Abstract
Cinnamoyl CoA reductase (CCR) carries out the first committed step in monolignol biosynthesis and acts as a first regulatory point in lignin formation. CCR shows multiple substrate specificity towards various cinnamoyl CoA esters. Here, in Silico mutagenesis studies of active site residues of Ll-CCRH1 were carried out. Homology modeling based modeled 3D structure of Ll-CCRH1 was used as template for in Silico mutant preparations. Docking simulations of Ll-CCRH1 mutants with CoA esters by AutoDock Vina tools showed altered substrate specificity as compared to wild type. The study evidences that conformational changes, and change in geometry or architecture of active site pocket occurred following mutations. The altered substrate specificity for active site mutants suggests the possible physiological role of CCR either in lignin formation or in defense system in plants.
Abbreviations
Ll-CCRH1 - Leucaena leucocephala cinnamoyl CoA reductase 1, OPLS - Optimized Potentials for Liquid Simulations, RMSD - Root Mean Square Deviation.
Keywords: Cinnamoyl CoA reductase 1, Mutagenesis, Homology Modeling, Docking Simulations, Substrate Specificity
Background
Lignin, an integral cell wall component of plants, is a phenolic heteropolymer of monolignols namely, p-coumaryl alcohol, coniferyl alcohol and sinapyl alcohol [1]. Among several genes involved in lignin biosynthesis, Cinnamoyl CoA Reductase (CCR, EC 1.2.1.44) plays a key regulatory role in lignin formation [2]. Hydroxycinnamoyl CoA esters of general phenylpropanoid pathway become destined to form respective monolignols after action of CCR. Being entry point enzyme in monolignol biosynthesis, CCR diverts phenylpropanoid derived metabolites towards lignin.
CCR exhibits substrate specificity for different hydroxycinnamoyl CoA esters (p-coumaroyl CoA, caffeoyl CoA, feruloyl CoA, 5-hydroxyferuloyl CoA and sinapoyl CoA); and reduce them to corresponding aldehydes. Cinnamoyl CoA esters are the common precursors of wide range of phenolic compounds and flavonoids. For example, coumaroyl CoA esters are the substrates for chalcone synthase enzyme, the first catalytic step towards secondary metabolites synthesis. Secondary metabolites are considered as the first line of defense against pathogens and diseases. Differential substrate specificity of CCR has been correlated to its exclusive or redundant function inside the cell either in lignification (feruloyl CoA/ sinapoyl CoA as most preferred substrate) during plant development or in defense mechanism (Coumaroyl CoA as favored substrate) [3–5].
The major limitation in understanding structure-function relationship of CCR is lack of its three dimensional structure till date by any experimental means. Homology modeling provides an alternative approach for constructing three dimensional structures if X-ray crystal structure data is not available. We constructed 3D model of Ll-CCRH1 using Dihydroflavanol Reductase from Vitis vinifera as template (PDB ID: 2c29). Putative active site residues involved in substrate binding, stabilization and catalysis were identified based on amino acid sequence analysis and docking simulations. These residues were further investigated and confirmed by site directed mutagenesis and chemical modification studies (data unpublished).
Here, the present study was aimed to investigate the effects of various substitution mutations (in Silico) of active site residues on substrate specificity of Ll-CCRH1. Five different in Silico mutants were prepared for each amino acid residue and subjected to docking simulations with different hydroxycinnamoyl CoA esters. Based on docking energies obtained, substrate preferences for each mutant were determined. These substrate specificities were used to predict the possible role of in Silico Ll-CCRH1 mutants either in lignin formation or in defense mechanisms.
Methodology
Starting molecule:
Three dimensional model of Ll-CCRH1, generated using MODELLER 9v9, was used as template for in Silico mutagenesis studies (Protein Model Database ID: PM0078699).
Preparation of the in Silico mutants:
Active site of Ll-CCRH1 is made up of ten residues, Phe30, Ile31, Arg51, Asp77, Ser136, Tyr170, Lys174, Val200, Ser200 and His215. Five one-point substitution mutants were prepared for each active site residue. The three dimensional structures of in Silico mutants were constructed by homology modeling, using the program TRITON interfaced with MODELLER [6–8]. Each mutant homology model was evaluated for its stereo chemical quality using PROCHECK [9] and also checked for environmental profile using ERRAT (Structure analysis and verification servers).
Substrate docking:
Docking studies were carried out with five different hydroxycinnamoyl CoA esters; 4-coumaroyl CoA, caffeoyl CoA, feruloyl CoA, 5- hydroxyferuloyl CoA, sinapoyl CoA. Substrate molecules were downloaded from Pubchem database on NCBI, and converted to 3D molecules using LigPrep module in the schrodinger suite (LigPrep, version 2.4; Schrodinger: New York, 2010). Protein-ligand complexes were minimized within an RMSD of 0.30 Å with force field OPLS2005 using MacroModel package (MacroModel, version 9.8; Schrodinger: New York, 2010). Protein- ligand docking simulations were conducted using AutoDock Vina tool to prepare the systems for calculations [10]. For each ligand, around 100 docking runs with default parameters were performed treating protein as rigid and the ligand as flexible. The results were visualized using PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrodinger, LLC.), wherein all the conformations for each of the ligand were found to be within the cavity of protein indicating that the docking run was free from errors. The conformational clusters with lowest binding energy (Ea) for each ligand were considered for further studies.
Results & Discussion
Wild type Ll-CCRH1:
Docking simulations of different hydroxycinnamoyl CoA esters with Ll-CCRH1 showed feruloyl CoA (-9.9 Kcal/mole) as most favored substrate over others (-8.8 to -9.7 Kcal/mole) (data unpublished). Better affinity of Ll-CCRH1 towards feruloyl CoA indicates its possible role in lignification during growth and development of plants (Figure 2).
Figure 2.
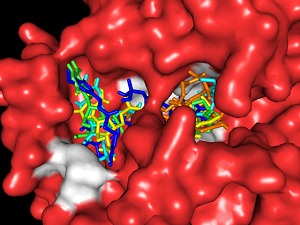
Surface representation of the catalytic active site in Ll- CCRH1 model (close up view) and docking of cinnamoyl CoA esters in substrate binding pocket of Ll-CCRH1; caffeoyl CoA (tv green), feruloyl CoA (yellow), hydroxyferuloyl CoA (blue), coumaroyl CoA (orange) and sinapoyl CoA (cyan). Active site residues are shown in white color.
Phenylalanine30:
Phe30 residue interacts with CoA esters via its main chain function and takes part in substrate binding or stabilization. Five mutants, namely F30C, F30L, F30S, F30V and F30Y were generated by homology modeling using TRITON software. Table 1 (see supplementary material) shows a comparison of the MODELLER produced mutants with respect to RMSD. Docking studies showed that F30S and F30V showed feruloyl CoA as favored substrate; while mutants F30L, F30S and F30Y showed preference for coumaroyl CoA Table 2 (see supplementary material). The main chain function of all mutants was expected to remain same even after mutation. Thus, there should be conformational alterations or change in geometry of active site following mutations resulting in differential substrate specificity (Figure 1 & Figure 3A,B). The RMSD values for phe30 mutants lay between 0.107-0.125 Å. Mutants F30C and F30V may play a role in lignin biosynthesis; while F30S, F30Y and F30L could be important for defense cascades [3–5].
Figure 1.
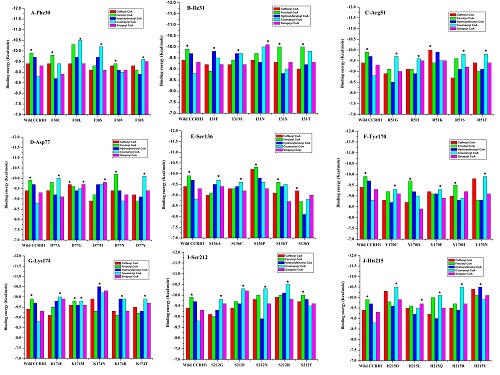
Cinnamoyl CoA esters binding energy change for in Silico active site mutants. X-axis represents Ll-CCRH1 amino acid mutants generated by homology modeling, using TRITON/MODELLER software, and Y-axis is respective binding energies for CoA esters docked with each individual mutant. The ‘*’ shows best docked substrate with most negative binding energy among particular mutant.
Figure 3.
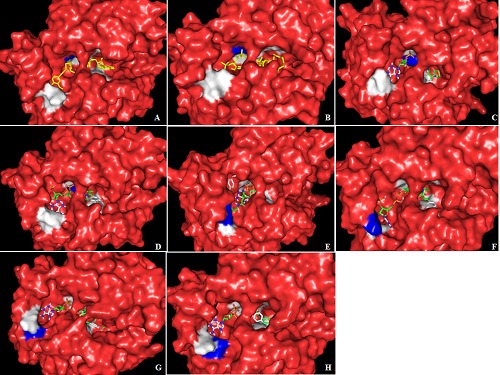
The best docked structures of cinnamoyl CoA esters into the active site of in Silico Ll-CCRH1 mutants at various positions. Mutated Ll-CCRH1 molecule (red) is represented as solid surface, whereas docked structure is shown as sticks. Mutated amino acid is displayed in blue color and remaining active site residues are shown in white. (A) F30C mutant, feruloyl CoA (B) F30Y mutant, coumaroyl CoA (C) I31F mutant, coumaroyl CoA (D) I31N mutant, sinapoyl CoA (E) R51I mutant, coumaroyl CoA (F) R51K mutant, caffeoyl CoA (G) D77H mutant, sinapoyl CoA (H) D77Y mutant, coumaroyl CoA.
Isoleucine31:
Ile31 residue also plays role in substrate binding through its main chain function. Mutant I31F showed more negative binding energy for hydroxyferuloyl CoA; while I31M exhibited equal affinity for coumaroyl and hydroxyferulol CoA. Mutant I31N demonstrated better affinity for sinapoyl CoA over others. Substitution with serine and threonine (I31S and I31T) resulted in feruloyl CoA as preferred substrate with equal binding energy (-10 Kcal/mole) (Figure 1, Table 2). in Silico mutations of Ile31 lead to overall change in architecture of active site pocket (Figure 3C, D). Slightly higher RMSD value was observed for mutant I31M compared to other Ile31 mutants (Table 1).
Arginine51:
In wild type Ll-CCRH1, Arg51 interacts with CoA esters via its main chain as well as side chain and plays important role in substrate binding or stabilization. In mutant R51G, coumaroyl CoA showed most favorable binding energy (-9.7 Kcal/mole). Substitution of Arg51 by glycine altered side chain from polar charged to small compact neutral residue, resulting in marked decrease in accessible surface area and leads to loss of interactions with substrate (Table 2). Similar decrease in surface area was observed with mutants R51I, R51S and R51T except for R51K, which showed similar area as that of wild type. Except R51K, all three remaining mutants have shown better preference for coumaroyl CoA; and thus could play role in defense. Mutant R51K showed affinity towards caffeoyl CoA (Table 2) (Figure 1 & Figure 3E, F). Thus, all mutants R51G, R51I, R51S, R51K and R51T may function in defense [3, 4].
Aspartate77:
No Asp77 interactions were observed during docking of Ll- CCRH1 with different CoA esters; but site directed mutagenesis and chemical modification confirmed its role in CCR catalyzed reaction (data not shown). D77 is present partially on surface and in proximity of R51. Substitution with alanine and glycine resulted in slight decrease in surface areas, but substrate affinities differ for both these mutants; D77A showed specificity towards coumaroyl CoA while D77G displayed equal affinity for Caffeoyl CoA and sinapoyl CoA (Figure 1, Table 2). This may be due to change in structure of binding pocket and allowing better conformations to interact with other amino acids. Mutant D77H showed more negative binding energy for sinapoyl CoA (-9.7 Kcal/mol). D77N has same substrate affinity (feruloyl CoA) as that of wild type. In mutation D77Y, change from D to Y altered small polar side chain to bulky hydrophobic aromatic ring, induced a significant increase in accessibility for surface area (Figure 3G, H). RMSD values for all mutants are very comparable to each other (0.104-0.122 Å) (Table 1).
Serine136:
In wild type Ll-CCRH1, Ser136 plays a key role in catalysis and is a part of reaction centre. Mutations in ser136 caused change in the geometry of active site and non specific interactions with substrates have been increased. Ser136 is buried in the substrate binding pocket and mutations resulted in partial or complete exposure of mutant residue. Coumaroyl CoA was found to be better substrate for mutant S136A and S136C. S136Y showed preference for caffeoyl CoA and, S136P along with S136T showed favored specificity for feruloyl CoA (Table 2) (Figure 1 and Figure 4A, B). Mutants S136P and S136T could have functional role in lignin formation; while mutants S136Y, S136A and S136C might be involved in defense [3–5].
Tyrosine170:
Tyr170 acts as catalytic base and accepts hydride from NADPH and transfers to Serine residues in catalysis reaction. Tyr170 is completely buried in active site pocket and is surrounded by Lys174, Ser212, His215 and Ser136 catalytic residues. Mutant Y170C displayed less number of interactions compared to wild type; still sufficient for displaying affinity towards coumaroyl CoA. Mutant Y170D exhibited better binding energy for feruloyl CoA (-9.7 Kcal/mol) (Figure 1). In case of mutant Y170D, increase in number of hydrogen bond interactions with Arg51 was observed. Arg51 is distantly present from Y170D. This indicates the drastic change in architecture of binding pocket and significant changes in pKa values of pocket. Y170F and Y170N showed preference for coumaroyl CoA; while Y170H demonstrated affinity for feruloyl CoA (Table 2 & Figure 4C, D).
Lysine174:
Lys174 residue promotes hydride transfer in CCR mediated reduction reactions. Docking simulations of K174E and K174T mutants showed coumaroyl CoA as preferred substrate (-10 Kcal/mole) (Figure 1 & Table 2). On the other hand, K174M mutant is specific for feruloyl CoA; while K174N and K174R have favorable binding energy for hydroxyferuloyl CoA (Figure 1). Thus, mutant K174R may play role in both, that is, either in lignin biosynthesis or defense system [3, 4]. Lys174 is deeply buried in active site pocket and same conformational profile was also observed in all mutants (Figure 4E, F). Mutant K174R shows the lowest RMSD (0.097 Å) among the all mutants generated (Table 1).
Figure 4.
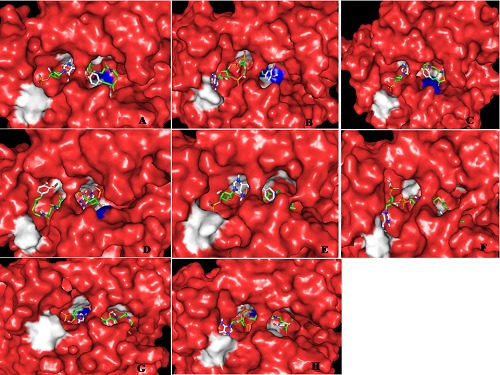
Cinnamoyl CoA esters conformations (sticks) in the active site of mutants Ll-CCRH1 (surface, red). Color code for mutated and remaining active site residue is same as mentioned in Figure 3 (A) S136C mutant, coumaroyl CoA (B) S136Y mutant, caffeoyl CoA (C) Y170F mutant, coumaroyl CoA (D) Y170H mutant, coumaroyl CoA (E) K174E mutant, coumaroyl CoA (F) K174N mutant, hydroxyferuloyl CoA (G) V200L mutant, hydroxyferuloyl CoA (H) V200M mutant, coumaroyl CoA. Lysine174 residue is deeply buried inside the binding pocket, thus it is not visible in E, F surface diagrams.
Valine200:
Replacement of Val200 with glycine and alanine displayed substrate specificity towards coumaroyl CoA. These substitutions have not altered aliphatic side chain profile of mutant residues. On the other hand, substitution of Valine by positively charged Aspargine resulted in charge redistribution along the active site pocket and allowed favorable conformational changes for substrate binding (feruloyl CoA). V200M residue exhibited increased affinity for coumaroyl CoA (Table 2 & Figure 4G, H). V200E mutant could possibly be involved in monolignol biosynthesis. Mutants V200G, V200M, V200A may take part in secondary metabolite synthesis [3, 4] (Supplementary Figure S1).
Serine212:
Ser212 residue is involved in proton shuttle mechanism and thus participates in CCR catalysis. Mutations in Ser residue (deeply buried) by Gly, Ile, Glu and Arg showed partial or complete exposure of mutated residues. These exposed mutated residues altered substrate binding conformation and favored coumaroyl CoA as substrate (Figure 1 & Supplementary FigureS2 A, B). Thus, these structural changes altered shape of active site pocket and assisted more number of interacting residues with maximum interactions. S212T has shown feruloyl CoA as promising substrate (Figure 1 & Supplementary FigureS2 C) (Table 2). All mutants of ser212 except S212T may function in secondary metabolism, ultimately in providing defense [3–5].
Histidine215:
His215 also takes part in CCR mediated reduction reactions either in substrate binding or catalysis. H215R and H215Y mutants showed significant increase in surface area due to bulky and long side chains. Both mutants showed same binding energy (-10.5 Kcal/mol), but for different substrates; that is, coumaroyl CoA is specific for H215R and hydroxyferuloyl CoA for H215Y (Figure 1 & Supplementary FigureS2 E, F). Replacement of His215 by Asp and Gln demonstrated affinity for coumaroyl CoA (Figure 1). Lastly, replacement of His with Leu displayed preference for sinapoyl CoA (Table 2) (Figure 1 & Supplementary FigureS2 D). Thus, only H215L mutant may possibly take part in lignification; while remaining mutants might prefer secondary metabolite pathway [3, 4].
Conclusion
In conclusion, in silico mutation analysis of active site residues of Ll-CCRH1 displayed differential substrate specificity. This differential substrate specificity was mainly due to conformational changes in substrate binding pocket or change in geometry/architecture/shape of active site or increase/decrease in number of interactions following mutations or charge redistribution along active site and physiochemical properties of mutated residues. On the basis of substrate specificity, possible physiological role of mutant CCRs could be predicted.
Supplementary material
Acknowledgments
This work was supported by Council of Scientific and Industrial Research (CSIR), New Delhi, India.
Footnotes
Citation:Sonawane et al, Bioinformation 9(5): 224-232 (2013)
References
- 1.Boerjan J, et al. Annu Rev Plant Biol. 2003;54:519. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 2.Lacombe E, et al. Plant J. 1997;11:429. doi: 10.1046/j.1365-313x.1997.11030429.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou R, et al. Proc Natl Acad Sci U S A. 2010;1071:17803. doi: 10.1073/pnas.1012900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escamilla-Trevino L, et al. New phytol. 2010;185:143. doi: 10.1111/j.1469-8137.2009.03018.x. [DOI] [PubMed] [Google Scholar]
- 5.Li L, et al. Plant Cell Physiol. 2005;46:1073. doi: 10.1093/pcp/pci120. [DOI] [PubMed] [Google Scholar]
- 6.Eswar N, et al. Curr Protoc Bioinformatics. 2006;15 doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 8.Prokop M, et al. Bioinformatics. 2008;24:1955. doi: 10.1093/bioinformatics/btn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskowski RA, et al. J Biomol NMR. 1996;8:477. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 10.Trott O, Olson A. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


