additional effects on signaling or recovery. Two mutants were more sensitive to alpha-factor than wild type (S145L, A229V). One mutant was more sensitive to alpha-factor-induced cell cycle arrest initially, but then recovered more efficiently (S145L-S219L). One mutant (L255S-S288P) conferred positive pheromone responses to alpha-factor as assayed by FUS1-lacZ reporter induction, but did not display growth arrest. The hydrophobic receptor core thus appears to control activation by some ligands and to play roles in aspects of signal transduction and recovery.
Full text
PDF




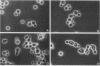
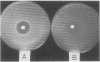
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burkholder A. C., Hartwell L. H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985 Dec 9;13(23):8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright C. P., Tipper D. J. In vivo topological analysis of Ste2, a yeast plasma membrane protein, by using beta-lactamase gene fusions. Mol Cell Biol. 1991 May;11(5):2620–2628. doi: 10.1128/mcb.11.5.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990 Nov 30;63(5):999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Cross F., Hartwell L. H., Jackson C., Konopka J. B. Conjugation in Saccharomyces cerevisiae. Annu Rev Cell Biol. 1988;4:429–457. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- Dietzel C., Kurjan J. The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell. 1987 Sep 25;50(7):1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Sigal I. S., Rands E., Register R. B., Candelore M. R., Blake A. D., Strader C. D. Ligand binding to the beta-adrenergic receptor involves its rhodopsin-like core. Nature. 1987 Mar 5;326(6108):73–77. doi: 10.1038/326073a0. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J., Caron M. G., Lefkowitz R. J. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- Egel-Mitani M., Hansen M. T. Nucleotide sequence of the gene encoding the Saccharomyces kluyveri alpha mating pheromone. Nucleic Acids Res. 1987 Aug 11;15(15):6303–6303. doi: 10.1093/nar/15.15.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Grisafi P. L., Fink G. R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990 Feb 23;60(4):649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- Findlay J. B., Pappin D. J. The opsin family of proteins. Biochem J. 1986 Sep 15;238(3):625–642. doi: 10.1042/bj2380625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig A., Holly J., Saari G., MacKay V. L. Multiple regulation of STE2, a mating-type-specific gene of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2106–2114. doi: 10.1128/mcb.6.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Konopka J. B., Hartwell L. H. S. cerevisiae alpha pheromone receptors activate a novel signal transduction pathway for mating partner discrimination. Cell. 1991 Oct 18;67(2):389–402. doi: 10.1016/0092-8674(91)90190-a. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Burkholder A. C., Hartwell L. H. Binding of alpha-factor pheromone to yeast a cells: chemical and genetic evidence for an alpha-factor receptor. Cell. 1983 Dec;35(2 Pt 1):521–529. doi: 10.1016/0092-8674(83)90186-1. [DOI] [PubMed] [Google Scholar]
- Konopka J. B., Jenness D. D. Genetic fine-structural analysis of the Saccharomyces cerevisiae alpha-pheromone receptor. Cell Regul. 1991 Jun;2(6):439–452. doi: 10.1091/mbc.2.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka J. B., Jenness D. D., Hartwell L. H. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988 Aug 26;54(5):609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Kurjan J., Hirsch J. P., Dietzel C. Mutations in the guanine nucleotide-binding domains of a yeast G alpha protein confer a constitutive or uninducible state to the pheromone response pathway. Genes Dev. 1991 Mar;5(3):475–483. doi: 10.1101/gad.5.3.475. [DOI] [PubMed] [Google Scholar]
- Marsh L., Herskowitz I. STE2 protein of Saccharomyces kluyveri is a member of the rhodopsin/beta-adrenergic receptor family and is responsible for recognition of the peptide ligand alpha factor. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3855–3859. doi: 10.1073/pnas.85.11.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L., Neiman A. M., Herskowitz I. Signal transduction during pheromone response in yeast. Annu Rev Cell Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- McCullough J., Herskowitz I. Mating pheromones of Saccharomyces kluyveri: pheromone interactions between Saccharomyces kluyveri and Saccharomyces cerevisiae. J Bacteriol. 1979 Apr;138(1):146–154. doi: 10.1128/jb.138.1.146-154.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I., Arai K., Matsumoto K. GPA1Val-50 mutation in the mating-factor signaling pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jun;9(6):2289–2297. doi: 10.1128/mcb.9.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I., Nakafuku M., Nakayama N., Brenner C., Miyajima A., Kaibuchi K., Arai K., Kaziro Y., Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987 Sep 25;50(7):1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Naider F., Becker J. M. Structure-activity relationships of the yeast alpha-factor. CRC Crit Rev Biochem. 1986;21(3):225–248. doi: 10.3109/10409238609113612. [DOI] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S. K., Naider F., Becker J. M. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem. 1988 Nov 25;263(33):17333–17341. [PubMed] [Google Scholar]
- Reneke J. E., Blumer K. J., Courchesne W. E., Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988 Oct 21;55(2):221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenbagamurthi P., Baffi R., Khan S. A., Lipke P., Pousman C., Becker J. M., Naider F. Structure-activity relationships in the dodecapeptide alpha factor of Saccharomyces cerevisiae. Biochemistry. 1983 Mar 1;22(5):1298–1304. doi: 10.1021/bi00274a047. [DOI] [PubMed] [Google Scholar]
- Stone D. E., Reed S. I. G protein mutations that alter the pheromone response in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4439–4446. doi: 10.1128/mcb.10.9.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader C. D., Candelore M. R., Hill W. S., Dixon R. A., Sigal I. S. A single amino acid substitution in the beta-adrenergic receptor promotes partial agonist activity from antagonists. J Biol Chem. 1989 Oct 5;264(28):16470–16477. [PubMed] [Google Scholar]
- Strader C. D., Sigal I. S., Register R. B., Candelore M. R., Rands E., Dixon R. A. Identification of residues required for ligand binding to the beta-adrenergic receptor. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Hougan L., Dignard D., Thomas D. Y., Bell L., Saari G. C., Grant F. J., O'Hara P., MacKay V. L. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989 Feb 10;56(3):467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]