The analysis of Alzheimer's disease is an important topic of research because the results and findings help elucidate problems associated with aging as well as other diseases including Downs' syndrome, Parkinson's disease, dementia with Lewy bodies, Multiple Sclerosis, Amyotrophic Lateral Sclerosis,and infectious disease including NeuroAIDS. Here we review just a few points from an ever growing and productive literature.
A recent article dealt with oxidation effects and neurodegeneration [1]. The hypothesis addressed was that oxidation of RNA affects miRNA and protein expression. In human as well as in rodent brain, neurodegeneration (neuronal) that is age-associated was demonstrated that is consequent to oxidation of nucleic acids and proteins. Such neuronal degenerative factors have also been analyzed in dementia with Lewy bodies, Amyotrophic Lateral Sclerosis, Parkinson disease, and Alzheimer disease. These findings occurred in cellular as well as animal models. Coding RNA as well as non-coding RNA were affected and deleterious effects on cell fate were analyzed. Such studies are needed for developing strategies of therapy.
A recent study also analyzed miRNAs in Alzheimer's disease pathogenesis [2]. The analysis of extracellular fluid from neocortex associated with temporal lobe from patients who had Alzheimer's disease indicated increased miRNAs. These miRNAs included miRNA-155 and miRNA-146a that are proinflammatory. Moreover, when cell culture medium was used with these miRNA added, Alzheimer's disease-like gene expression occurred. This profile was associated with downregulation of complement factor H (CFH), a repressor of the innate immune response. Anti-miRNA approaches reduced the pro-inflammatory effects. Additional miRNAs, miRNA-9 and miRNA-125bwere involved as well [3]. These miRNAs were regulated by NF-кB andbound independently to the mRNA 3'- untranslated region to reduce CFH translation.
The authors of an article in 2011 [4] stated that transcription factor, TAp73, drove expression of miRNA-34a (but not miRNA-34b or miRNA-34c). This was associated with decreased expression of synaptic proteins synaptotagmin-1and syntaxin-1A. These controlled molecular reactions generally occurred in retinoid-stimulated differentiation of neuroblastoma cells and rodents. The synaptotagmin-1 protein expression changes also occurred in Alzheimer's disease brain tissue.
These observations were extended to non-classical infectious disease related to prion infections. Several neurotrophic RNA and DNA viruses induced miRNA-146a in human brain cells. In addition, oxidative stress, neurotoxic metals,pro-inflammatory cytokines,as well as Aβ42 peptideupregulated miRNA-146a as wellin primary cell human glial-neuronal cocultures. These findings were in Alzheimer's disease and murine scrapie studies. miRNA-146a quantitieswere related to disease severity and inflammatory neuropathology in Alzheimer's disease. Moreover, findings were similar in Gerstmann-Straussler- Scheinker syndrome and sporadicCreutzfeldt-Jakob disease [5].
In addition, Aβ oligopeptide down regulated miRNA-9 and miRNA-181c in earlier studies. This occurred in hippocampal cultures. In addition, APP23 transgenicmice and human Alzheimer's disease tissue showed deposition of Aβ. Moreover, APP is a target of miRNA regulation and additional 3'- untranslated regions were identified from several mRNAs. These are down regulatedbymiR-9 and -181c, separately and contemporaneously and includedTGFBI, TRIM2, SIRT1 and BTBD3 [6].
NF-kB has a very large well-known number of protein-protein interactions and is not included here. Based on the literature, the protein Tap73 is more often recognized as TP73. Tap73 was absent in the GNCPro database and so the term TP73 was used. The figure illustrates various gene interactions among the proteins mentioned above:complement factor H (CFH), synaptotagmin-1 (SYT1), syntaxin-1A (STX1A), tumor protein p73 transcription factor (TP73), transforming growth factor (TGFBI), tripartite motif containing 2(TRIM2),sirtuin 1(SIRT1), andBTB (POZ) domain containing 3(BTBD3). It is left as a puzzle for the interested reader to identify the various genes and their functions in the Figure 1 [7–9].
Figure 1.
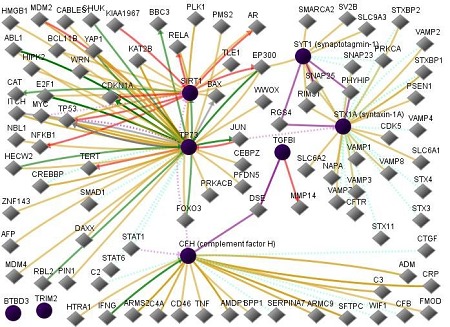
Network of input proteins (complement factor H, synaptotagmin-1, syntaxin-1A, TP73, TGFBI, TRIM2, SIRT1, andBTBD3) and the input neighbors of these proteins. In this figure, line-colors and various interactions with other genes are red Down-regulation, green Up-regulation, beige Regulation, purple Co-expression, brown Physical Interaction, turquoise dotted Predicted Protein Interaction, and mauve dotted Predicted TFactor Regulation (www. sabiosciences.com).
Acknowledgments
There are no financial conflicts.
Footnotes
Citation:Shapshak, Bioinformation 9(5): 222-223 (2013)
References
- 1.Nunomura A, et al. Neurotox Res. 2012;22:231. doi: 10.1007/s12640-012-9331-x. [DOI] [PubMed] [Google Scholar]
- 2.Lukiw WJ, et al. Neuroreport. 2012;2:621. doi: 10.1097/WNR.0b013e32835542b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukiw WJ, et al. Int J Biochem Mol Biol. 2012;3:105. [PMC free article] [PubMed] [Google Scholar]
- 4.Agostini M, et al. Proc Natl Acad Sci U S A. 2011;108:21093. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukiw WJ, et al. J Toxicol Environ Health A. 2011;74:1460. doi: 10.1080/15287394.2011.618973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schonrock N, et al. J Mol Neurosci. 2012;46:324. doi: 10.1007/s12031-011-9587-2. [DOI] [PubMed] [Google Scholar]
- 7. http://www.sabiosciences.com/
- 8. http://www.genecards.org/
- 9. http://www.ncbi.nlm.nih.gov/


