Abstract
Root-knot nematodes (RKNs), Meloidogyne spp, are found in all temperate and tropical areas, and are among the most damaging plant pathogens worldwide. M. graminincola is an economically important root parasite on upland, lowland and deepwater rice. FMRFamide-like peptides (FLPs) play significant role as neurotransmitters or neuromodulators in the nervous system and proposed as one of the important targets for the plant parasitic nematode management. Therefore, for the first time, we have cloned and characterized two neuropeptide genes (flp-1 and flp-12) from the cDNA of preparasitic second stage juveniles of M. graminicola. The flp-12 contains putative 22 residue long signal peptide at N-terminal suggesting function as an extra-cellular protein. We have found highly conserved motif LFRGR in flp-1. These two flp genes could be interesting and potential targets for functional validation to explore their utility for designing management strategies.
Background
Meloidogyne graminicola is one of the major biotic stresses in rice production. Reported losses to Meloidogyne vary with ranges of 12-33%; 28-87% 11-73% and 20-80% (mean 43 ± 7.5%) [1]. Approximately 57% of Indian rice is produced in the Indo- Gangetic plain, comprising nearly 12 million ha, which can be sub-divided on physiographic and bio-climatic considerations into four transects in India as well as one in the Pakistan Punjab. The four Indian transects have both individual and shared cropping problems. Each transect suffers a range of biotic stresses of which root-knot nematodes (Meloidogyne) are a key issue for three of the Indian transects i.e. a) Indian Punjab, Himachal Pradesh and Haryana (b) Eastern Uttar Pradesh, and Bihar and c) West Bengal. M. graminicola is also an important rice pest in several major rice growing areas of South and South-east Asia, causing estimated yield losses of between 20% and 80% [1–3]. Due to the inadequacy of the existing management approaches, there is a need to develop economically and environmentally sustainable approaches for which nematode genomics could be highly promising by providing novel targets.
Neuropeptide signaling system has been proposed to be the potential target for the management of plant parasitic nematodes [4, 5] due to its critical role in various parasitic activities like host recognition, migration and penetration, secretory activities, alimentation, and reproduction. FMRFamide-like peptides (FLPs) have shown to be widely expressed in the nervous system of root knot nematodes, Meloidogyne spp [6]. The largest families of neuropeptides in nematodes are the FLPs, which possess a C-terminal Arg-Phe- NH2 signature and play a central role in motor activities. There are 28 flp genes reported in Caenorhabditis elegans encoding at least 72 distinct peptides [7]. Likewise, 21 FLPs have been identified in economically important plant parasitic root knot nematode, M. incognita [8]. One or more FLPs are known to coexpress indicating that there is an interaction between them. Previous reports in C. elegans have indicated that, flp-1, 12 and 8 are co expressed and a mutation in flp-8 did not have any phenotypic aberration in C. elegans. But disruption of flp-1 and 12 resulted in various neuro muscular dysfunctions [9]. Further functions of flp genes are regulated by microRNAs (miRNAs) and flp-12 was identified as a potential target in pine wood nematode, Bursaphelenchus xylophilus [10]. Although studies on role of FLPs on behavior of plant parasitic nematodes are limited, a series of RNA interference studies of flp genes in M. incognita [6] and Globodera pallida [5] revealed aberrant behavioral phenotypes and migrational abilities. flp-12 is predicted to be crucial for the normal muscular function in G. pallida and M. incognita as it has interfered with the nematode migration in a sand column in response to the root defusates [11]. A similar effect can be envisaged in M. graminicola that will be very useful in designing an efficient management strategy. In view of this, for the first time, we have cloned and characterized flp-1 and flp-12 from the Indian isolates of M. graminicola using orthologous sequences present in M. incognita.
Methodology
Nematode collection:
Nematode infected roots were collected from the nematode culture pots and root galls were separated and used for hatching the 2nd stage juveniles (J2s) of M. graminicola.
RNA Isolation, quality controls and cDNA Synthesis:
Total RNA was extracted from J2s of M. graminicola using NucleoSpin total RNA Kit (Macherey-Nagel, Germany). The quality and quantity of each RNA sample was confirmed by using Nanodrop (Thermo Scientific). The RNA samples with 260/280 ratio from 1.9 to 2.1, 260/230 ratio from 2.0 to 2.5 were used for the analysis. One µg of the RNA sample was reverse transcribed to cDNA by using cDNA synthesis Kit (Superscript VILO, Invitrogen).
Cloning and Sequencing:
flp-1 and flp-12 were amplified from the M. graminicola cDNA, using the primers designed based on the published nucleotide sequence of flp-1 and flp-12 genes of M. incognita, (Accession Nos. AAW56944 and AY804187 respectively). The sequences of the primer sets for flp-1 were forward primer: 5' ATTCCTGGTCAGATGCAGACCCAA3'; reverse primer: 5'CCACTACTTCGGCCAAATCGAAGA3' while flp-12 was amplified using forward primer: 5'CCCAAGTTTGAGCTCTAAAAACAC3' and reverse primer: 5'TCATCGTCCAAATCGAATGA3'. PCR amplification reactions were performed in 50 µl reaction volume containing 5 µl 10 x assay buffer, 200 µM each of dATP, dCTP, dGTP and dTTP (Fermentas), 0.5 µM each primer, 0.5-2 units of Taq polymerase (Sigma-Aldrich) and 1 µl cDNA. The PCR amplification consisted of initial denaturation at 94°C for 4 min, followed by 35 cycles of amplification, denaturation at 94° C for 60 s, annealing at 60°C for 30 s and extension at 72°C for 1 min with a final extension at 72°C for 10 min. The amplified product was separated on 1.2% Agarose gel electrophoresis to confirm the size of the amplified product.
Fresh PCR product was cloned into pGEM-T easy cloning vector (Promega, USA) using standard protocol. Freshly prepared competent cells of Escherichia coli DH5α were transformed with the recombinant plasmids. Positive clones were selected by blue white colony screening along with ampicillin and colony PCR. Recombinant colonies were used for plasmid extraction. Inserts in the clones were confirmed by restriction digestion with EcoRI. The positive clones were custom sequenced by ABI SOLiD sequencing system [12].
Bioinformatics Analysis:
The amino acid sequences were deduced from the corresponding nucleotide sequences by using the OrfPredictor (ORF-Predictor) server, which is designed for ORF prediction and translation of a batch of EST or cDNA sequences. Database searches were performed with the BLAST Network Service (NCBI, National Center for Biotechnology Information), using the BLASTX and BLASTN algorithm [13]. Comparison with the homologous sequences was done with ClustalW [14]. Gene ontology term was assigned through AmiGO BLAST [15]. Phylogenetic trees were plotted by the maximum likelihood method using MEGA5 software package [16, 17].
Result & Discussion
Cloning of flp genes from cDNA of M. graminicola and deduced amino acid sequence:
Partial cDNA fragments of 214 bp and 299 bp, named Mg-flp-1 and Mg-flp-12 respectively, were generated by PCR amplification from M. graminicola (Figure 1A) cloned into pGEM-T TA cloning vector and confirmed the recombinant colonies. The inserts in the plasmids for the positive recombinants were confirmed by restriction digestion with EcoRI (Figure 1B). The cloned genes were sequenced using ABI solid sequencing platform and sequences along with the predicted ORF obtained from ORF-predictor.
Figure 1.
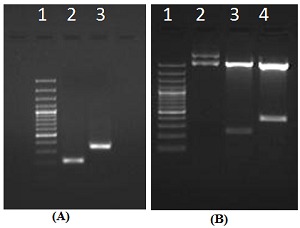
(A) PCR amplification of flp-1 and flp-12 genes from cDNA of M. graminicola. Lane 1:100 bp DNA marker, Lane2: flp- 1 and Lane 3: flp-12; (B) EcoRI digestion of pGEMT vector containing flp-1 and flp-12. Lane 1:100 bp DNA marker, Lane 2: undigested plasmid, Lane 3: flp-1 and Lane 4: flp-12.
Characterization of the sequences:
Sequences of the partial cDNAs of flp-1 (214 bp) and flp-12 (299 bp) obtained from M. graminicola were submitted to Genbank sequence database (Accession Nos. KC250005 and KC250006).The percentage of the GC content was 43% and 35% for flp-1 and flp-12 respectively. BLASTN was carried out to determine the homology against non-redundant Genbank database. flp-1 showed best hit with the M. incognita having 70% identities (E value 3e-26) and 17% Gaps. Whereas, database searches for the flp-12 revealed 99% identity with Heterodera avenae [18] and 95% with M. incoginta. Conceptually translated nucleotide sequences into the corresponding amino acids resulted in 70 (flp-1) and 91 (flp-12) amino acids.
The BLASTX algorithm was also used to compare the M. graminicola sequence with protein sequences in database. flp-1 showed the most significant match with (Score = 236, Expect = 1e-23) with that of M. incognita while flp-12 showed most significant (Score = 472, Expect = 6e-59) match with FMRFamide-like protein 12 of cereal cyst nematode, H.avenae. SignalP server was used to identify the signal peptide in both the FLPs. SignalP result did not produce significant hits for flp- 1. However, flp-12 was predicted to contain a putative Nterminal short secretion signal peptide of 22 amino acids. This indicates that flp-12 could be involved in extra cellular function like signal transduction. AmiGO BLAST hit was performed to assign the putative function of flp-1 and flp-12 from M. graminicola and suggested their potential role in neuropeptide signaling pathway (GO: 0007218) and locomotory behavior (GO: 0007626).
Putative function for the flp-12 sequence was identified as G protein coupled receptor activity (GO: 0004930). Previous studies on C.elegans showed that flp-1 can encode up to seven distinct, yet highly similar, FMRF amide-related peptides (FaRPs), small neuromodulators peptides that are characterized by a C-terminal Arg-Phe-amide motif. flp-1 peptides are required for the regulation of several behaviors, including wellcoordinated, sinusoidal movement and the transition between active and inactive states of egg laying. Receptors for the flp-1 peptides have not yet been identified, but genetic studies have suggested that flp-1 peptides may act through G-protein coupled receptors. flp-1 mRNAs are detected at all developmental stages, and a flp-1 translational reporter fusion detects expression in the anteriorly positioned neurons AVK, AVA, AVE, RIG, RMG, AIY, AIA, and M5. Based on spring model, generated interaction network image by the STRING database suggested that flp-1, flp-8 and flp-12 are co-expressed (Figure 2). flp-8 encodes three copies of a FMRFamide-related short peptide neurotransmitter. Although the flp-8 peptide can increase pharyngeal action potential frequency, loss of flp-8 function does not result in a mutant phenotype, suggesting that flp-8 may function redundantly with other FMRFamide-like peptides in the nervous system. flp-12 gene encodes a predicted FMRFamide-like peptide neurotransmitter that affects locomotion when injected into A. suum [19]. Similarly, silencing of flp-1 and flp-12 of M. incognita in our laboratory interfered with host recognition and infection (unpublished data).
Figure 2.
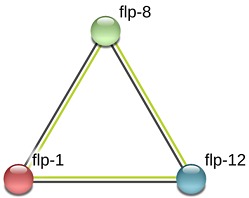
Interaction network generated by the STRING database based on spring model, generated for flp-1, flp-8 and flp-12. Confidence scored for co-expression of flp-1 and flp-12 (0.730) and confidence score for co-expression of flp-1 and flp- 8(0.822).
Sequence comparison of M. graminicola neuropeptides (flp-1, flp-12) with other nematodes:
The conceptually translated Mg-flp-1 protein revealed highest similarity with that of Mi-flp-1 while the Mg-flp-12 exhibited closeness with H.avenae. BLASTX identified homologous sequences for flp-1 from the free-living nematode C. elegans, animal parasitic nematode, A. suum and plant parasitic nematodes, M. incognita, D. destructor, G. pallida. On the other hand, homologous sequences for flp-12 were only found in plant parasitic nematodes, H. avenae, M. incognita and M. minor. They were aligned with the deduced amino acid sequence of the M. graminicola flp-1 and flp-12 respectively, as shown in (Figure 3). Multiple sequence alignment result suggested the diversity of flp-1 among the homologous sequences. NH2 terminal of flp-1 seems to appear poorly conserved than COOH terminal and the LFRGR motif present in three repeats is highly conserved across the nematode species (Figure 3A). This strongly suggests that these conserved motifs could be essential in signal transduction mediated by GPCR (G-protein coupled receptor) [20, 21]. Alignment of flp-12 revealed high sequence conservation only among the plant parasitic nematodes. Figure 3B clearly indicated that C- terminal FMRF signature motif was not amplified during the PCR experiment (As shown in the rectangular box). Phylogenetic analysis was carried out for both Mg-flp-1 and Mg-flp-12 respectively. Protein sequence for M. graminicola and M. incognita were clustered together but appeared as an out group. Interestingly, flp-1 of A. suum, an animal parasitic nematode was grouped with C. elegans, a free living nematode. flp-1 of Ditylenchus distructor and G. pallida appeared different from the other sequences (Figure 4A). flp-12 of H. avenae, M. graminicola and M. incognita could be clustered together as they were highly similar whereas, G. pallida was as an out-group. The sequence from M. minor was slightly diverse from the two root knot and one cyst nematode species (Figure 4B).
Figure 3.
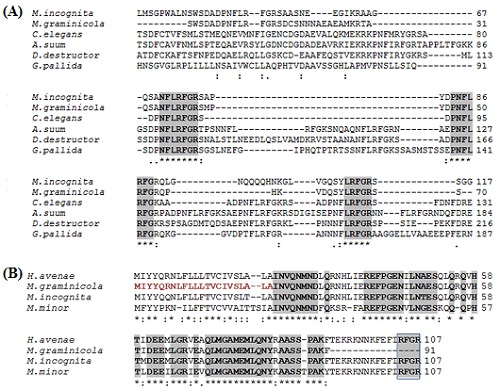
Multiple sequence alignment of the predicted amino acid sequence of the (A) flp-1 and flp-12 (B) from M. graminicola with flp sequences from other nematodes. Genbank identifier for the sequences used of flp-1 as follows, M. incognita (AAW56944); G. pallida (CAC36149); Ditylenchus destructor (ACZ97403); Ascaris suum (AEX09241); C. elegans (NP_872078) and for flp-12 genebank identifiers as H. avenae (AEN71840); M. incognita (AAX19364); M. minor (ABB49052) and G. pallida (CAC32452) and Signal peptide shown in red.
Figure 4.
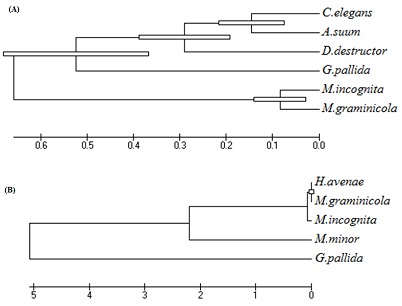
Phylogenetic relationships of the predicted (A) flp-1 and (B) flp-12 from M. graminicola and flp sequences from other nematodes. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. Initial tree for the heuristic search were obtained automatically as follows. When the number of common sites was < 100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise BIONJ method with MCL distance matrix was used. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA5. The homologous sequences genebank accessions were given in Figure 4 were used in this analysis.
Conclusion
Neuromuscular system of nematodes has been established as an important target for nematode control efforts and majority of the leading anthelminthic drugs act on targets within the neuromuscular system. M. graminicola is an obligate parasite of rice; despite of its high economical importance, very less genetic information is available. In this regard, we have cloned and characterized the two partial cDNA sequences of M. graminicola neuropeptide, flp-1 and flp-12 genes. flp-1 predicted to have role in neuropeptide signaling pathway and locomotory behavior and putative function of flp-12 is indicated its association with G protein coupled receptor activity. These two neuropeptides can be used for future molecular and genetic studies for confirming its practical utility for the management of M. graminicola.
Acknowledgments
We would like to thank Indian Agricultural Research Institute, New Delhi and Department of Biotechnology, Government of India for the financial support to carry out the Research work.
Footnotes
Citation:Rao et al, Bioinformation 9(4): 182-186 (2013)
References
- 1.Padgham JL, et al. J Nematol. 2004;36:42. [PMC free article] [PubMed] [Google Scholar]
- 2.Pokharel RR, et al. J Nematol. 2007;39:221. [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta TK, et al. Afr J Microbiol Res. 2012;6:6016. [Google Scholar]
- 4.Maule AG, et al. Curr Top Med Chem. 2002;2:733. doi: 10.2174/1568026023393697. [DOI] [PubMed] [Google Scholar]
- 5.Kimber MJ, et al. FASEB J. 2007;21:1233. doi: 10.1096/fj.06-7343com. [DOI] [PubMed] [Google Scholar]
- 6.Johnston MJ, et al. J Helminthol. 2010;84:253. doi: 10.1017/S0022149X09990630. [DOI] [PubMed] [Google Scholar]
- 7.Husson SJ, et al. Commun Agric Appl Biol Sci. 2005;70:153. [PubMed] [Google Scholar]
- 8.Abad P, et al. Nat Biotechnol. 2008;26:909. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- 9.Masler EP. J Helminthol. 2008;82:279. doi: 10.1017/S0022149X08982596. [DOI] [PubMed] [Google Scholar]
- 10.Huang QX, et al. PLoS One. 2010;5:e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalzell JJ, et al. Int J Parasitol. 2010;40:91. doi: 10.1016/j.ijpara.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Ondov BD, et al. Bioinformatics. 2008;24:2776. doi: 10.1093/bioinformatics/btn512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Larkin MA, et al. Bioinformatics. 2007;23:2947. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Carbon S, et al. Bioinformatics. 2009;25:288. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DT, et al. Comput Appl Biosci. 1992;8:275. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K, et al. Mol Biol Evol. 2011;28:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakur P, et al. Bioinformation. 2012;8:617. doi: 10.6026/97320630008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris TW, et al. Nucleic Acids Res. 2010;38:463. [Google Scholar]
- 20.Geary TG, et al. Trends Pharmacol Sci. 2005;26:56. doi: 10.1016/j.tips.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Kubiak TM, et al. Biochem Biophys Res Commun. 2003;301:456. doi: 10.1016/s0006-291x(02)03054-1. [DOI] [PubMed] [Google Scholar]


