Abstract
Purpose
Hypospadias is a common congenital malformation of the male external genitalia. Association studies for single nucleotide polymorphisms (SNPs) in genes encoding steroid-5-alpha-reductase (SRD5A2), estrogen receptors 1 (ESR1) and 2 (ESR2), and activating transcription factor 3 (ATF3) have been equivocal. The aim of this study was to examine whether non-replication of findings for four SNPs in these genes could be due to interaction with environmental exposures.
Materials and Methods
We genotyped 712 Dutch hypospadias case-parent triads for the four SNPs, used questionnaire information to determine exposures, and performed association tests using the log-linear approach. We studied gene-environment interactions for the four SNPs with exposure to estrogens, cytokines or cigarette smoke, multiple pregnancy, being born small for gestational age, and maternal hypertension or preeclampsia, high BMI, or primiparity. In addition, the presence of maternal genetic and parent-of-origin effects was tested.
Results
Gene-environment interactions were identified for rs523349 in SRD5A2 with estrogen exposure and maternal hypertension or preeclampsia, as well as for rs11119982 in ATF3 with exposure to cytokines. Both SNPs only seemed to influence hypospadias risk in exposed cases. For rs6932902 in ESR1, only maternally derived alleles appeared to increase hypospadias risk in offspring.
Conclusions
This study shows that interactions between genetic and environmental factors may help to explain non-replication in genetic studies of hypospadias.
Keywords: Case-parent triad study, Gene-environment interaction, Genetic Association Studies, Hypospadias, Parent-of-origin effects
Introduction
Hypospadias is a congenital hypoplasia of the penis, resulting from developmental arrest of urethral fusion. This leads to displacement of the urethral opening along the ventral side of the penis. Hypospadias is one of the most common birth defects among boys, affecting 0.3-0.7% of newborn boys in Europe1. It shows familial clustering and segregation analyses suggest that the majority of cases have a multifactorial etiology2, involving both genes and environmental factors.
Some environmental factors have consistently been associated with hypospadias. Hypospadias occurs more often in children born small for gestational age (SGA), and in first, intracytoplasmic sperm injection (ICSI)-induced, or multiple pregnancies. In addition, maternal hypertension, preeclampsia, high body mass index (BMI), pre-existing diabetes, and use of anti-epileptic drugs increase hypospadias risk, as does maternal intra-uterine exposure to diethylstilbestrol (DES)1.
Genetic associations with hypospadias have also been reported, mainly for single nucleotide polymorphisms (SNPs) in endocrine-related genes, such as those encoding estrogen receptors 1 (ESR1)3 and 2 (ESR2)4, activating transcription factor 3 (ATF3)5, and steroid-5-alpha-reductase (SRD5A2)6.7. ATF3 is an estrogen-responsive gene showing upregulation in hypospadias8, while SRD5A2 encodes an enzyme that converts circulating testosterone in the genital tubercle to the more potent androgen dihydrotestosterone.
The numbers of samples analyzed in these genetic studies were relatively small, and most associations could not be replicated in a much larger study by our group9.
This lack of consistency might reflect differences in environmental exposures between populations9. Several reviews have called for studies simultaneously examining genes and environment in relation to hypospadias1,10, but so far, such studies have rarely been performed. Therefore, we set out to examine whether the lack of replication could be due to gene-environment interactions between the four SNPs described above and risk factors for hypospadias.
In addition to gene-environment interactions, other (epi)genetic mechanisms may be involved in the etiology of hypospadias. Maternal genotype may affect the intra-uterine environment, thus modulating hypospadias risk, and gene imprinting may cause the copy derived from one parent to be more fully expressed than the copy derived from the other parent11. Therefore, we also examined the maternal genotype and imprinting effects.
Materials and methods
Cases and parents
AGORA (Aetiologic research into Genetic and Occupational/environmental Risk factors for Anomalies in children) is a large data- and biobank at the Radboud University Nijmegen Medical Centre in the Netherlands, in which questionnaire data and DNA samples are collected from patients with congenital malformations or childhood cancer and their parents. For the current study, DNA was available from 796 hypospadias cases born between 1980 and 2008 and 1,422 parents. Medical records of all cases were reviewed to identify syndromic hypospadias cases, collect clinical characteristics, and obtain information about anatomical location of the urethral opening. The regional Committee on Research Involving Human Subjects approved the study protocol and all parents and children over 11 years of age gave written informed consent.
Environmental risk factor data
Questionnaires were sent to the parents of all patients, containing a variety of questions on health and lifestyle just before and during pregnancy, which were used to define environmental risk factors. Although exogenous exposure to estrogens is not a known risk factor for hypospadias1, we included it in the gene-environment interaction analyses, because SRD5A2, ESR1, and ESR2 are involved in endocrine processes and ATF3 is an estrogen-responsive gene. Exogenous exposure to estrogens was defined as continued use of oral contraceptives during early pregnancy or consumption of soy or linseed products, which contain high amounts of phytoestrogens12, at least once a week in the first 14 weeks after conception. Women with a hormonal coil implanted who became pregnant were excluded because of weak estrogen exposure. Women exposed to pesticides at work were also excluded because pesticides can have either estrogenic or anti-estrogenic effects.
In addition, we studied interactions with factors associated with hypospadias occurrence: SGA (defined as birth weight<10th percentile for that gestational age, using Dutch reference curves13), mothers with hypertension or preeclampsia, high BMI (defined as BMI>25 kg/m2), primiparity, and multiple pregnancy.
In most tissues, ATF3 mRNA can be induced by various stress signals, such as cytokines and chemicals from cigarette smoke14,15. Therefore, we also included these exposures in the gene-environment interaction analyses for ATF3. Because the placental barrier may be permeable to cytokines16,17 and chemicals from cigarette smoke18, we categorized cases whose mothers smoked at least one cigarette per day during some time in the first 14 weeks after conception as exposed to cigarette smoke, and cases whose mothers reported the presence of an infection and/or inflammation in this period as exposed to cytokines.
Genotyping
Blood was collected in EDTA containing tubes (n=1,405) or saliva using Oragene containers (n=687; DNA Genotek Inc., Ottawa, Canada). DNA extraction and genotyping was performed as described previously9.
Statistical analyses
We used the case-parent triad design. The most frequent homozygous genotypes in parents served as reference genotypes in the log-linear approach19 that was applied to assess genetic associations. Log-linear models were fitted without assumption of Hardy-Weinberg equilibrium (HWE). Information on families with one missing parental genotype was included in the analyses using the expectation-maximization algorithm20. Likelihood ratio tests (LRT), comparing full models including both maternal and offspring genotypes to reduced models including either maternal or offspring genotype only, were computed to determine the relevance of maternal and offspring genotypes for hypospadias risk. We also conducted these analyses separately for the groups of anterior, middle, and posterior hypospadias cases, because different risk factors may be responsible for the different phenotypes21,22. Although the case-parent triad design is robust to population-stratification when testing genetic effects, effects of environmental exposures cannot be estimated.
Parent-of-origin analyses were conducted in two steps. As an initial screening, we used the transmission asymmetry test (TAT)19. This approach provides insights into the data, but is invalid when maternal effects exist. Therefore, the parent-of-origin LRT (PO-LRT), was used to confirm the results23.
Interactions between environmental exposures and offspring genotypes were tested using log-linear models with the LRT comparing a full model including gene-environment interactions to a reduced model including only the offspring genotypic effect24. We used a dominant interaction parameter, assuming that the environmental factor affects carriers with one or two copies of the variant allele similarly. We did not correct the critical P-value for multiple testing, as we only tested a limited number of well-founded hypotheses. If the LRT indicated the presence of an interaction (PLRT<0.05), relative risks (RR) and 95% confidence intervals (95% CI) were calculated separately for the different strata of the exposure variable using the variance calculated with the LEM program, which takes into account missing genotypes25. All other analyses were performed using the SAS System for Windows, release 8.02 (SAS Institute, Cary, North Carolina).
Results
Of the 796 available hypospadias cases, 38 patients were excluded due to lack of parental DNA. To ensure independence, we excluded the youngest brother from 22 sib-pairs, while from three twin-pairs, one brother was excluded at random. We excluded 19 patients because of syndromic hypospadias, chromosome abnormalities, or a known cause of hypospadias. Finally, two triads were excluded because of Mendelian errors. The final data set consisted of 712 cases. For 668 cases, DNA of both parents was available, while for 44 cases, we only had DNA from one parent. Environmental data were missing for 70 families. The majority of cases were of European Caucasian descent (91%), and the remaining were of non-European (5%) or unknown descent (4%). Almost 60% of cases had an anterior hypospadias, while 20% and 13% had middle and posterior urethral openings, respectively. Table 1 shows the distribution of the environmental risk factors studied. Exogenous exposure to estrogens, multiple pregnancies and fetal exposure to cytokines were relatively rare (<10%), whereas the other factors were more common.
Table 1.
Distribution of environmental risk factors for hypospadias patients.
| yes | no | unknowna | ||||
|---|---|---|---|---|---|---|
| Exposure | n | (%) | n | (%) | n | (%) |
| Exogenous exposure to estrogens | 29 | (4%) | 580 | (81%) | 103 | (14%) |
| Use of oral contraceptives during pregnancy | 8 | (1%) | 626 | (88%) | 78 | (11%) |
| Consumption of soy products during pregnancy | 16 | (2%) | 620 | (87%) | 76 | (11%) |
| Consumption of linseed products during pregnancy | 7 | (1%) | 626 | (88%) | 79 | (11%) |
| Known hypospadias risk factors | ||||||
| Small for gestational age | 125 | (18%) | 505 | (71%) | 82 | (12%) |
| Hypertension or preeclampsia | 107 | (15%) | 533 | (75%) | 72 | (10%) |
| BMI > 25 kg/m2 | 140 | (20%) | 468 | (66%) | 104 | (15%) |
| First pregnancy | 361 | (51%) | 277 | (39%) | 74 | (10%) |
| Multiple pregnancy | 49 | (7%) | 590 | (83%) | 73 | (10%) |
| Fetal exposure to cytokines | 67 | (9%) | 499 | (70%) | 146 | (21%) |
| From a severe cold | 15 | (2%) | 574 | (81%) | 123 | (17%) |
| From other viral, bacterial or fungal infections | 28 | (4%) | 563 | (79%) | 121 | (17%) |
| From chronic inflammatory diseases | 29 | (4%) | 605 | (85%) | 78 | (11%) |
| Fetal exposure to cigarette smoke | 117 | (16%) | 505 | (71%) | 90 | (13%) |
Percentages do not add up to 100% due to rounding and overlapping categories; n, number;
for 70 patients, environmental data were completely missing because parents did not fill out the questionnaires.
Genotyping of the SNPs was completed with a success rate of more than 98.5%. All genotype frequencies in parents were in HWE (P≥0.28). Genetic association results showed that offspring genotype of the variant in ESR1 was associated with hypospadias, as reported earlier in a partly overlapping sample9, whereas results for the variant in ATF3 were suggestive of an association. Maternal genotypes were not associated with hypospadias in offspring (Table 2). Repeating the analyses separately for subgroups of anterior, middle, and posterior hypospadias cases showed comparable results.
Table 2.
Genetic association results for offspring and maternal genotypes of single nucleotide polymorphisms in SRD5A2, ESR1, ESR2 and ATF3 with hypospadias.
| Offspring genotype |
Maternal genotype |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single nucleotide polymorphism |
Geno- type |
Cases |
P- value |
Mothers |
P- value |
||||||||
| n | (%) | RR | 95% CI | P LRT a | n | (%) | RR | 95% CI | P LRT b | ||||
| rs523349 in SRD5A2 | CC | 333 | (48%) | ref | 0.87 | 326 | (47%) | ref | 0.64 | ||||
| CG | 298 | (43%) | 1.1 | 0.9, 1.3 | 0.60 | 313 | (45%) | 1.0 | 0.8, 1.3 | 0.69 | |||
| GG | 70 | (10%) | 1.0 | 0.7, 1.5 | 0.83 | 57 | (8%) | 0.9 | 0.6, 1.3 | 0.46 | |||
| rs6932902 in ESR1 | GG | 513 | (72%) | ref | 3×10−3 | 523 | (75%) | ref | 0.42 | ||||
| AG | 177 | (25%) | 1.5 | 1.2, 2.0 | 1×10−3 | 159 | (23%) | 1.1 | 0.8, 1.4 | 0.61 | |||
| AA | 18 | (3%) | 2.0 | 1.1, 3.8 | 0.03 | 16 | (2%) | 1.7 | 0.7, 3.7 | 0.21 | |||
| rs2987983 in ESR2 | AA | 345 | (49%) | ref | 0.13 | 328 | (47%) | ref | 0.98 | ||||
| AG | 293 | (42%) | 0.8 | 0.7, 1.0 | 0.06 | 297 | (43%) | 1.0 | 0.8, 1.3 | 0.97 | |||
| GG | 67 | (10%) | 0.8 | 0.5, 1.1 | 0.12 | 72 | (10%) | 1.0 | 0.7, 1.4 | 0.86 | |||
| rs11119982 in ATF3 | CC | 163 | (23%) | ref | 0.07 | 180 | (26%) | ref | 0.09 | ||||
| CT | 354 | (50%) | 1.2 | 1.0, 1.6 | 0.07 | 332 | (48%) | 0.9 | 0.7, 1.1 | 0.23 | |||
| TT | 188 | (27%) | 1.4 | 1.0, 1.9 | 0.02 | 187 | (27%) | 1.1 | 0.8, 1.6 | 0.37 | |||
Percentages do not add up to 100% due to rounding; CI, confidence interval; LRT, likelihood ratio test; n, number; RR, relative risk;
P-value of the likelihood ratio test comparing a full model including maternal and offspring genotypes to a reduced model including only maternal genotypes;
P-value of the likelihood ratio test comparing a full model including maternal and offspring genotypes to a reduced model including only offspring genotypes.
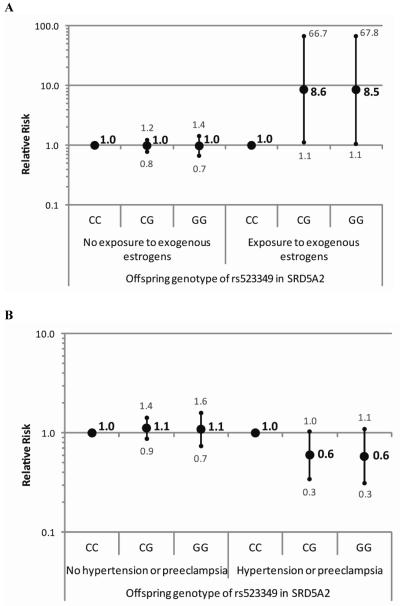
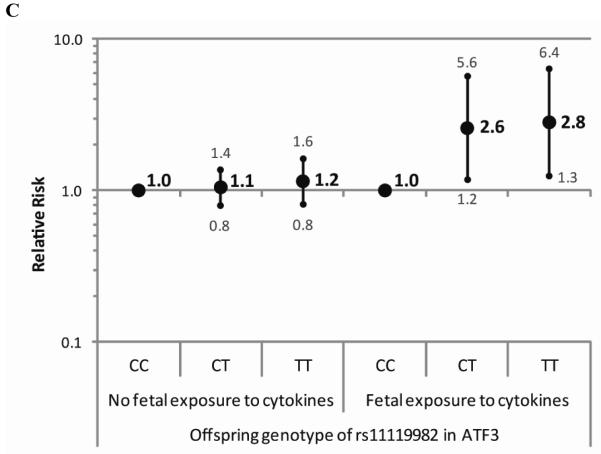
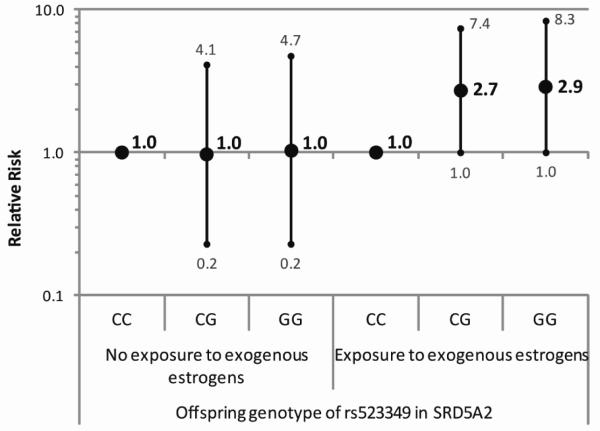
The results of the gene-environment interaction analyses pointed towards interactions between offspring genotype of rs523349 in SRD5A2 and exogenous estrogen exposure and maternal hypertension or preeclampsia (Table 3). Offspring carrying the variant allele seemed to be at increased risk of hypospadias when estrogen exposure occurred and at decreased risk when the mother had hypertension or preeclampsia. Furthermore, an interaction was observed between rs11119982 in ATF3 and exposure to cytokines, with an increased risk of hypospadias for offspring carrying the variant allele only when the mother reported an infection and/or inflammation (Figure 1). Due to small numbers of cases with certain exposures, we also considered a reduced model assuming HWE, which handles small sample size situations better. The risk estimates from this model showed the same direction of gene by exposure interaction for the SNP in SRD5A2 and exogenous estrogen exposure, albeit less strongly (Figure 2). For the other interactions, similar results were obtained as in the full model.
Table 3.
Results of the tests for gene-environment interactions for single nucleotide polymorphisms in SRD5A2, ESR1, ESR2 and ATF3.
| Single nucleotide polymorphism |
Environmental risk factor | P LRT a |
|---|---|---|
| rs523349 in SRD5A2 | Exogenous exposure to estrogens | 7×10−3* |
| Small for gestational age | 0.92 | |
| Hypertension or preeclampsia | 0.04* | |
| BMI > 25 kg/m2 | 0.17 | |
| First pregnancy | 0.42 | |
| Multiple pregnancy | 0.44 | |
| rs6932902 in ESR1 | Exogenous exposure to estrogens | 0.45 |
| Small for gestational age | 0.11 | |
| Hypertension or preeclampsia | 0.06 | |
| BMI > 25 kg/m2 | 0.89 | |
| First pregnancy | 0.10 | |
| Multiple pregnancy | 0.72 | |
| rs2987983 in ESR2 | Exogenous exposure to estrogens | 0.42 |
| Small for gestational age | 0.92 | |
| Hypertension or preeclampsia | 0.86 | |
| BMI > 25 kg/m2 | 0.28 | |
| First pregnancy | 0.27 | |
| Multiple pregnancy | 0.11 | |
| rs11119982 in ATF3 | Exogenous exposure to estrogens | 0.78 |
| Small for gestational age | 0.11 | |
| Hypertension or preeclampsia | 0.21 | |
| BMI > 25 kg/m2 | 0.82 | |
| First pregnancy | 0.40 | |
| Multiple pregnancy | 0.17 | |
| Fetal exposure to cytokines | 0.02* | |
| Fetal exposure to cigarette smoke | 0.60 |
LRT, likelihood ratio test;
P-value of the likelihood ratio test comparing a full model including gene-environment interactions to a reduced model including only offspring genotypes;
indication of gene-environment interaction.
Figure 1.
Relative risks of hypospadias with 95% confidence intervals for genotypes of (a) rs523349 in SRD5A2 within strata of exogenous estrogen exposure (b) rs523349 in SRD5A2 within strata of maternal hypertension or preeclampsia and (c) rs11119982 in ATF3 within strata of fetal exposure to cytokines.
Figure 2.
Relative risks of hypospadias with 95% confidence intervals for genotypes of rs523349 in SRD5A2 within strata of exogenous estrogen exposure using a reduced model assuming Hardy Weinberg equilibrium.
The results of the parent-of-origin effects analyses are shown in Table 4. For rs6932902 in ESR1, the estimated PO-LRT relative risk for an imprinting effect was 1.61 (95% CI=1.02-2.53), indicating that a maternally derived copy seemed to be associated with a greater risk of hypospadias than a paternally derived copy. The TAT showed that only the maternally derived copy increased the risk of hypospadias (RR=1.8, 95% CI=1.3-2.7).
Table 4.
Results of the parent-of-origin analyses.
| Transmission asymmetry test (TAT) |
PO-LRT |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mothers |
Fathers |
|||||||||||||
| Single nucleotide polymorphism |
Minor allelea |
T | NT | RR | 95% CI |
P- value |
T | NT | RR | 95% CI |
P- value |
P TAT | RR |
P- value |
| rs523349 in SRD5A2 | G | 81 | 80 | 1.0 | 0.7, 1.4 | 0.94 | 66 | 81 | 0.8 | 0.6, 1.1 | 0.22 | 0.34 | 1.18 | 0.27 |
| rs6932902 in ESR1 | A | 79 | 43 | 1.8 | 1.3, 2.7 | 1×10−3 | 58 | 53 | 1.1 | 0.8, 1.6 | 0.64 | 0.05 | 1.61 | 0.07 |
| rs2987983 in ESR2 | G | 59 | 95 | 0.6 | 0.5, 0.9 | 4×10−3 | 64 | 82 | 0.8 | 0.6, 1.1 | 0.14 | 0.33 | 0.76 | 0.27 |
| rs11119982 in ATF3 | T | 83 | 71 | 1.2 | 0.9, 1.6 | 0.33 | 111 | 70 | 1.6 | 1.2, 2.1 | 3×10−3 | 0.17 | 0.79 | 0.25 |
CI, confidence interval; NT, minor allele not transmitted; PO-LRT, parent-of-origin likelihood ratio test; RR, relative risk; T, minor allele transmitted; TAT, transmission asymmetry test;
the least frequent allele in the parents.
Discussion
This study is a follow-up to our earlier association study of genetic variants in SRD5A2, ESR1, ESR2, and ATF3 and hypospadias risk in which 620 cases were included. For the current study, we excluded 37 cases because DNA of both parents was not available or a brother was present in the dataset, and included 129 cases not included in the earlier study because of non-Caucasian or unknown ethnicity or because they were collected after 2007. We included gene-environment interactions as well as maternal and parent-of-origin effects in an attempt to reconcile our findings with those of others.
The estimated interaction between offspring genotype of the SNP in SRD5A2 and exogenous estrogen exposure during early pregnancy suggests that offspring carrying the variant have a more than eight fold increased risk of hypospadias only in case of exogenous estrogen exposure in the full log-linear model, and an almost three times increased risk in the reduced model. The interaction between this variant in SRD5A2 and exogenous estrogen exposure seems biologically plausible, as it causes a valine to leucine substitution (V89L) resulting in an approximately 30% decrease in enzyme activity26 and thus in less dihydrotestosterone. Additional estrogen exposure might cause an androgen-estrogen imbalance in carriers of this variant, resulting in hypospadias. The gene-environment interaction observed could help to explain differences in findings for this SNP between our study and studies from Sweden and China6,7,9. The latter two observed associations with the malformation, not taking environmental parameters into account, while we did not. However, phytoestrogen exposure is known to be higher in Chinese and Swedish populations compared to the Dutch. Chinese people consume more soy products, while in Nordic countries more rye bread and berries are consumed. These food products contain large amounts of isoflavonoids and lignans, respectively, whereas in a typical Western diet, both lignans and isoflavonoids are almost completely lacking27.
The SNP in SRD5A2 seemed to decrease hypospadias risk in case of maternal hypertension or preeclampsia, which may result from placental insufficiency. The latter may also lead to decreased provision of human chorionic gonadotropin (hCG) to the foetus. As hCG stimulates foetal testicular steroidogenesis before the foetus’s own pituitary-gonadal axis is established, and the SNP in SRD5A2 may result in even less DHT being formed, we would expect the SNP to increase hypospadias risk in case of maternal hypertension or preeclampsia.
The SNP in ATF3 seemed to be associated with an increased hypospadias risk only when the mother reported an infection and/or inflammation during pregnancy. ATF3 shows strong upregulation in hypospadias patients8 and is upregulated in response to cytokines14. While the rs11119982 variant has not been functionally characterized, a working hypothesis could be that the variant underlying the association with hypospadias causes an increased expression of ATF3 in response to cytokines. However, this finding does not reconcile our results with those reported earlier, describing a decreased hypospadias risk in the presence of the variant5.
The parent-of-origin analyses indicated that only a maternally derived copy of the variant in ESR1 seems to be associated with an increased hypospadias risk. This suggests that the maternally derived allele of ESR1 is more fully expressed than the paternally derived allele. Although ESR1 is not one of the currently known imprinted genes (www.geneimprint.com), the experimental identification of imprinted genes is challenging, because monoallelic expression of imprinted genes may occur only in particular tissues, at particular stages of development, or in one of the isoforms28. Therefore, it is unlikely that all human imprinted genes are already known. However, the observed parent-of-origin effect could also have arisen from an effect of maternal-fetal genotype interaction. Unfortunately, we do not have enough statistical power to disentangle these two possible effects.
We have to acknowledge some limitations. For all factors studied, we relied on information from questionnaires which may result in misclassification due to recall problems, especially since the average time between birth and filling out the questionnaires was 10.2 years, ranging from 0 to 27 years. However, most factors studied are relatively easy to remember. Also, this misclassification probably does not depend on genotype and would have resulted in attenuation of the results only, which may have obscured some effects. Misclassification due to measurement error could also account for not finding gene-environment interactions for well-known factors such as SGA or primiparity, which may be proxies for underlying causes that are difficult to measure, such as placental insufficiency. To our knowledge, we investigated the largest sample of hypospadias cases thus far reported in genetic studies, while the power of our study was further increased by including information on families with one missing parental genotype using the expectation-maximization algorithm. Nevertheless, numbers of cases having a specific genotype and being exposed to a particular environmental risk factor were still small, resulting in large confidence intervals for the effect estimates. Our definition of exogenous estrogen exposure, for example, assured a selective group of women experiencing high levels of exposure, but resulted in low numbers of exposed women. This indicates that very large samples are needed to study gene-estrogen-exposure interactions. In addition, the gene-environment interaction test assumes that the SNP under study is a disease-causing mutation. If a marker in linkage disequilibrium with the causative mutation is studied instead, the test is susceptible to exposure-related population-stratification29.
Conclusions
We showed that parent-of-origin effects and gene-environment interactions contribute to the etiology of hypospadias, and that environmental factors can explain genetic non-replication between studies. Our results warrant further research directed at elucidating combined effects of genetic and environmental factors for this frequently occurring urological birth defect.
Acknowledgements
We are grateful to everyone involved in the data collection: Saskia van der Velde-Visser, Christel Beumer, Karen Kwak, Jacqueline Knoll, Dr. Robert de Gier, Dr. Barbara Kortmann, Astrid Paauwen, Dr. Hing Gwan Kho, Dr. Jacques Driessen, and the anesthesiologists of OR 18. We would also like to thank Dr. Marieke Coenen, Mascha Schijvenaars, Remco Makkinje, and Johanne Groothuismink for helpful discussions and practical guidance.
Funding This work was supported by the Radboud University Nijmegen Medical Centre as part of a PhD project; and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Disclosure summary The authors have nothing to disclose
References
- 1.van der Zanden LFM, van Rooij IALM, Feitz WFJ, et al. Aetiology of hypospadias: a systematic review of genes and environment. Hum Reprod Update. 2012;18:260. doi: 10.1093/humupd/dms002. [DOI] [PubMed] [Google Scholar]
- 2.Fredell L, Iselius L, Collins A, et al. Complex segregation analysis of hypospadias. Hum Genet. 2002;111:231. doi: 10.1007/s00439-002-0799-y. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe M, Yoshida R, Ueoka K, et al. Haplotype analysis of the estrogen receptor 1 gene in male genital and reproductive abnormalities. Hum Reprod. 2007;22:1279. doi: 10.1093/humrep/del513. [DOI] [PubMed] [Google Scholar]
- 4.Beleza-Meireles A, Kockum I, Lundberg F, et al. Risk factors for hypospadias in the estrogen receptor 2 gene. J Clin Endocrinol Metab. 2007;92:3712. doi: 10.1210/jc.2007-0543. [DOI] [PubMed] [Google Scholar]
- 5.Beleza-Meireles A, Töhönen V, Söderhäll C, et al. Activating transcription factor 3: a hormone responsive gene in the etiology of hypospadias. Eur J Endocrinol. 2008;158:729. doi: 10.1530/EJE-07-0793. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Li Q, Xu J, et al. Mutation analysis of five candidate genes in Chinese patients with hypospadias. Eur J Hum Genet. 2004;12:706. doi: 10.1038/sj.ejhg.5201232. [DOI] [PubMed] [Google Scholar]
- 7.Thai HTT, Kalbasi M, Lagerstedt K, et al. The valine allele of the V89L polymorphism in the 5-alpha-reductase gene confers a reduced risk for hypospadias. J Clin Endocrinol Metab. 2005;90:6695. doi: 10.1210/jc.2005-0446. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Liu BC, Lin GT, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177:1939. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 9.van der Zanden LFM, van Rooij IALM, Feitz WFJ, et al. Genetics of hypospadias: are single-nucleotide polymorphisms in SRD5A2, ESR1, ESR2, and ATF3 really associated with the malformation? J Clin Endocrinol Metab. 2010;95:2384. doi: 10.1210/jc.2009-2101. [DOI] [PubMed] [Google Scholar]
- 10.Kalfa N, Philibert P, Sultan C. Is hypospadias a genetic, endocrine or environmental disease, or still an unexplained malformation? Int J Androl. 2009;32:187. doi: 10.1111/j.1365-2605.2008.00899.x. [DOI] [PubMed] [Google Scholar]
- 11.Pfeifer K. Mechanisms of genomic imprinting. Am J Hum Genet. 2000;67:777. doi: 10.1086/303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson LU, Boucher BA, Liu Z, et al. Phytoestrogen content of foods consumed in Canada, including isoflavones, lignans, and coumestan. Nutr Cancer. 2006;54:184. doi: 10.1207/s15327914nc5402_5. [DOI] [PubMed] [Google Scholar]
- 13.Visser GHA, Eilers PHC, Elferink-Stinkens PM, et al. New Dutch reference curves for birthweight by gestational age. Early Hum Dev. 2009;85:737. doi: 10.1016/j.earlhumdev.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Hai T, Wolfgang CD, Marsee DK, et al. ATF3 and stress responses. Gene Expr. 1999;7:321. [PMC free article] [PubMed] [Google Scholar]
- 15.Bosio A, Knörr C, Janssen U, et al. Kinetics of gene expression profiling in Swiss 3T3 cells exposed to aqueous extracts of cigarette smoke. Carcinogenesis. 2002;23:741. doi: 10.1093/carcin/23.5.741. [DOI] [PubMed] [Google Scholar]
- 16.Zaretsky MV, Alexander JM, Byrd W, et al. Transfer of inflammatory cytokines across the placenta. Obstet Gynecol. 2004;103:546. doi: 10.1097/01.AOG.0000114980.40445.83. [DOI] [PubMed] [Google Scholar]
- 17.Dahlgren J, Samuelsson AM, Jansson T, et al. Interleukin-6 in the maternal circulation reaches the rat fetus in mid-gestation. Pediatr Res. 2006;60:147. doi: 10.1203/01.pdr.0000230026.74139.18. [DOI] [PubMed] [Google Scholar]
- 18.Lackmann GM, Salzberger U, Töllner U, et al. Metabolites of a tobacco-specific carcinogen in urine from newborns. J Natl Cancer Inst. 1999;91:459. doi: 10.1093/jnci/91.5.459. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberg CR. Allowing for missing parents in genetic studies of case-parent triads. Am J Hum Genet. 1999;64:1186. doi: 10.1086/302337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brouwers MM, van der Zanden LFM, de Gier RPE, et al. Hypospadias: risk factor patterns and different phenotypes. BJU Int. 2010;105:254. doi: 10.1111/j.1464-410X.2009.08772.x. [DOI] [PubMed] [Google Scholar]
- 22.van der Zanden LFM, van Rooij IALM, Feitz WFJ, et al. Common variants in DGKK are strongly associated with risk of hypospadias. Nat Genet. 2010;43:48. doi: 10.1038/ng.721. [DOI] [PubMed] [Google Scholar]
- 23.Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umbach DM, Weinberg CR. The use of case-parent triads to study joint effects of genotype and exposure. Am J Hum Genet. 2000;66:251. doi: 10.1086/302707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermunt JK. LEM: a general program for the analysis of categorical data. Tilburg University; Tilburg, the Netherlands: 1997. [Google Scholar]
- 26.Makridakis NM, di Salle E, Reichardt JKV. Biochemical and pharmacogenetic dissection of human steroid 5 alpha-reductase type II. Pharmacogenetics. 2000;10:407. doi: 10.1097/00008571-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Adlercreutz H. Epidemiology of phytoestrogens. Baillieres Clin Endocrinol Metab. 1998;12:605. doi: 10.1016/s0950-351x(98)80007-4. [DOI] [PubMed] [Google Scholar]
- 28.Luedi PP, Dietrich FS, Weidman JR, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi M, Umbach DM, Weinberg CR, et al. Family-based gene-by-environment interaction studies: revelations and remedies. Epidemiology. 2011;22:400. doi: 10.1097/EDE.0b013e318212fec6. [DOI] [PMC free article] [PubMed] [Google Scholar]