Abstract
Substrate activities of various linear polyamines to human spermine oxidase (hSMO) were investigated. The activities were evaluated by monitoring the amount of H2O2 released from sample polyamines by hSMO. H2O2 was measured by a HPLC method that analyzed fluorescent dimers derived from the oxidation of homovanillic acid in the presence of horseradish peroxidase. Six triamines were tested and were found not to be hSMO substrates. Of sixteen tetramines tested, spermine (Spm) was the most active substrate, followed by homospermine and N-butylated Spm. Pentamines showed a characteristic pattern of substrate activity. Of thirteen pentamines tested, 3343 showed higher substrate activity than Spm, and 4343 showed similar activity to Spm. The activities of the other pentamines were as follows: 3443, 4443, 4344, 3344, 4334, 4444, and 3334 (in decreasing order). Product amines released from these pentamines by hSMO were then analyzed by HPLC. Triamine was the only observed product, and the amount of triamine was nearly equivalent to that of released H2O2. A marked difference in the pH dependency curves between tetramines and pentamines suggested that hSMO favored reactions with a non-protonated secondary nitrogen at the cleavage site. The Km and Vmax values for Spm and 3343 at pH 7.0 and 9.0 were consistent with the higher substrate activity of 3343 compared to Spm, as well as with the concept of a non-protonated secondary nitrogen at the cleavage site being preferred, and 3343 was well degraded at a physiological pH by hSMO.
Keywords: Spermine oxidase, Polyamine, Pentamine, Hydrogen peroxide, Homovanillic acid
Introduction
The polyamines spermine (Spm), spermidine (34), and their precursor putrescine are important in cell proliferation, differentiation, and survival.1) Recently, there has been increasing interest in polyamine catabolism. Polyamine catabolism is mediated by three enzymes2). Spermidine/spermine N1-acetyltransferase (SSAT) acetylates Spm and 34 to produce N1-acetylated compounds, which are exported from cells or oxidized by the peroxisomal enzyme N1-acetylpolyamine oxidase (APAO) to yield 34 or putrescine, respectively, with H2O2 and 3-acetamidopropanal. The cytosolic enzyme Spm oxidase (SMO) can catalyze the oxidation of Spm directly to 34, bypassing the necessity for acetylation. The human SMO (hSMO) cDNA was first cloned and characterized by Wang et al3), and the recombinant SMO protein has been used to elucidate the properties of this enzyme.4-7) The substrate specificity of SMO appears to be limited and distinct from APAO, which catalyzes a number of polyamines and their analogues, some of which are inhibitors of SMO. A few compounds have been reported to exhibit substrate activity for SMO. N1-Ethyl-Spm (Et343) is degraded very efficiently by SMO to produce 34, while N1-Acetyl-Spm (Ac343) is degraded weakly.4) (S,S)-α,ω-Dimethyl-Spm served as an excellent substrate for SMO, compared to Spm, based on kcat/Km values.8) These findings using N,N’-dialkylated polyamines suggest SMO is capable of oxidizing other polyamine analogues. In this study, the substrate activities of a series of linear polyamines with a terminal primary amine were examined.
Materials and Methods
Chemicals
Bis(3-aminopropyl)amine (33, norspermidine) and N,N’-Bis(3-aminopropyl)-1,3-diaminopropane (333) were purchased from Aldrich (Tokyo, Japan) and their hydrochloride salts were prepared and used after recrystallization from aqueous ethanol. Spm tetrahydrochloride was purchased from Tokyo Chemical Industries (Tokyo, Japan). Spermidine (34) trihydrochloride was purchased from Aldrich. The following compounds were prepared according to previously described methods9-11) : Homospermidine (44) trihydrochloride, 1,9-Diamino-4-azanonae (35) trihydrochloride, 1,10-Diamino-4-azadecane (36) trihydrochloride, 1,12-Diamino-4-azadodecane (38) trihydrochloride, 1,13-Diamino-4,10-diazatridecane (353) tetrahydrochloride, 1,14-Diamino-4,11-diazatetradecane (363) tetrahydrochloride, 1,16-Diamino-4,13-diazahexadecane (383) tetrahydrochloride, 1,18-Diamino-4,15-diazaoctadecane (3103) tetrahydrochloride, 1,20-Diamino-4,17-diazaeicosane (3123) tetrahydrochloride, Thermospermine (334) tetrahydrochloride, 1,13-Diamino-4,9-diazatridecane (344, homospermine) tetrahydrochloride, 1,13-Diamino-5,9-diazatridecane (434) tetrahydrochloride, 1,14-Diamino-5,10-diazatetradecane (444) tetrahydrochloride, N1-Acetylspermine (Ac343) trihydrochloride, Diacetylspermine (DA343) dihydrochloride, N1-Butylspermine (Bu343) tetrahydrochloride, N1,N12-Diethylspermine (DiEt343) tetrahydrochloride, N1,N14-Diethyl-1,14-diamino-5,10-diazatetradecane (DiEt444) tetrahydrochloride, 1,15-Diamino-4,8,12-triazapentadecane (3333) pentahydrochloride, 1,16-Diamino-4,8,12-triazahexadecane (3334) pentahydrochloride, 1,16-Diamino-4,8,13-triazahexadecane (3343) pentahydrochloride, 1,17-Diamino-4,8,13-triazaheptadecane (3344) pentahydrochloride, 1,17-Diamino-4,9,13-triazaheptadecane (3434) pentahydrochloride, 1,17-Diamino-4,9,14-triazaheptadecane (3443) pentahydrochloride, 1,17-Diamino-5,9,13-triazaheptadecane (4334) pentahydrochloride, 1,18-Diamino-4,9,14-triazaoctadecane (3444) pentahydrochloride, 1,18-Diamino-5,9,14-triazaoctadecane (4344) pentahydrochloride, 1,19-Diamino-5,10,15-triazanonadecane (4444) pentahydrochloride, N1,N16-Diacetyl-1,16-diamino-4,8,13-triazahexadecane (DiAc3343) trihydrochloride, N1,N19-Diethyl-1,19-diamino-5,10,15-triazanonadecane (DiEt4444) pentahydrochloride, N1,N17-Diethyl-1,17-diamino-5,9,13-triazaheptadecane (DiEt4334) trihydrochloride, 1,19-Diamino-4,8,12,16-tetraazanonadecane (33333) hexahydrochloride, 1,20-Diamino-4,8,13,17-tetraazaeicosane (33433) hexahydrochloride, 1,21-Diamino-4,9,13,18-tetraazahenicosane (34343) hexahydrochloride. All other reagents and organic solvents were of commercial analytical grade.
Preparation of hSMO enzyme solution
The BL21 (DE3) strain of Escherichia coli. containing the pET15b/PAOh1/SMO plasmid5) was cultured. Following IPTG induction of PAOh1/SMO protein expression, the cells were sonicated in a 10 mM Tris-HCl buffer (pH 7.8) containing 1 mM ethylenediaminetetraacetic acid (EDTA) and 20 μM dithiothreitol (DTT). After 105,000 x g centrifugation at 4°C for 1 hr, the supernatant was dialyzed against the same buffer. Aliquots were stored at −80°C and used as the hSMO enzyme source. Protein was determined using Coomassie Brilliant Blue and bovine serum albumin for calibration.
Determination of hSMO activity
hSMO activity was assayed by measuring the amount of H2O2 generated by the reaction. The standard incubation mixture (final volume, 100 μl) contained the enzyme solution, 0.25 mM Spm, 0.56 mM aminoguanidine, 0.036 mM pargyline and 1 mM EDTA, 0.04 μg horseradish peroxidase, 0.1 mg homovanillic acid in 0.1 M Tris-HCl buffer (pH 9.0). Before the addition of homovanillic acid and Spm, the mixtures were preincubated for 5 min at 37°C to remove endogenous substrates of H2O2-producing enzymes, and where used, MDL72527 was added at 0.25 mM. After preincubation, homovanillic acid and Spm were added, the mixtures were incubated (0 – 120 min) at 37°C and the reaction was stopped by the addition of 100 μl of 20% trichloroacetic acid solution. The reaction mixtures were centrifuged and the resulting fluorescence of homovanillic acid dimer was analyzed by ion-pair reversed phase HPLC. The ion-pair reversed phase HPLC conditions were as follows: column, TOSOH ODS-80TM (4.6 mm ϕ × 150 mm); isocratic elution solution, acetonitrile : water (25 : 75) containing 5 mM tetrabutylammonium bromide and 0.1% trifluoroacetic acid; flow rate, 0.7 ml/min; post-column mixing solution, acetonitrile : water (5 : 95) containing 0.6% 2-aminoethanol; flow rate, 0.5 ml/min and fluorescence detection, Ex 315 nm and Em 425 nm. Aliquots of sample solutions were injected, and quantitation was performed based on peak heights in comparison with the homovanillic acid dimer standard.
The measurement of product amines in the reaction mixture was performed according to a method for polyamine analysis using o-phthalaldehyde (OPA)-post label ion-exchange HPLC.12) Measurement of substrate activity and pH dependency were performed by the substitution of 0.25 mM tested compound with Spm and Tris-HCl buffer at pH 9.0, 8.5, 8.0, 7.5, 7.0 in the standard reaction mixture described above. The Km and Vmax values for the enzyme with the indicated substrates were estimated using the Lineweaver-Burk transformation of the Michaelis-Menten kinetic equation.
Results and Discussion
Measurement of hSMO activity using an HPLC-based method
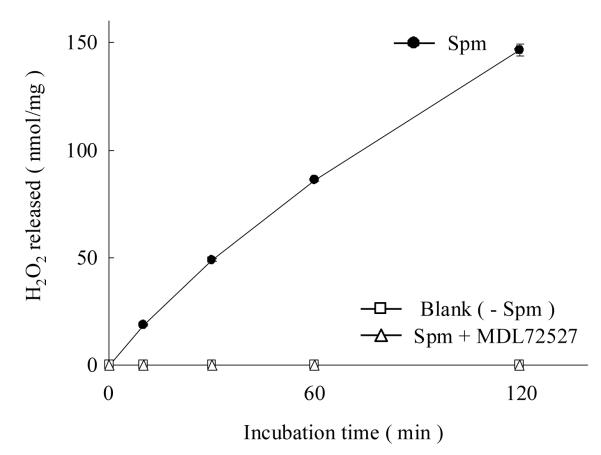
For the measurement of hSMO activity, the H2O2 stoichiometrically released by the polyamine-hSMO reaction was monitored. H2O2 is usually measured fluorometrically using homovanillic acid.3). However, methods using fluorescence spectrophotometers often require relatively high amounts of sample and encounter interference by contamination with other fluorescent substances. In this study, the H2O2 released by reactions with homovanillic acid and HRP, and the resulting fluorescent dimer, were separated and determined by HPLC. Temporal changes in the H2O2 released by hSMO oxidation of Spm are shown in Fig. 1. Only minimal amounts of activity were observed in the absence of Spm or after preincubation with MDL72527, which inhibited the release of H2O2 perfectly. Extracts from E. coli without the pET15b/PAOh1/SMO plasmid had no hSMO-like activity (data not shown). These results demonstrate that this HPLC method is useful for screening potential hSMO substrates.
Fig 1. Measurement of H2O2 release from Spm by hSMO.
The experiment was performed in triplicate with the data presented as mean ± standard deviation. The experiment was performed in the absence and presence of 0.25 mM Spm or preincubated with MDL72527.
Substrate activity of polyamines for hSMO
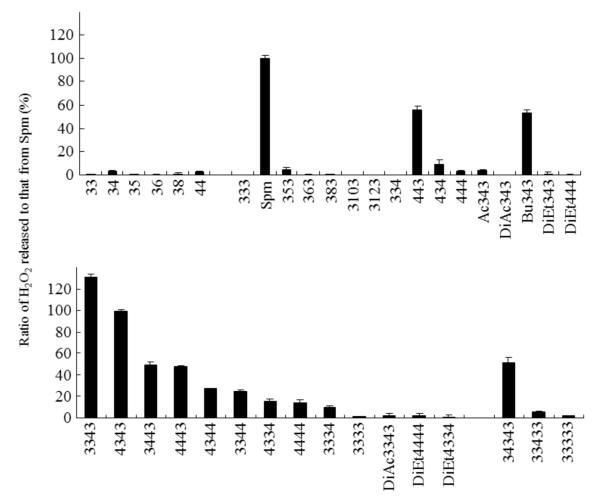
A series of triamines, tetramines, pentamines, hexamines and their derivatives, with different methylene chain intervals, were tested for their substrate activities for hSMO under the conditions described in Materials and Methods. The results are summarized in Fig. 2. All triamines were found to be poor hSMO substrates, resulting in the release of scarce amounts of H2O2. With respect to tetramines, the natural substrate Spm was the most active substrate (100 %), followed by homospermine (443) and N1-butylated Spm (Bu343), for which the activity was ≈ 60 %. The other tetramines (434, 353, and 444) exhibited low activity (< 10 %). Ac343 activity was very low compared to Spm, which is consistent with a previous report by Vujcic et al.4) N1,N12-diacetylSpm (DiAc343) did not exhibit substrate activity for hSMO. Diethylated tetramines of terminal primary amines (DiEt343, DiEt444) also exhibited very low activity, similar to the report by Vujcic et al (2002). In contrast, pentamines, with the exception of 3333, exhibited marked activity: 3343 (> 120 %), 4343 (100 %), 3443 (≈ 50%), 4443 (≈ 50 %), 4344 and 3344 (≈ 30 %), 4334, 4444 and 3334 (≈ 20 %). Diethylated or diacetylated pentamines of terminal primary amines exhibited very low activity. Of the three hexamines tested, only 34343 exhibited a significant amount of activity (≈ 50 %). In summary, hSMO appeared to recognize pentamines with terminal aminopropyl as well as aminobutyl groups, suggesting an unknown pentamine cleavage reaction catalyzed by hSMO.
Fig. 2. Substrate activities of polyamines.
Polyamines (0.25 mM) were incubated with hSMO for 10 min under the conditions described in Materials and Methods. The experiment was performed in triplicate, the data were normalized to that of Spm, and presented as mean ± standard deviation.
Measurement of amines released from pentamines by hSMO
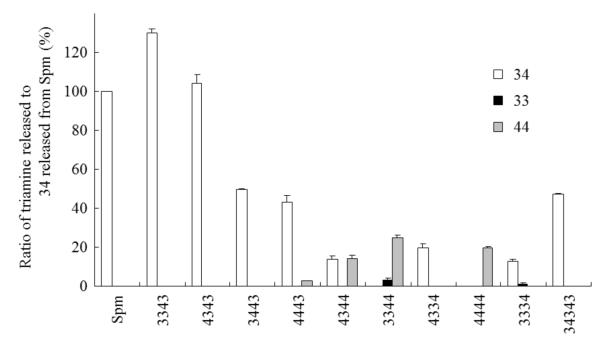
To elucidate the cleavage reaction, product amines were analyzed by OPA-post label ion-exchange HPLC method. Only triamines (no diamines or tetramine) were detected in enzyme reactions with pentamines, and the amounts of the resulting triamines were nearly equal to that of released H2O2. The results are summarized in Fig. 3. A larger amount of 34 was liberated from 3343 than 34 from Spm itself, indicating the production of 34 through oxidative degradation at the central aza and probably an aminopropanal compound corresponding to 33. The amount of 34 liberated from 4343 was similar to that from Spm, indicating degradation at the central aza to produce 34 and probably an aminopropanal and/or an aminobutanal corresponding to 34. The amount of 34 liberated from 3443 was approximately half of that from Spm, indicating degradation at the central aza to produce 34 and probably a butanal corresponding to 34. The products from 4443 were 34 and a small amount of 44, indicating cleavage at the central aza to preferentially form 34. The products from 4344 were nearly equal amounts of 34 and 44, and those from 3344 were mainly 44 with a small amount of 33. The similar amounts of 34 from 4334 to 44 from 4444 suggest that hSMO recognized terminal 34 and 44 equally. Products from 3334 were 34 and a small amount of 33. The product from 34343 was a significant amount of 34 only. These results demonstrated that hSMO catalyzed the oxidative degradation of pentamines at a preferred side bond of the central aza to produce triamines depending on the pentamine sequence. Pentamines with a sequence of xx43 preferentially released 34, with triamine release decreasing in the following order: 34 from xx43, 44 from xx44 or 34 from xx34, and 33 from 3344.
Fig 3. Measurement of triamines released from indicated polyamines by hSMO.
Indicated polyamines (0.25 mM) were incubated with hSMO for 30 min. The experiment was performed in triplicate, the data were normalized to 34 released from Spm, and presented as mean ± standard deviation.
Significant difference in pH dependency of hSMO degradation of tetramines and pentamines
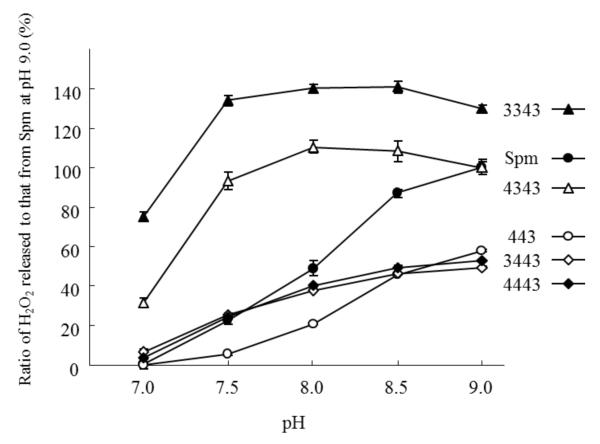
The effect of pH on H2O2 release by hSMO was examined using tetramines (Spm and 443) and pentamines (3343, 4343, 3443 and 4443) (Fig. 4). The two tetramines showed similar pH dependency curves, with the peak H2O2 release at pH 9.0 followed by a marked decrease until pH 7.0. On the other hand, the four pentamines exhibited similar pH dependency curves, with 3343 and 4343 plateauing between pH 7.5 to 9.0 and moderately decreasing over this pH range. The differences in pH dependency curves are potential indicators of the basicity of the nitrogen at the hSMO cleavage site of polyamines. hSMO seemed to favor a non-protonated secondary nitrogen at the catalytic site. Additional information on the activity of hSMO towards Spm and 3343, kinetic data at pH 7.0 and 9.0, is provided in Table 1. Both the Km and Vmax values for Spm were markedly different at pH 7.0 and 9.0, whereas those for 3343 were largely unchanged. The Km values for Spm were higher than those for 3343 at both pH levels, and were consistent with 3343 being a better hSMO substrate than Spm. At pH 7.0, the ratio of Kms (Spm / 3343) was about twice that observed at pH 9.0, suggesting that the affinity of Spm for hSMO decreases more rapidly than 3343.
Fig. 4. Tetramines and pentamines exhibit distinct patterns of pH-dependency.
H2O2 release was determined using the indicated polyamines (0.25 mM) as a substrate at pH 7.0 - 9.0. The experiment was performed in triplicate, the data were normalized to that of Spm at pH 9.0, and presented as mean ± standard deviation.
Table 1.
| pH 9.0 | pH 7.0 | |||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μM | nmol/min/mg | μM | nmol/min/mg | |
| Spm | 18 | 2.5 | 1200 | 0.2 |
| 3343 | 1.3 | 3.3 | 45 | 2.1 |
Limited data on pKa values for linear polyamines have been reported, however, there is no published data on the pentamines used here. pKa1 values (the weakest basic amine) for a series of linear polyamines were reported as 4.23 for triamine 22, 3.27 for tetramine 222, 2.92 for pentamine 2222, 7.69 for triamine 33, 7.21 for tetramine 333, 7 for hexamine 33333.13) The data indicated that there is an inverse relationship between polyamines size and pKa1. The pKa1 of the secondary amine of Spm has been reported to be 8.014, 15), however, the pKa1 values of the pentamines used here may be lower than 8.0. Adachi et al. reported that the optimal pH for the oxidation of Spm by SMO was 8.3, based on the kcat/Km-pH profile, suggesting a tri-protonated form of Spm around the pH. 16) These data are consistent with the contention that hSMO catalyzes the oxidative cleavage of the C-N bond at a non-protonated nitrogen.
The cellular distribution of SMO was reported to be present in the cytosol and nucleus.17) If 3343 exists in cells, hSMO can degrade 3343 at cytosolic pH and produce H2O2 and aldehydes, which may act as oxidative signaling molecules in the cells. The existence of 3343 in bacteria18) and spider venoms19) has been reported. It will be interesting to determine if 3343 is present in mammalian cells in future studies.
References and Notes
- 1).Tabor CW. Tabor H. Polyamines. Annu. Rev. Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2).Casero RA. Pegg AE. Polyamine catabolism and disease. Biochem. J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Wang Y, Devereux W, Woster PM, Murray-Stewart T, Hacker A, Casero RA. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370–5373. [PubMed] [Google Scholar]
- 4).Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem. J. 2002;367:665–675. doi: 10.1042/BJ20020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA. Properties of purified recombinant human polyamine oxidase PAOh1/SMO. Biochem. Biophys. Res. Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 6).Devereux W, Wang Y, Murray-Stewart T, Hacker A, Smith R, Frydman B, Valasinas AL, Reddy VK, Marton LJ, Ward TD, Woster PM, Casero RA. Induction of the PAOh1/SMO polyamine oxidase by polyamine analogues in human lung carcinoma cells. Cancer Chemother. Pharmacol. 2003;52:383–390. doi: 10.1007/s00280-003-0662-4. [DOI] [PubMed] [Google Scholar]
- 7).Häkkinen MR, Hyvönen MT, Auriola S, Casero RA, Vepsäläinen J, Khomutov AR, Alhonen L, Keinänen TA. Metabolism of N-alkylated spermine analogues by polyamine and spermine oxidase. Amino Acids. 2010;38:369–381. doi: 10.1007/s00726-009-0429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Hyvönen MT, Keinänen TA, Cerrada-Gimenez M, Sinervirta R, Grigorenko N, Khomtov AR, Vepsäläinen J, Alhonen L, Jänne J. Role of hypusinated eukaryotic translation initiation factor 5A in polyamine depletion-induced cytostasis. J. Biol. Chem. 2007;282:34700–34706. doi: 10.1074/jbc.M704282200. [DOI] [PubMed] [Google Scholar]
- 9).Samejima K, Takeda Y, Kawase M, Okada M, Kyogoku Y. Synthesis of 15N-enriched polyamines. Chem. Pharm. Bull. 1984;32:3428–3435. doi: 10.1248/cpb.32.3428. [DOI] [PubMed] [Google Scholar]
- 10).Niitsu M, Samejima K. Synthesis of a series of linear pentaamines with three and four methylene chain intervals. Chem. Pharm. Bull. 1986;34:1032–1038. [Google Scholar]
- 11).Takao K, Ozawa T, Shibata S, Wada M, Sugita Y, Shirahata A, Samejima K. Formation of spermidine or norspermidine from synthetic diacetylpolyamines by acetylpolyamine oxidase in cultured cells. Biol. Pharm. Bull. 2007;30:2389–2393. doi: 10.1248/bpb.30.2389. [DOI] [PubMed] [Google Scholar]
- 12).Shirahata A, Takahashi N, Beppu T, Hosoda H, Samejima K. Effects of inhibitors of spermidine synthase and spermine synthase on polyamine synthesis in rat tissues. Biochem. Pharmacol. 1993;45:1897–1903. doi: 10.1016/0006-2952(93)90449-7. [DOI] [PubMed] [Google Scholar]
- 13).Bencini A, Bianchi A, Garcia-Espaňa E, Micheloni M, Ramirez JA. Proton coordination by polyamine compounds in aqueous solution. Coord. Chem. Rev. 1999;188:97–156. [Google Scholar]
- 14).Takeda Y, Samejima K, Nagano K, Watanabe M, Sugeta H, Kyogoku Y. Determination of protonation sites in thermospermine and in some other polyamines by 15N and 13C nuclear magnetic resonance spectroscopy. Eur. J. Biochem. 1983;130:383–389. doi: 10.1111/j.1432-1033.1983.tb07164.x. [DOI] [PubMed] [Google Scholar]
- 15).Weisell J, Hyvönen MT, Vepsäläinen J, Alhonen A, Keinänen TA, Khomtov AR, Soininen P. Novel isosteric charge-deficient spermine analogue – 1,12-diamino-3,6,9-triazadodecane: synthesis, pKa measurement and biological activity. Amino Acids. 2010;38:501–507. doi: 10.1007/s00726-009-0409-6. [DOI] [PubMed] [Google Scholar]
- 16).Adachi MS, Juarez PR, Fitzpatrick PF. Mechanistic studies of human spermine oxidase: kinetic mechanism and pH effects. Biochemistry. 2010;49:386–392. doi: 10.1021/bi9017945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Murray-Stewart T, Wang Y, Goodwin A, Hacker A, Meeker A, Casero RA. Nuclear localization of human spermine oxidase isoforms – possible implications in drug response and disease etiology. FEBS J. 2008;275:2795–806. doi: 10.1111/j.1742-4658.2008.06419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Oshima T. Unique polyamines produced by an extreme thermophile, Thermus thermophiles. Amino Acids. 2007;33:367–372. doi: 10.1007/s00726-007-0526-z. [DOI] [PubMed] [Google Scholar]
- 19).Tzouros M, Chesnov S, Bienz S, Hesse M, Bigler L. New linear polyamine derivatives in spider venoms. Toxicon. 2005;46:350–354. doi: 10.1016/j.toxicon.2005.04.018. [DOI] [PubMed] [Google Scholar]