Abstract
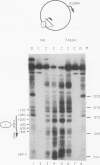
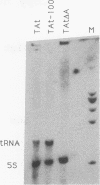
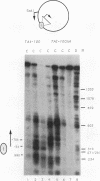
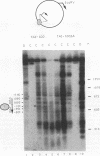
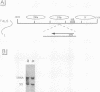
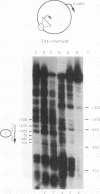
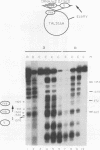
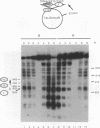
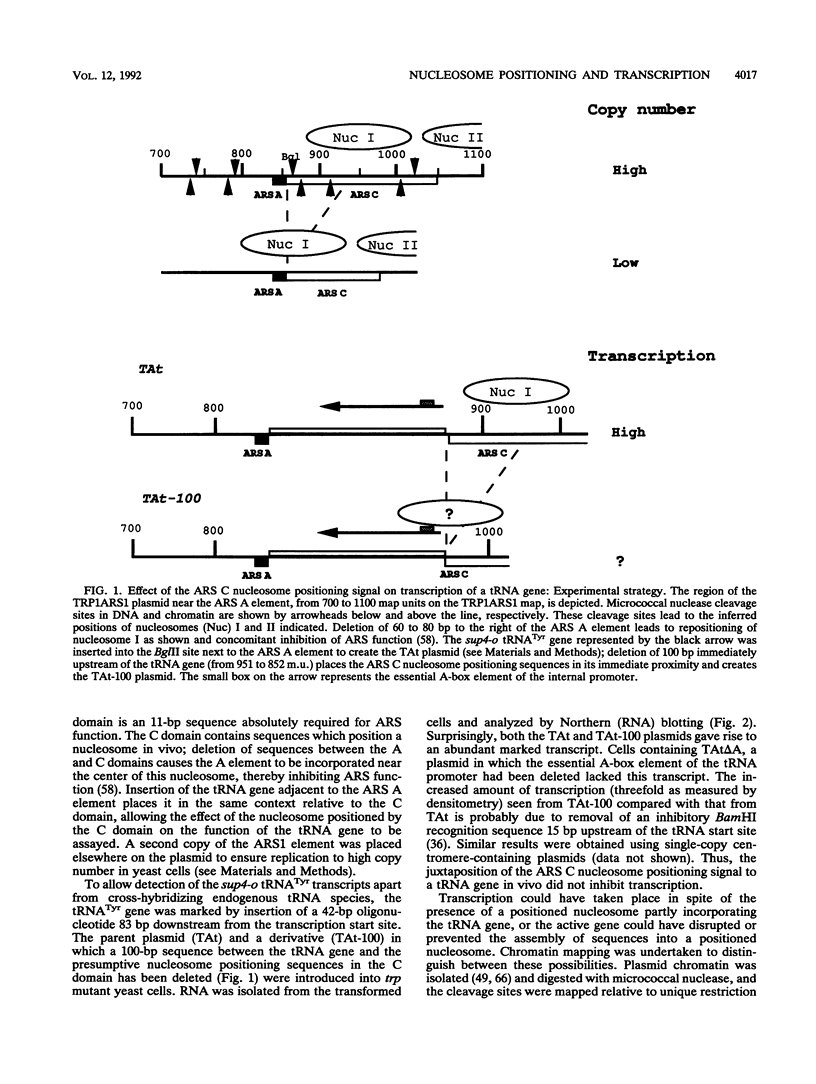
Incorporation into a positioned nucleosome of a cis-acting element essential for replication in Saccharomyces cerevisiae disrupts the function of the element in vivo [R. T. Simpson, Nature (London) 343:387-389, 1990]. Furthermore, nucleosome positioning has been implicated in repression of transcription by RNA polymerase II in yeast cells. We have now asked whether the function of cis-acting elements essential for transcription of a gene transcribed by RNA polymerase III can be similarly affected. A tRNA gene was fused to either of two nucleosome positioning signals such that the predicted nucleosome would incorporate near its center the tRNA start site and essential A-box element. These constructs were then introduced into yeast cells on stably maintained, multicopy plasmids. Competent tRNA genes were transcribed in vivo and were not incorporated into positioned nucleosomes. Mutated, inactive tRNA genes were incorporated into nucleosomes whose positions were as predicted. This finding demonstrates that the transcriptional competence of the tRNA gene determined its ability to override a nucleosome positioning signal in vivo and establishes that a hierarchy exists between cis-acting elements and nucleosome positioning signals.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman E. J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2(8):1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Méchali M., Wolffe A. P. Competition between transcription complex assembly and chromatin assembly on replicating DNA. EMBO J. 1990 Feb;9(2):573–582. doi: 10.1002/j.1460-2075.1990.tb08145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Méchali M., Wolffe A. P. Transcription complex disruption caused by a transition in chromatin structure. Mol Cell Biol. 1991 Feb;11(2):655–665. doi: 10.1128/mcb.11.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer T. K., Cordingley M. G., Wolford R. G., Hager G. L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991 Feb;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer T. K., Lefebvre P., Wolford R. G., Hager G. L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992 Mar 20;255(5051):1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Majors J. An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res. 1989 Jan 11;17(1):171–183. doi: 10.1093/nar/17.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P. B., Rabindran S. K., Wu C. Heat shock-regulated transcription in vitro from a reconstituted chromatin template. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4109–4113. doi: 10.1073/pnas.88.10.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman L. W., Kramer R. A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Jun 10;258(11):7223–7227. [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992 Jan 25;20(2):273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988 Feb;2(2):150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigel A., Feldmann H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982;1(10):1245–1250. doi: 10.1002/j.1460-2075.1982.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Felts S. J., Weil P. A., Chalkley R. Transcription factor requirements for in vitro formation of transcriptionally competent 5S rRNA gene chromatin. Mol Cell Biol. 1990 May;10(5):2390–2401. doi: 10.1128/mcb.10.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988 Dec 23;55(6):1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5'-flanking region. Mol Cell Biol. 1989 Aug;9(8):3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988 Jul 25;16(14A):6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Hamer D. H., Roeder R. G. Stable transcription complex on a class III gene in a minichromosome. Mol Cell Biol. 1985 Jan;5(1):40–45. doi: 10.1128/mcb.5.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lowary P. T., Widom J. Higher-order structure of Saccharomyces cerevisiae chromatin. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8266–8270. doi: 10.1073/pnas.86.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Brechner T., Amon-Böhm E., Dingermann T. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989 Jun 23;244(4911):1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
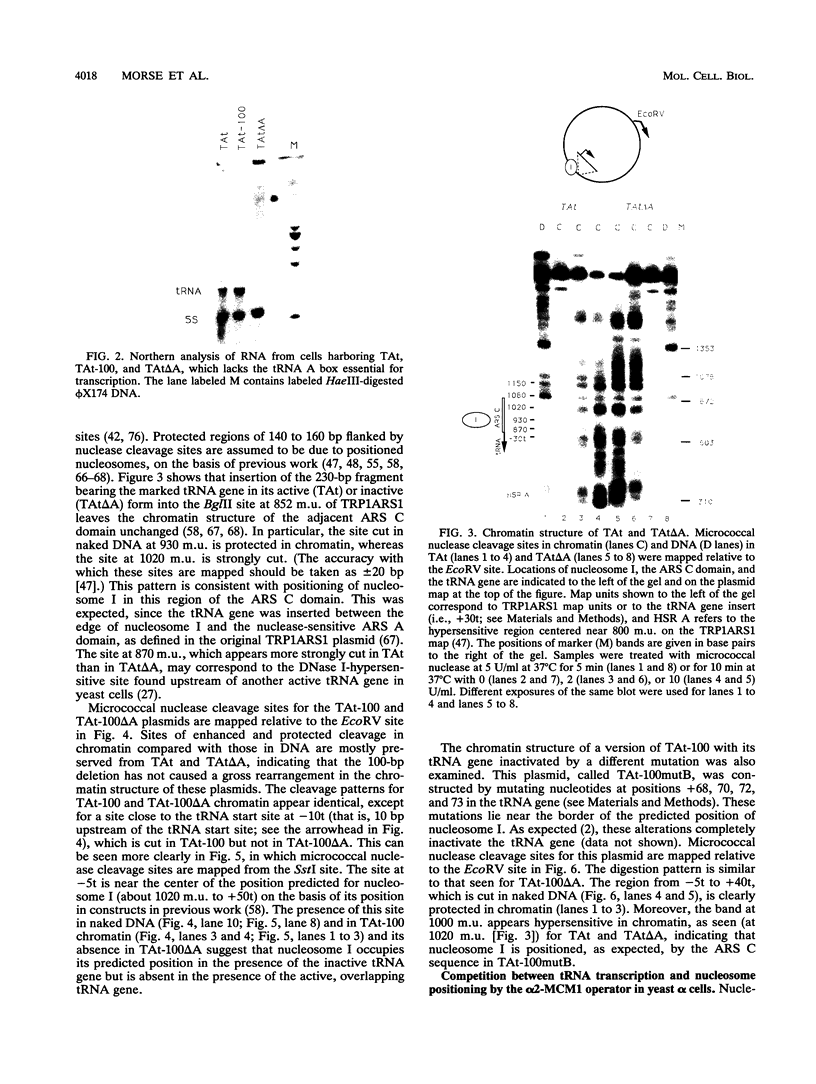
- Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987 Apr;7(4):1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989 Aug;8(8):2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Simpson R. T. DNA in the nucleosome. Cell. 1988 Jul 29;54(3):285–287. doi: 10.1016/0092-8674(88)90190-0. [DOI] [PubMed] [Google Scholar]
- Morse R. H. Topoisomer heterogeneity of plasmid chromatin in living cells. J Mol Biol. 1991 Nov 20;222(2):133–137. doi: 10.1016/0022-2836(91)90198-f. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988 Oct;7(10):3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Simpson R. T. Yeast minichromosomes. Methods Cell Biol. 1991;35:289–314. [PubMed] [Google Scholar]
- Schlissel M. S., Brown D. D. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984 Jul;37(3):903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Schnell R., Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183(1):59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Smerdon M. J., Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990 May 18;61(4):675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Hagenbüchle O., Zuniga M. C. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977 Jul;11(3):561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- Straka C., Hörz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991 Feb;10(2):361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Gouilloud E., Clarkson S. G. Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes Dev. 1989 Aug;3(8):1190–1198. doi: 10.1101/gad.3.8.1190. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Taylor M. J., Segall J. Characterization of factors and DNA sequences required for accurate transcription of the Saccharomyces cerevisiae 5 S RNA gene. J Biol Chem. 1985 Apr 10;260(7):4531–4540. [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Thoma F., Zatchej M. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell. 1988 Dec 23;55(6):945–953. doi: 10.1016/0092-8674(88)90240-1. [DOI] [PubMed] [Google Scholar]
- Tremethick D., Zucker K., Worcel A. The transcription complex of the 5 S RNA gene, but not transcription factor IIIA alone, prevents nucleosomal repression of transcription. J Biol Chem. 1990 Mar 25;265(9):5014–5023. [PubMed] [Google Scholar]
- Wilson C., Bellen H. J., Gehring W. J. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 1989 Feb;8(2):527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]