Abstract
Fetal prostate development is initiated by androgens and patterned by androgen dependent and independent signals. How these signals integrate to control epithelial cell differentiation and prostatic bud patterning is not fully understood. To test the role of beta-catenin (Ctnnb1) in this process, we used a genetic approach to conditionally delete or stabilize Ctnnb1 in urogenital sinus (UGS) epithelium from which the prostate derives. Two opposing mechanisms of action were revealed. By deleting Ctnnb1, we found it is required for separation of UGS from cloaca, emergence or maintenance of differentiated UGS basal epithelium and formation of prostatic buds. By genetically inducing a patchy subset of UGS epithelial cells to express excess CTNNB1, we found its excess abundance increases Bmp expression and leads to a global impairment of prostatic bud formation. Addition of NOGGIN partially restores prostatic budding in UGS explants with excess Ctnnb1. These results indicate a requirement for Ctnnb1 in UGS basal epithelial cell differentiation, prostatic bud initiation and bud spacing and suggest some of these actions are mediated in part through activation of BMP signaling.
Keywords: prostate, Bmp, Ctnnb1, urothelium, urogenital sinus, cloaca, rectourethral fistula, basal epithelium, noggin, androgen, lower urogenital tract
Introduction
Mouse prostate development initiates from a subcompartment of the lower urogenital tract known as the definitive urogenital sinus (UGS). The UGS must complete a series of developmental events to initiate a normal prostate. One requirement is appropriate UGS epithelial differentiation. The mouse UGS forms from cloacal endoderm at around mouse embryonic day (E)13. Primitive UGS epithelium then differentiates into a multilayered transitional epithelium consisting of basal, intermediate and superficial layers (Abler et al., 2011a). It is generally believed formation of histologically normal prostatic ducts in mice requires basal epithelium, as evidenced by the abnormal prostate ductal phenotype in mice deficient in the basal epithelial cell transcription factor transformation related protein 63 (Trp63) (Kurita et al., 2004; Signoretti et al., 2005; Signoretti et al., 2000). However, there is conflicting evidence about whether basal epithelium is required for the earliest stage of prostate ductal development, prostatic bud formation. Kurita et al. (2004) observed in the formation of epithelial evaginations that histologically resembled prostatic buds in mice lacking basal epithelium. However, Signoretti et al (2005; 2000) reported that prostatic buds do not form in the absence of basal epithelium. It is further unknown how primitive UGS epithelium, marked by TRP63 protein undergoes maturation to form a mature keratin 14 (KRT14)-positive UGS basal epithelium.
Prostatic development also requires appropriate androgen receptor (AR) activation. Testicular androgens activate AR signaling in UGS mesenchyme, which induces prostatic bud formation in UGS epithelium (Cunha et al., 1987; Lasnitzki and Mizuno, 1980). Mouse prostatic buds emerge as solid basal epithelial outgrowths and later arborize and differentiate into a pseudostratified prostate ductal epithelium consisting of neuroendocrine, basal and luminal epithelial cells. Mouse and rat prostatic buds are patterned along two asymmetric axes (dorsoventral, craniocaudal) and one symmetrical axis (mediolateral) to give rise to a ductal network organized into ventral, anterior, dorsal and lateral prostate lobes (Cunha et al., 1987; Timms et al., 1994). In turn, UGS epithelium reciprocally interacts with UGS stroma to pattern stromal architecture (Cunha et al., 1992; Hayward et al., 1998). How prostatic buds are patterned is not fully known.
Though many questions remain regarding mechanisms of normal prostate development, there is evidence that CTNNB1 is likely to participate in prostatic bud formation. Ctnnb1 has been identified in almost every structure that undergoes a budding program; its activation is necessary and/or sufficient for specification of hair follicle buds (Gat et al., 1998; Lo Celso et al., 2004), mammary gland buds (Faraldo et al., 2006), feather buds (Noramly et al., 1999; Widelitz et al., 2000) and tooth buds (Liu et al., 2008). Potential CTNNB1-stabilizing WNT ligands are abundant in male mouse UGS during epithelial differentiation, prostatic bud formation and prostatic branching morphogenesis (Mehta et al., 2011; Yu et al., 2009; Zhang et al., 2006). CTNNB1–responsive Axin2 and lymphoid enhancer binding factor 1 (Lef1) mRNAs are present in basal epithelial cell precursors of the definitive UGS (Abler et al., 2012) and later localize to developing prostatic bud tip epithelium (Mehta et al., 2011; Wu et al., 2011). While these data indicate a role for CTNNB1 in prostate development, there is growing controversy over its exact function. Some evidence supports a role for CTNNB1 in promoting prostatic budding and branching (Joesting et al., 2008), especially during bud specification when CTNNB1 is required (Simons et al., 2012). Other evidence supports an inhibitory role, especially during prostatic ductal branching morphogenesis (Wang et al., 2008; Yu et al., 2009).
In this manuscript, we examined the impact of genetically deleting or inducing excess CTNNB1 expression in UGS epithelium. We specifically sought to identify the CTNNB1 role in UGS epithelium because we previously identified a sexually dimorphic and male dominant pattern of CTNNB1-responsive gene expression in this tissue compartment (Mehta et al., 2011). We show that Ctnnb1 is needed in UGS epithelium for prostatic bud formation and for formation of UGS basal epithelium. In contrast, we found that expression of excess CTNNB1 in a patchy subset of UGS epithelial cells increases BMP ligand expression in the same cells, increases BMP signaling in surrounding cells, and globally inhibits prostatic bud formation. The latter role positions CTNNB1 to participate in the mechanism by which prostatic bud spacing intervals are established. Because it fulfills at least two roles during prostate development, titration of CTNNB1 signaling is crucial for establishing the normal pattern and number of mouse prostatic buds.
Materials and Methods
Mice
Mice were housed as described previously (Mehta et al., 2011). All procedures were approved by the University of Wisconsin Animal Care and Use Committee. Mice carrying the R26R allele (Gt(ROSA)26Sortm1Sor), Ctnnb1 exon 2–6 targeted deletion loss-of-function (LOF) allele (Ctnnb1tm2Kem) or the Ctnnb1 exon 3 targeted deletion gain-of-function (GOF) allele (Ctnnb1tm1Mmt) were mated to wild type mice or to mice carrying Shhcre (Shhtm1(EGFP/cre)Cjt) or ShhcreERT2 (Shhtm2(cre/ERT2)Cjt). To induce ShhcreERT2, dams were injected with sterile corn oil (2.5 mL/kg i.p. maternal dose) containing 10% ethanol, tamoxifen (25 mg/kg maternal dose, Sigma #T56482, St. Louis MO) and progesterone (18.75 mg/kg maternal dose, Watson #NDC0591-3128-79, Corona CA) and dams were euthanized by CO2 asphyxiation. Embryos of the following genotypes were assessed alone or together with their phenotypically normal paired littermate controls: Shhcre/+;R26R, Shh+/+;R26R, ShhcreERT2/+;R26R, Shh+/+;R26R, conditional Ctnnb1 loss-of-function (cLOF) Ctnnb1cLOF (Shhcre/+;Ctnnb1tm2Kem/tm2Kem) and its paired control (Shhcre/+;Ctnnb1tm2Kem/+), inducible Ctnnb1 loss-of-function (iLOF) Ctnnb1iLOF (ShhcreERT2/+;Ctnnb1tm2Kem/tm2Kem) and its paired control (Shh+/+;Ctnnb1tm2Kem/tm2Kem) or Ctnnb1iLOF;R26R (ShhCreERT2/+;Ctnnb1tm2Kem/tm2Kem;R26R+) and its paired control (ShhCreERT2/+; Ctnnb1tm2Kem/+R26R+), inducible Ctnnb1 gain-of-function (iGOF) Ctnnb1iGOF (ShhcreERT2/+;Ctnnb1 tm1Mmt/tm1Mmt) and its paired control (Shh+/+; Ctnnb1tm1Mmt/tm1Mmt) (Brault et al., 2001; Harada et al., 1999; Harfe et al., 2004; Soriano, 1999). Ctnnb1 tm1Mmt mice were from Dr. Makoto Mark Taketo, Kyoto University, and all other mice were from The Jackson Laboratory. The morning of copulatory plug identification was considered E0.5. Genotyping primer sequences are listed in Supplementary Material Table 1.
Assay for β-galactosidase activity
LacZ-dependent β-galactosidase activity was assessed as described previously (Cheng et al., 1993) with modifications described byMehta et al. (2011).
Immunohistochemistry (IHC)
For most IHC analyses, UGS tissues were fixed in 4% paraformaldehyde, dehydrated in ethanol, cleared in xylene, infiltrated with paraffin and stained as described previously (Mehta et al., 2011). To preserve color intensity in LacZ-stained samples, tissues were cleared in XS-3 xylene substitute (Statlab, McKinney, TX) prior to paraffin infiltration. Antibodies are listed in Supplementary Material Table 2. Labeling of TRP63 and CTNNB1 in the same tissue section required using primary antibodies from the same mouse host. The first primary and secondary antibody was added in sequence, tissues were then blocked with unlabeled monovalent anti-mouse IgG and the second primary and secondary antibody were added in sequence. Immunolabeled tissues were counterstained with 4',6-diamidino-2-phenylindole, dilactate (DAPI) and mounted in anti-fade media (phosphate-buffered saline containing 80% glycerol and 0.2% n-propyl gallate).
In situ hybridization (ISH)
Detailed protocols are available at www.gudmap.org and were described previously (Abler et al., 2011b). Primer sequences for generating PCR-amplified probe templates are listed in Supplementary Material Table 3, except for Bmp4 (Feng et al., 1995) and Fgf10 (Bellusci et al., 1997), which were generated from plasmid DNA. Some ISH-stained tissues were also immunolabeled using previously described protocols (Abler et al., 2011b; Keil et al., 2012). The staining pattern for each hybridized riboprobe was assessed in at least three litter-independent mice per genotype. Control and mutant tissues were processed together in the same tubes and as a single experimental unit to allow for qualitative comparisons among biological replicates and between genotypes or treatment groups.
UGS organ culture
Dissected UGSs were placed on 0.4 µm Millicell-CM filters and cultured as described previously (Vezina et al., 2008) in media containing 5α-dihydrotestosterone (DHT, diluted from an ethanol stock solution to a 10 nM final media concentration). The following supplements were added alone or in combination to organ culture media: 4-hydroxytamoxifen (Sigma #H6278, >70% Z-isomer, dissolved in ethanol and diluted in media to 1 µM 4-hydroxytamoxifen and 0.1% ethanol), or recombinant NOGGIN protein (R&D Systems, dissolved in saline and diluted in media to 1 µg/ml). Media and supplements were changed every 2 d.
Scanning Electron Microscopy
Scanning electron microscopy was conducted as described previously (Lin et al., 2003) at the University of Wisconsin Biological and Biomaterials Preparation, Imaging and Characterization Laboratory. Specimens were imaged from multiple angles to visualize budding across the entire surface of the UGS epithelium.
Statistics
All experimental groups consisted of three to five UGSs from at least three independent litters. Results are reported as mean ± standard error of the mean (SE). Prostatic bud number per UGS was calculated by averaging counts from three individuals blinded to experimental conditions. Only prostatic bud stalks were counted; a bud with one or more branches was counted as one bud. The percentage of UGS cells expressing CTNNB1, Ki67, KRT14 or combinations thereof was determined by counting the labeled cell fraction in a region bounded cranially by the bladder neck and caudally by the ejaculatory duct in three nonadjacent sections per UGS. The Companion to Applied Regression (CAR) package for R (version 2.13.1) was used for statistical analyses (Fox and Weisberg, 2011). Student’s t-test was conducted on untransformed data that passed Levene’s test for homogeneity of variance and appeared to be normally distributed.
Results
Ctnnb1 deletion from early-stage mouse UGS epithelium (UGE) impairs cloacal division, prostate field specification, and formation of prostatic buds and KRT14+ basal epithelium
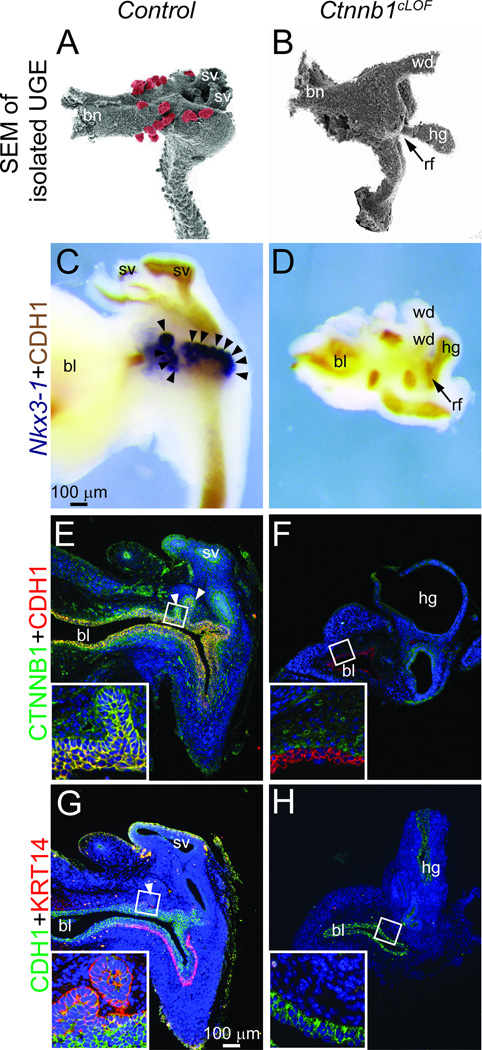
In an earlier study, we observed that CTNNB1-responsive Axin2 and Lef1 mRNAs are more abundant in male compared to female UGE and are particularly abundant in male prostatic bud tip epithelium (Mehta et al., 2011). This fact led us to hypothesize that prostatic bud formation requires CTNNB1 signaling specifically in UGE. To test this possibility, we generated a conditional Ctnnb1 loss of function (cLOF) mutant using the Shhcre knock-in allele (Harfe et al., 2004) which is selectively expressed in UGE but not elsewhere in the UGS (supplementary material Fig. S1A–C). On its own, a Shh knock-in allele does not interfere with prostatic bud formation (supplementary material Fig. S2). Effective Shhcre-mediated recombination is supported by the loss of detectable CTNNB1 protein in E17.5 Ctnnb1cLOF mutant UGE (Fig. 1). Control and Ctnnb1cLOF mutant male UGE morphology was assessed by enzymatically digesting UGSs with trypsin, removing mesenchyme and visualizing the underlying UGE by scanning electron microscopy (Fig. 1A–B). Ctnnb1cLOF mouse UGEs are smaller than those of control littermates, often lack a pelvic urethra and are attached to the hindgut by a fistulous connection (a rectourethral fistula or anal atresia phenotype). A requirement for Ctnnb1 in prostate specification, prostatic bud initiation or both is suggested by an epithelial bud deficiency in Ctnnb1cLOF UGEs at a stage when numerous buds are visible in control UGEs. Intact E17.5 control and Ctnnb1cLOF UGSs were then stained for Nkx3-1 to determine whether Ctnnb1cLOF mouse prostate development arrests at the prostate specification or bud initiation stage. Nkx3-1 marks the prostate field at E15.5, approximately one day prior to prostatic bud formation in control UGS (Keil et al., 2012; Sciavolino et al., 1997) but is undetectable in Ctnnb1cLOF UGSs (Fig. 1C–D), suggesting a failure to undergo prostate specification.
Fig. 1. Ctnnb1 deletion from early-stage mouse UGS epithelium (UGE) impairs cloacal division, prostate field specification and formation of prostatic buds and KRT14+ basal epithelium.
(A–B) Electron micrographs of E17.5 Shhcre/+;Ctnnb1tm2Kem/+ (control) and Shhcre/+; Ctnnb1tm2Kem/tm2Kem (Ctnnb1cLOF) UGE after removal of overlying UGS stroma. Prostatic buds are pseudocolored red. Arrow indicates an inappropriate hindgut-UGE connection (rectourethral fistula, rf) in Ctnnb1cLOF mice. (C–D) E17.5 male UGSs were stained by ISH to visualize the prostatic bud marker Nkx3-1 (purple) and counterstained by IHC to visualize UGE marked by cadherin 1 (CDH1). (E–H) E17.5 UGS sections were immunofluorescently labeled to detect CTNNB1, CDH1 and the UGS basal epithelial cell marker KRT14. Nuclei were stained with DAPI. Staining patterns in each panel represent three males. Abbreviations are bl: bladder, bn: bladder neck, hg: hindgut, sv: seminal vesicle, wd: Wolffian duct. Arrowheads indicate prostatic buds.
The Ctnnb1cLOF mutant UGE is thinner than control UGE (Fig. 1E–F), suggestive of possible changes in epithelial composition. We specifically examined whether Ctnnb1cLOF mutants form UGS basal epithelium. Ctnnb1cLOF mutant UGE lacks detectable KRT14+ basal epithelium (Fig. 1G–H) even though this multi-cell layer forms at least two days earlier in control UGS (supplementary material Fig. S3). Together, these results indicate Ctnnb1 requirements in cloacal division, specification of the prostate field, formation of prostatic buds and formation or maintenance of KRT14+ UGS basal epithelium.
Ctnnb1 deletion from late-stage mouse UGE impairs prostatic bud formation but not formation of mature KRT14+ basal epithelium
Rectourethral fistula and hypoplastic UGE in Ctnnb1cLOF mice support a direct Ctnnb1 role in UGS morphogenesis from cloaca. Yet, these results do not shed light on whether Ctnnb1 is implicated directly in UGS cell differentiation or prostatic bud formation because these events occur after and likely depend on normal cloacal division. We therefore utilized an inducible cre driver, ShhcreERT2, as a means to delete Ctnnb1 from UGE at a later developmental stage, after cloacal division but prior to and during prostatic bud formation.
We optimized the tamoxifen dosing regimen to activate ShhcreERT2 near the end of cloacal division (Seifert et al., 2008). We tested two tamoxifen doses (25 and 50 mg/kg maternal doses) and two dosing periods (once daily intraperitoneal injections to the dam on E13.5 and E14.5 or on E14.5 and E15.5). Progesterone (18.75 mg/kg, maternal dose) was co-administered as an anti-abortifacient. We determined that the 25 mg/kg tamoxifen maternal dose, given in combination with 18.75 mg/kg progesterone at E13.5 and E14.5, was the highest and latest tolerable in vivo tamoxifen dose which consistently maintained pregnancy until E18.5, the embryonic stage when prostatic bud formation is completed in male embryos. This dosing paradigm activates ShhcreERT2 in a patchy subset (mosaic) of UGE cells beginning at E15.5 and persisting until at least E18.5 (supplementary material Fig. S1D–F, G) and does not interfere with cloacal division or alter the total number of prostatic buds formed per UGS, although it does cause a small but significant (p = 0.046) decrease in ventral prostatic bud number (supplementary material Fig. S4).
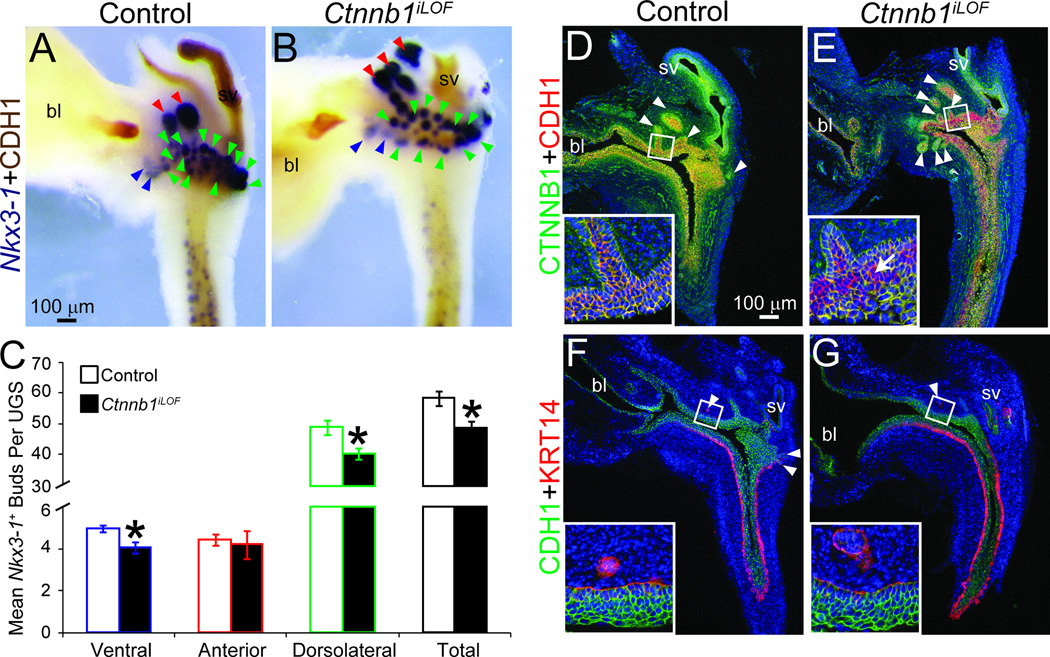
We generated Ctnnb1iLOF (ShhcreERT2/+;Ctnnb1tm2Kem/tm2Kem) embryos and exposed them to tamoxifen and progesterone on E13.5 and E14.5. At E18.5, Ctnnb1iLOF UGSs are comparable in size to ShhcreERT2/+;Ctnnb1tm2Kem/+ littermate controls but have fewer ventral, dorsolateral and total prostatic buds (Fig. 2A–C). There were more prostatic buds in Ctnnb1iLOF mice (Fig. 2) than Ctnnb1cLOF mice (Fig. 1). We suspect that the latter stage of cre activation (E15.5 in Ctnnb1iLOF mice versus E9.5 in Ctnnb1cLOF mice) and pharmacokinetic limitations of tamoxifen-dependent cre activation in Ctnnb1iLOF mice (limited tamoxifen distribution, incomplete metabolic activation and sub-effective dosing) are largely responsible for the milder prostatic budding impairment phenotype in Ctnnb1iLOF compared to Ctnnb1cLOF embryos.
Fig. 2. Ctnnb1 deletion from late-stage mouse UGE impairs prostatic bud formation but not KRT14+ mature UGS basal epithelium formation.
Male Shh+/+;Ctnnb1tm2Kem/tm2Kem (control) and ShhcreERT2/+;Ctnnb1tm2Kem/tm2Kem (Ctnnb1iLOF) embryos were exposed to tamoxifen and progesterone as described. (A–B) E18.5 male UGSs were stained by ISH to visualize Nkx3-1 (purple) and counterstained by IHC to visualize UGE marked by CDH1. Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. (C) Nkx3-1+ bud numbers are mean ± SE of five independent samples per group from at least three litters. Asterisks indicate significant differences from controls (p < 0.05). (D–G) Near mid-sagittal sections from three UGSs per genotype UGS sections were immunofluorescently labeled to visualize CTNNB1, CDH1 and KRT14. Cell nuclei are stained with DAPI (blue). Arrowheads indicate prostatic buds. The arrow in panel E (inset) indicates a representative CTNNB1 immunonegative cell. Abbreviations are bl: bladder, sv: seminal vesicle.
We next investigated UGS histological phenotype of E18.5 male control and Ctnnb1iLOF UGSs (Fig. 2D–G). The most noteworthy phenotypic difference is a ubiquitous CTNNB1 protein expression in control UGE compared to a loss of CTNNB1 protein expression in a mosaic of Ctnnb1iLOF UGE (Fig. 2D–E). UGE thickness is comparable and a well organized KRT14+ basal epithelium is present in both genotypes (Fig. 2F–G). Remarkably, when we determined the percent of CTNNB1-immunonegative cells in Ctnnb1iLOF UGE in Fig. 2E (15.5±2.2%), it closely matches the percent reduction in total Nkx3-1+ prostatic bud number in these mice compared to controls in Fig. 2C (16.6%). Even in the presence of a histologically normal UGS basal epithelium, these results suggest a tight relationship between the number of CTNNB1+ epithelial cells and prostatic buds formed.
Ctnnb1-immunopositive cells are more likely than Ctnnb1-immunonegative UGE cells to become KRT14+ basal epithelium
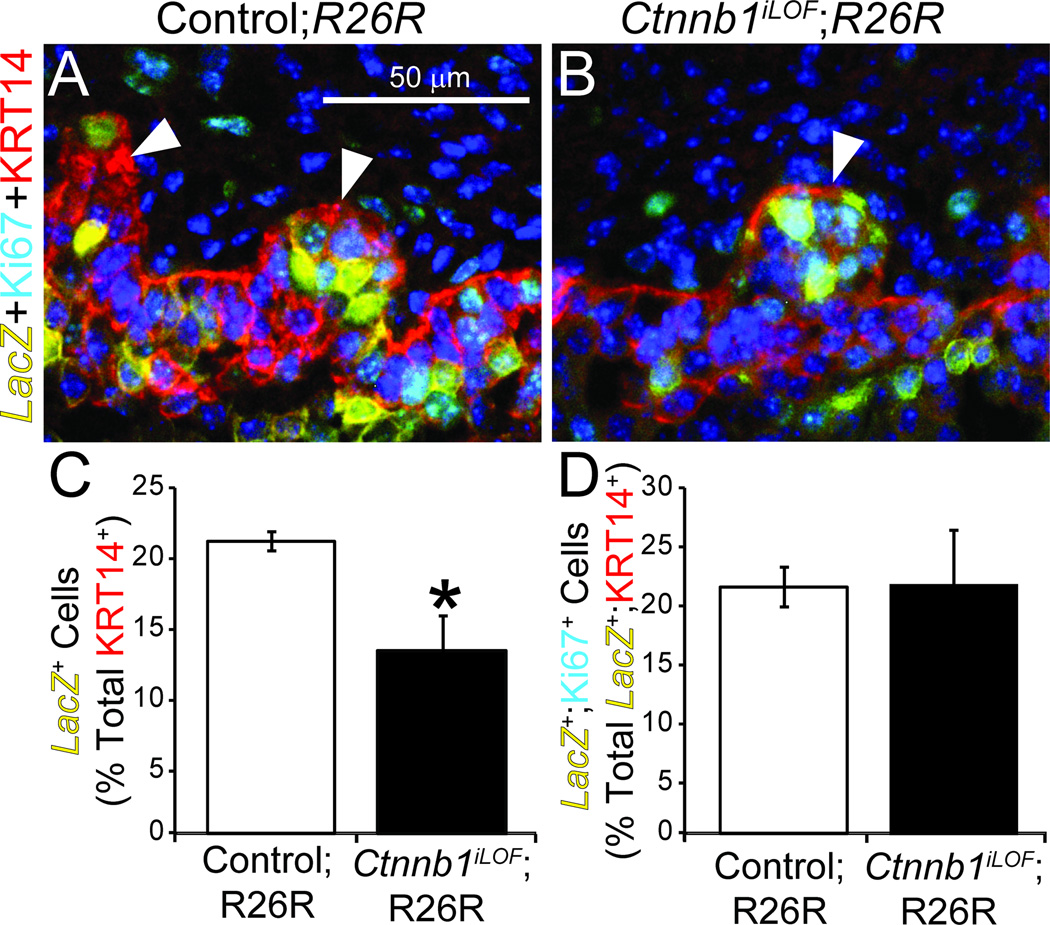
We next determined whether Ctnnb1 confers an advantage to UGE cells to integrate into or be maintained in the KRT14+ multi-cell layer. We generated Ctnnb1iLOF;R26R and control mice and used the β-galactosidase assay to label E18.5 UGE cells in which ShhcreERT2 had been activated. We then calculated the LacZ+ cell percentage contained within the KRT14+ UGE multi-cell layer. This cell population is significantly smaller in Ctnnb1iLOF;R26R compared to control mice (Fig. 3A–C).
Fig. 3. Ctnnb1-positive cells are more likely than Ctnnb1-negative cells to become UGS basal epithelium.
Male ShhcreERT2/+;Ctnnb1tm2Kem/+;R26R+ (control;R26R) and ShhcreERT2/+;Ctnnb1tm2Kem/tm2Kem;R26R+ (Ctnnb1iLOF;R26R) embryos were exposed to tamoxifen and progesterone as described. (A–B) E18.5 UGSs were stained to visualize LacZ activity, embedded in paraffin, sectioned and immunofluorescently labeled to detect the cell proliferation marker Ki67 and KRT14. Brightfield LacZ staining images were captured, inverted, pseudocolored yellow and merged with fluorescent images. Nuclei were stained with DAPI. Staining patterns represent at least three UGSs per genotype. Arrowheads indicate prostatic buds. (C) The LacZ+ cell percentage of KRT14+ cells and (D) the Ki67+ cell percentage of LacZ+;KRT14+ cells are mean ± SE of three independent samples per group from at least three litters. Asterisks indicate significant differences from controls (p < 0.05).
To determine whether Ctnnb1 is required or advantageous for KRT14+ cell proliferation, we assessed in each genotype the Ki67+ cell percentage within the LacZ+;KRT14+ cell population (Fig. 3A–B). Surprisingly, there were no differences in this proliferative index between Ctnnb1iLOF;R26R and control mice (Fig. 3D). Together, these results suggest CTNNB1 participates in the differentiation or maintenance of KRT14+ UGS basal epithelial cells but is not required for their proliferation.
UGE cells with excess Ctnnb1 act non-autonomously to impair prostatic bud formation
Results from Ctnnb1cLOF and Ctnnb1iLOF mice suggest Ctnnb1 is necessary for formation of basal epithelium and prostatic buds. We tested whether excess CTNNB1 expression in UGE would independently drive either of these processes. We generated mice harboring ShhcreERT2 and the inducible conditional dominant stable Ctnnb1tm1Mmt gain of function (GOF) allele (Ctnnb1iGOF mice). These mice harbor a floxed Ctnnb1 exon 3 which when subjected to cre-mediated recombination encodes a functional and highly stable CTNNB1 protein form that accumulates in cells. Cre activity was induced, as described above, by tamoxifen:progesterone administration to the dam on E13.5 and 14.5. All assessments were made at E18.5, after completion of prostatic bud formation in control mice.
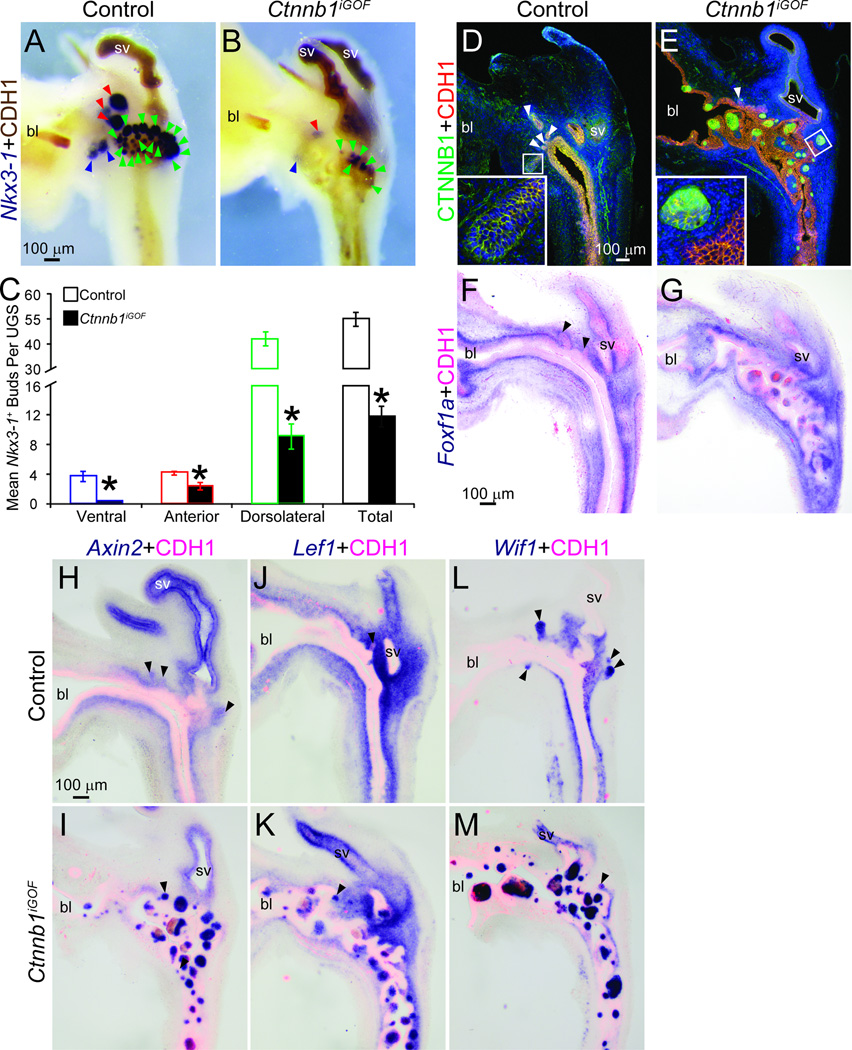
We expected Ctnnb1iGOF UGSs to form at least as many, if not more prostatic buds compared to control UGSs. We instead observed fewer ventral, dorsolateral, anterior and total Nkx3-1+ prostatic buds in Ctnnb1iGOF UGSs compared to control UGSs (Fig. 4A–C). In fact, to our surprise there were fewer buds in Ctnnb1iGOF than in Ctnnb1iLOF mice. We also identified an unusual histological feature in Ctnnb1iGOF mice: dense cadherin 1 (CDH1)-positive epithelial cell islands, 4–8 cells in diameter and with varying degrees of organization, interrupting the otherwise normal urothelial histotype of Ctnnb1iGOF UGS and bladder trigone (Fig. 4). Excess CTNNB1 protein is present in membranes, cytoplasm and nuclei of all Ctnnb1iGOF epithelial island cells (Fig. 4D–E); whereas normal CTNNB1 protein levels are present and localized only to cell membranes of control mouse UGE cells. Nearly all Ctnnb1iGOF UGE cell islands are accompanied by stromal invasion into the UGE which was visualized by marking UGS stroma with the submucosa/lamina propria marker (Abler et al., 2011a) forkhead box F1a, Foxf1a (Fig. 4F–G). We also identified inappropriate expression of CTNNB1-responsive Axin2, Lef1 and Wif1 mRNAs in the cell islands (Fig. 4H–M) and an inappropriate reduction of these markers in associated UGS stroma. In age-matched control mouse UGE, epithelial expression of Axin2, Lef1 and Wif1 mRNAs is normally restricted to prostatic buds and each mRNA is abundant in associated UGS stroma.
Fig. 4. Excess CTNNB1 in UGE impairs prostatic bud formation and causes an inappropriate pattern of CTNNB1-responsive mRNA expression.
Male Shh+/+; Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+;Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) embryos were exposed to tamoxifen and progesterone as described. (A–B) E18.5 male UGSs were stained by ISH to visualize Nkx3-1 (purple) and counterstained by IHC to visualize UGE marked by CDH1. Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. (C) Nkx3-1+ bud numbers are mean ± SE of five independent samples per group from at least three litters. Asterisks indicate significant differences from controls (p < 0.05). (D–M) Sections from three UGSs per genotype were immunofluorescently labeled to visualize CTNNB1 and CDH1 proteins or stained by ISH to visualize mRNA expression (purple) of the urethra lamina propria mesenchyme Foxf1a or the CTNNB1 responsive genes Axin2, Lef1 and Wif1. Sections were immunofluorescently counterstained to visualize UGE marked by CDH1. Abbreviations are bl: bladder, sv: seminal vesicle. Arrowheads indicate prostatic buds.
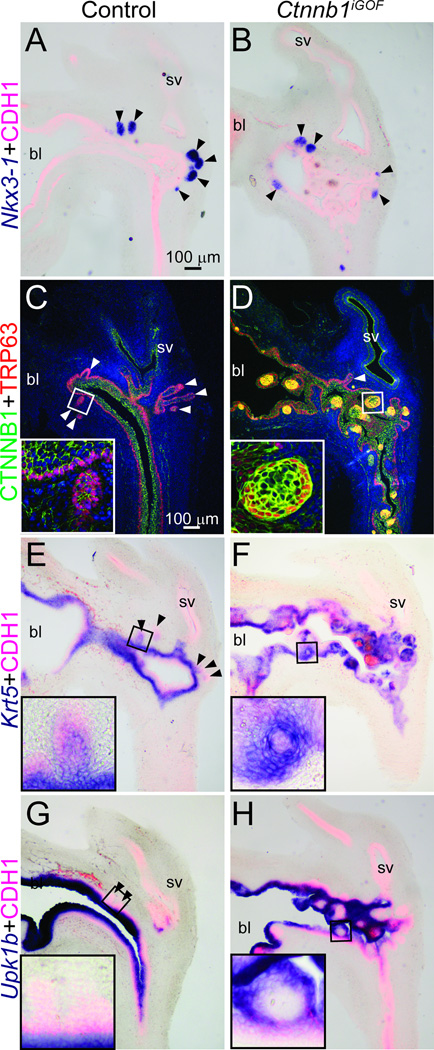
We next tested whether expression of excess Ctnnb1 was sufficient for imposing prostate identity onto UGE. Nkx3-1, the earliest known marker of prostatic epithelial identity (Sciavolino et al., 1997), localizes to prostatic buds in Ctnnb1iGOF and control UGSs but is not visible in Ctnnb1iGOF cell islands with excess CTNNB1 protein expression (Fig. 5A–B). These results indicate that expression of excess CTNNB1 is insufficient for imposing Nkx3-1+ prostatic identity onto UGE.
Fig. 5. Excess CTNNB1 in UGE is not sufficient for prostate identity but is sufficient for basal epithelial cell differentiation.
Male Shh+/+; Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+; Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) embryos were exposed to tamoxifen and progesterone as described. (A-B, E-H) At E18.5, UGS sections were stained by ISH to visualize mRNAs (purple) for the prostatic bud marker Nkx3-1, the basal epithelial marker Krt5 and the intermediate and superficial urothelial marker Upk1b. Sections were immunofluorescently counterstained to visualize UGE marked by CDH1. (C–D) E18.5 male UGS sections were immunofluorescently labeled to visualize CTNNB1 and the basal epithelial marker TRP63. Staining patterns in each panel represent three males. Abbreviations are bl: bladder, sv: seminal vesicle. Arrowheads indicate prostatic buds.
Since Ctnnb1cLOF UGSs do not form a well developed basal epithelium (Fig. 1G–H) and since there are fewer Ctnnb1+ UGE basal epithelial cells (Fig. 3C), we assessed whether expression of excess CTNNB1 in UGE is sufficient for imposing a basal cell identity on UGE. Ctnnb1iGOF cell islands feature a TRP63+;Krt5+ basal epithelial sheath (Fig. 5C–F) and a TRP63−;Krt5- cell core that lacks detectable uroplakin 1b (Upk1b)–a marker of the only other known urothelial cell type (Abler et al., 2011a) present in E18.5 control mouse urethra (Fig. 5G–H). Together, these results provide evidence for self-organizing activity in Ctnnb1iGOF cell islands, reveal that some cells with excess CTNNB1 express basal epithelial cell markers and that others inappropriately retain a primitive, undifferentiated UGE cell histotype.
Expression of excess Ctnnb1 in UGE impairs prostatic bud formation by a paracrine mechanism involving bone morphogenetic protein signaling
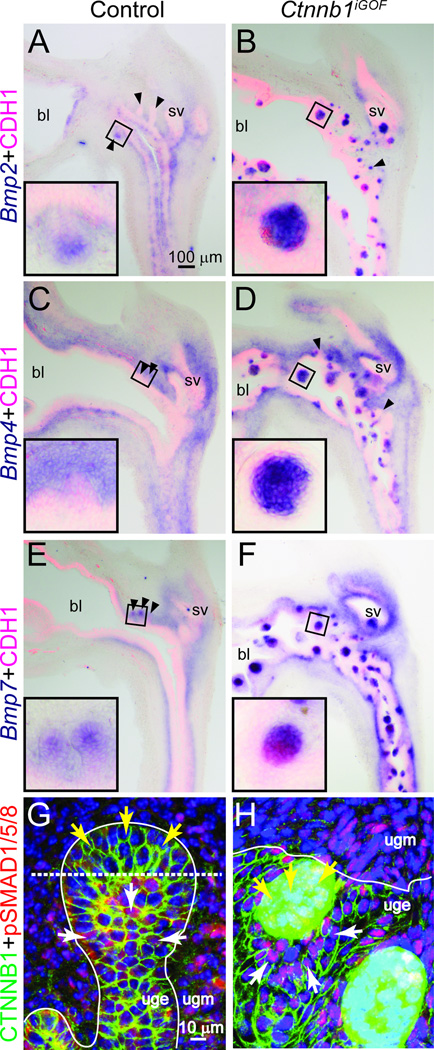
CTNNB1 influences skin appendage patterning by inducing formation of hair and feather placodes and by establishing placode spacing intervals (Chang et al., 2004; Zhang et al., 2009). We considered the possibility that CTNNB1 within prostatic bud tips may control prostatic bud spacing by suppressing bud formation elsewhere in UGS epithelium. In the case of Ctnnb1iGOF UGSs, such a bud spacing mechanism would be precociously or prematurely activated in cell islands to suppress prostatic buds in a much larger domain—throughout the entire UGE, and would provide a rationale for how prostatic bud formation is inhibited in this mouse strain. Bone morphogenetic proteins (BMPs) are known inhibitors of prostatic bud formation. Bmp4 and Bmp7 mRNAs are expressed in the UGS during prostatic bud formation, addition of exogenous BMP4 or BMP7 protein to organ culture media inhibits prostatic bud formation in vitro, and genetic deletion of the BMP antagonist Noggin inhibits prostatic bud formation in vivo (Cook et al., 2007; Grishina et al., 2005; Lamm et al., 2001; Vezina et al., 2008). However, mechanisms regulating Bmp expression in UGS epithelium have not been determined. We tested the hypothesis that CTNNB1 induces BMP ligand expression in UGE. We observe low-level expression of Bmp 2, 4 and 7 mRNAs where CTNNB1-responsive Axin2 and Lef1 mRNAs are localized in prostatic bud tips of control UGSs (Fig. 6A, C, E). Each Bmp is highly abundant in Ctnnb1iGOF cell islands with excess CTNNB1 (Fig. 6B, D, F) suggesting CTNNB1-dependent activation of BMP ligand expression.
Fig. 6. Excess CTNNB1 in UGE induces BMP expression and increases BMP signaling.
E18.5 Male Shh+/+;Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+;Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) embryos were exposed to tamoxifen and progesterone as described. (A–F) Sections stained by ISH to visualize mRNA expression (purple) of Bmp 2, 4 and 7 Black arrowheads indicate prostatic bud tips. (G–H) Sections immunofluorescently labeled to visualize CTNNB1 (green), phospho-SMAD (pSMAD) 1/5/8 (red) and nuclei stained with DAPI (blue). White lines demarcate the interface between UGE and UGM. Yellow arrows indicate cells in which CTNNB1 is known to be high (prostatic bud tips in G and CTNNB1 cell islands in H). White arrows indicate cells surrounding those with high CTNNB1. Abbreviations are bl: bladder, sv: seminal vesicle.
BMPs elicit their activity in part by binding BMP receptors and inducing SMAD protein phosphorylation to prompt assembly of a heterodimeric transcription factor complex. We used an antibody that specifically recognizes BMP-responsive phosphorylation of SMAD (pSMAD) 1/5/8 to determine whether BMPs were activating downstream signaling in the UGS. In control UGS, pSMAD1/5/8 immunostaining was present in UGM and UGE (Fig. 6G). We noted a specific trend in pSMAD1/5/8 distribution in UGE: it was more abundant in the base of prostatic buds (Fig. 6G, white arrows) than in prostatic bud tips (Fig. 6G, yellow arrows). This pattern is the inverse of CTNNB1-responsive Axin2, Lef1 and Wif1 mRNA expression in control UGE, which are more abundant in prostatic bud tips than elsewhere in prostatic buds (Fig. 4H,J,L). pSMAD1/5/8 was noticeably more abundant in Ctnnb1iGOF UGS than in control UGS (Fig. 6H), and particularly in UGE surrounding Ctnnb1iGOF cell islands with excess CTNNB1 (Fig. 6H, white arrowheads) than in the islands themselves (Fig. 6H, yellow arrowheads). Again, this pattern is the inverse of Ctnnb1-responsive mRNA expression in the Ctnnb1iGOF UGS (Fig. 4I,K,M). These results reveal an important trend: active BMP signaling in fetal mouse UGE is typically localized to a cell field surrounding, but not within cells where CTNNB1 signaling exists.
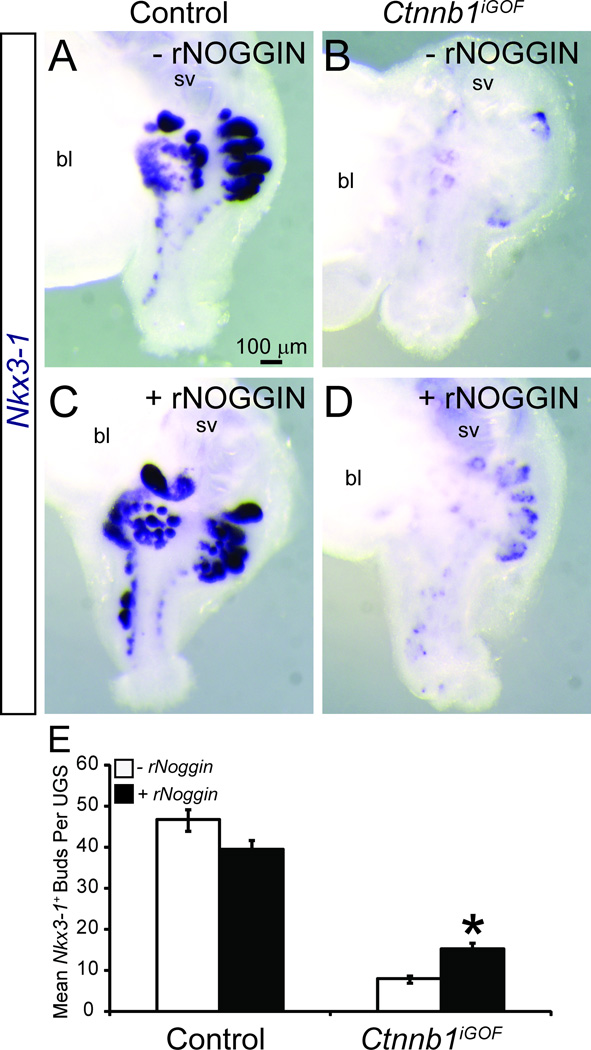
If an overabundance of BMP signaling contributes to prostatic bud impairment in Ctnnb1iGOF UGSs, we hypothesized that the BMP inhibitor NOGGIN would at least partially restore prostatic bud formation. E14.5 male Ctnnb1iGOF and control UGSs were grown in organ culture media containing 4-hydroxytamoxifen, DHT and either vehicle or recombinant NOGGIN protein. We confirmed this 4-hydroxytamoxifen concentration activates creERT2 (supplementary material Fig. S1H) and does not alter prostatic bud number in control UGS explants (supplementary material Fig. S6). The concentration of recombinant NOGGIN protein (1 µg/mL) used in this study was used previously to antagonize BMP signaling in control mouse UGS explants (Cook et al., 2007; Vezina et al., 2008). In the absence of exogenous NOGGIN, Ctnnb1iGOF UGSs form fewer Nkx3-1 positive prostatic buds compared to control cultured UGSs (Fig. 7A,B,E). Exogenous NOGGIN increased Nkx3-1-positive bud number in Ctnnb1iGOF UGSs (Fig. 7C,D,E) but did not significantly change prostatic bud number in control UGSs (Fig. 7A,C,E). These results indicate a genetic sensitivity of Ctnnb1iGOF UGSs to NOGGIN and support the notion that excess BMP signaling is a potential mechanism for prostatic bud impairment in Ctnnb1iGOF UGSs. Taken together, our results support the hypothesis that CTNNB1 activation in prostatic bud tips participate in prostatic bud spacing by activating BMP signaling to inhibit inappropriate prostatic bud formation in surrounding UGE.
Fig. 7. BMP signaling inhibition in cultured UGS with excess CTNNB1 partially restores prostatic bud formation.
E14.5 male Shh+/+;Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+;Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) UGSs were incubated for 4 d in organ culture medium containing 1 µm 4-hydroxytamoxifen, 10nM 5α dihydrotestosterone and either vehicle or recombinant NOGGIN protein (rNOGGIN, 1 µg/ml). Media and supplements were replenished after the second day. (A–D). Whole UGSs were stained by ISH to visualize Nkx3-1 mRNA (purple). Abbreviations are bl: bladder, sv: seminal vesicle. (E) Nkx3-1+ bud numbers are mean ± SE of five independent samples per group from at least three litters. Asterisks indicate a significant difference (p < 0.05) in prostatic bud number compared to Ctnnb1iGOF UGS grown in the absence of rNOGGIN.
Discussion
Two different Ctnnb1 actions are revealed in the developing prostate
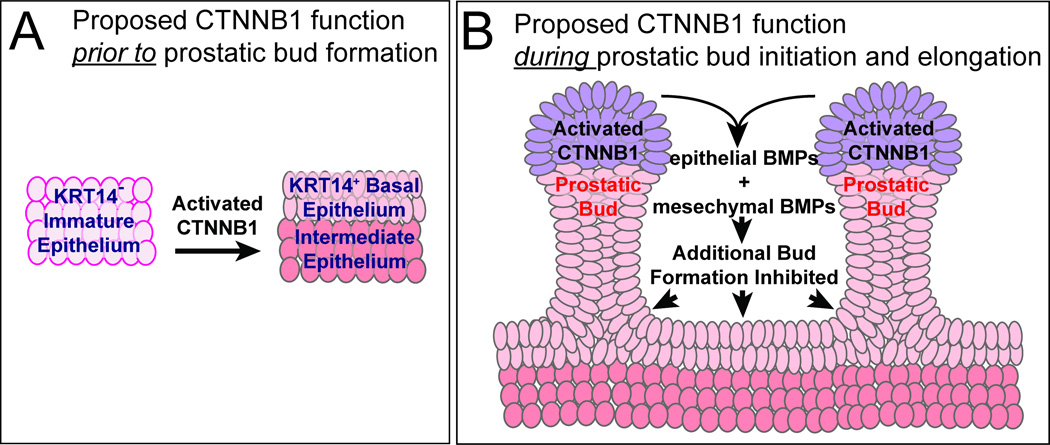
A recent report indicated a required role for CTNNB1 in prostatic bud initiation (Simons et al., 2012). Our results extend those findings by showing that prostatic bud formation specifically requires CTNNB1 in UGS epithelium. We also identified two crucial Ctnnb1 roles in prostate development that have not been reported previously. Ctnnb1 is first required in UGS epithelium prior to prostatic budding where it is hypothesized to promote formation of KRT14+ UGS basal epithelium (Fig. 8A). Ctnnb1 is then activated in prostatic bud tips where it is hypothesized to inhibit additional bud formation by activating BMP signaling (Fig. 8B). Together, these Ctnnb1 actions provide a rationale for how modification of CTNNB1 signaling in either a positive (increased signaling) or negative (decreased signaling) direction can elicit deleterious effects on early prostate development. This also suggests a necessity for strict CTNNB1 regulation during prostate development to give rise to an appropriately patterned ductal network.
Fig. 8. Proposed dichotomous CTNNB1 activity during mouse prostate development.
(A) During early prostate development, functional CTNNB1 in primitive UGS epithelium supports formation or maintenance of mature KRT14+ basal epithelium, which is needed for prostatic bud formation. (B) During prostatic bud formation and elongation, CTNNB1 is activated in prostatic bud tips where it initiates BMP 2, 4 and 7 synthesis, which act upon UGS epithelium to prevent additional buds from forming. We propose that these CTNNB1 mechanisms in part establish prostatic bud spacing intervals.
Ctnnb1 promotes UGS basal epithelium
We provide multiple lines of evidence to support a Ctnnb1 role in formation of UGS basal epithelium. First, we found that Ctnnb1 deletion from all UGS epithelial cells at an early stage of prostate development prevents formation of KRT14+ UGS basal epithelium. Second, we observed that cells with functional CTNNB1 are more likely than cells without functional CTNNB1 to establish or maintain KRT14 expression. Third, we found that UGS epithelial cell islands expressing excess CTNNB1 develop their own basal cell layer. Taken together, these results directly implicate Ctnnb1 in promoting or maintaining basal epithelial identity in developing prostate.
Though we did not identify the specific mechanism by which CTNNB1 supports UGS basal epithelial development, our results argue against a pro-proliferative action. We found that once KRT14+ cells had formed, Ctnnb1 was not needed or advantageous for their proliferation. Instead, Ctnnb1 is likely to either initiate or maintain basal cell differentiation. The former possibility is consistent with the established CTNNB1 function in adult prostate progenitor cells. Activation of LRP5, a WNT co-receptor, expands the proportion of TRP63+ basal epithelial cells during in vitro prostate sphere formation (Shahi et al., 2012). In vitro treatment of the same enriched prostate progenitor cell population with WNT3A conditioned media re-patterns TRP63+ cells within, forming epithelial cell colonies (Shahi et al., 2011). It will be important in future studies to determine specifically how Ctnnb1 promotes basal epithelial cell identity within the UGE progenitor population, whether this process is reversible and whether the progenitors are multipotent and capable of giving rise to non-basal lineages (intermediate or superficial epithelium).
Ctnnb1 participates in prostatic bud spacing
Ctnnb1 gain-of-function mutations cause supernumerary bud formation in several developing organs (Faraldo et al., 2006; Gat et al., 1998; Liu et al., 2007; Lo Celso et al., 2004; Noramly et al., 1999; Widelitz et al., 2000). Yet, intriguingly in prostate, CTNNB1 stabilization is not sufficient for inducing supernumerary prostatic bud formation, at least not in the specific UGS epithelial cell type and developmental stages examined herein. Instead, we found that prostatic bud formation requires Ctnnb1 in combination with other factors.
Because expression of excess Ctnnb1 in a mosaic of UGE cells globally inhibited prostatic bud formation, we further hypothesized that Ctnnb1 impaired prostatic bud formation through a paracrine or juxtacrine mechanism. We linked CTNNB1 to prostatic bud inhibitory actions by involving increased BMP signaling as indicated in the discussion below.
We identified Bmp signaling as a downstream mediator of CTNNB1 function in UGE. We found that Ctnnb1iGOF cell islands express Bmp 2, 4 and 7 mRNAs and that BMP-responsive phospho-SMAD1/5/8 is expressed in adjacent UGE but not in UGE island cells excess CTNNB1. We also found that BMP signaling antagonism by NOGGIN partially restored prostatic bud formation in Ctnnb1iGOF UGS explants and Ctnnb1iGOF UGSs are more sensitive than control UGSs to exogenous NOGGIN-induced increases in prostatic bud formation. These results identify BMP signaling as one mechanism by which prostatic budding is impaired in Ctnnb1iGOF UGSs. It is important to note that the NOGGIN concentration used in this study was not capable of fully rescuing prostatic bud formation in Ctnnb1iGOF UGSs. Whether a greater NOGGIN concentration would have caused a greater rescue of prostatic bud formation is not known and whether other CTNNB1-responsive signaling pathways contribute to prostatic budding impairment in Ctnnb1iGOF UGSs was not determined. Furthermore, additional studies are needed to determine how Ctnnb1iGOF cell islands are protected from the BMPs they synthesize.
Little is known about how prostatic bud spacing intervals are achieved. In the absence of androgens or intact AR signaling, small epithelial buds form in an inappropriate pattern in the ventral aspect of the mouse UGS (Allgeier et al., 2010). Though these small buds do not elongate, their androgen-independent formation is important because it suggests prostatic bud initiation can occur independently of androgens and that androgens participate in prostatic bud patterning (Allgeier et al., 2010). Prostatic bud patterning mechanisms are not fully known. Results from this and other studies support CTNNB1 as a regulatory node for controlling prostatic bud patterning. We previously showed that high levels of androgen signaling in male mouse UGS are associated with CTNNB1-responsive Axin2 and Lef1 expression in prostatic bud tips (Mehta et al., 2011). Bmp mRNAs are also expressed in prostatic bud tips; in this study, we provide evidence that CTNNB1 induces Bmp expression in this location. The BMP signaling pathway is one of few identified endogenous antagonists of prostatic bud formation (Cook et al., 2007; Grishina et al., 2005; Lamm et al., 2001). Fine tuning of BMP signaling has been reported as a CTNNB1 action in skin appendage spacing (Zhang et al., 2008). Likewise, results from this study support the hypothesis that CTNNB1 activity acts as a mechanism for fine tuning of BMP expression in UGS epithelium during the emergence of prostatic buds. Other mechanisms are also likely to contribute to prostatic bud spacing, especially during prostatic bud initiation when BMPs dominate in UGS mesenchyme (Grishina et al., 2005; Lamm et al., 2001).
It is unclear how nascent prostatic buds overcome bud inhibitory BMP signals from UGS stroma and how prostatic bud tips are protected from the bud inhibitory BMPs they produce. One potential protective mechanism for prostatic buds is Noggin, which is highly expressed in UGS mesenchyme near prostatic bud tips and is least abundant in mesenchyme near interbud UGE (Cook et al., 2007). This particular expression pattern would allow for maximal sequestration of BMPs near prostatic bud tips to permit their outgrowth and maximal BMP activity in interbud UGS epithelium to limit/restrict additional/new bud formation.
Supplementary Material
(A–B) In vivo cre activation prior to and during prostatic bud formation in mice carrying the Shhcre allele was demonstrated by revealing β-galactosidase activity (blue) in E12.5 whole-mount cloaca and in E18.5 whole-mount Shhcre;R26R UGS. To demonstrate epithelial specificity of β- galactosidase activity (inset images), whole-mount stained tissues were cut into sections and counterstained with DAPI to label all cells. Note that β-galactosidase activity was confined to all E12.5 cloacal epithelial cells and E18.5 UGE. (C) Timing and distribution of in vivo Shhcre activation is summarized with respect to other known events important for lower urogenital tract development. (D–F) In vivo cre activation prior to and during prostatic bud formation in mice carrying the ShhcreERT2 allele was demonstrated by treating pregnant dams with tamoxifen (Tam) and progesterone as described and visualizing β-galactosidase activity in whole-mount ShhcreERT2;R26R UGS at E14.5, 15.5 and E18.5. Whole-mount stained tissues were cut into sections (inset image) and counterstained with DAPI. Black line marks the mesenchymal-epithelial interface. Note that β-galactosidase activity was detected in some but not all E18.5 UGE. (G) Timing and distribution of in vivo ShhcreERT2 activation is summarized with respect to other known events important for lower urogenital tract development. (H) In vitro cre activation during prostatic bud formation in cultured UGSs carrying the ShhcreERT2;R26R allele was demonstrated by growing E14.5 male UGSs in media containing DHT (10 nM) and 4-hydroxytamoxifen (4OHTam, 1 µm) and revealing β-galactosidase activity after 4 d in culture. Results represent at least 3 UGSs per genotype and per experimental condition. Abbreviations are bl: bladder, cl: cloaca, sv: seminal vesicle; wd: Wolffian duct. Arrowheads indicate prostatic buds. Scale bars are 500 µM.
E18.5 male Shh+/+ (control) and ShhcreERT2/+ mouse UGSs were stained by ISH to visualize the prostatic bud marker Nkx3-1 (purple). Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. Abbreviations are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers were determined as mean ± SE of five independent samples per group from at least three litters.
E14.5, E15.5 and E16.5 wild type control mouse near mid-sagittal UGS sections were immunofluorescently labeled to visualize CDH1 and UGS basal epithelial markers KRT14 and TRP63. Cell nuclei stained with DAPI. Results in each panel represent three males. Abbreviations are bl: bladder, sv: seminal vesicle, wd: Wolffian duct.
Pregnant wild type control dams were administered a solution containing tamoxifen (25 mg/kg maternal dose) and progesterone (18.75 mg/kg maternal dose) or corn oil (vehicle control, 2.5 ml/kg maternal dose) via once daily intraperitoneal injections on E13.5 and E14.5. (A–B) E18.5 whole-mount male UGSs from treated dams were stained by ISH to visualize Nkx3-1 (purple). Staining patterns represents five males. Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. Abbreviations are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers are mean ± SE of five independent samples per group from at least three litters. An asterisk indicates a significant difference (p <0.05) from vehicle control.
E18.5 male Shh+/+;Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+;Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) UGS sections. (A–B) Sections were immunofluorescently labeled to detect the smooth muscle marker ACTA2. Nuclei stained with DAPI (blue). (C–D) UGS sections were stained by ISH to visualize mRNA expression (purple) of the urethral mesenchymal pad marker Fgf10 and counterstained by IHC to visualize UGE marked by CDH1. Arrowheads indicate prostatic buds. Staining patterns in each panel represent three males. Abbreviations are bl: bladder, sv: seminal vesicle.
E14.5 male C57BL/6J wild type control mouse UGSs were incubated for 4 d in organ culture medium containing 10 nM 5α dihydrotesterosterone and vehicle (0.1% ethanol) or 4-hydroxytamoxifen (1 µM). Media and supplements were replenished after the second day. (A–B) UGSs were stained by ISH to visualize Nkx3-1 mRNA (purple). Staining patterns in each panel represent five males. Abbreviations used are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers are the mean ± SE of five independent samples per group from at least three litters.
Acknowledgements
We would like to thank Dr. Ruth Sullivan for her intellectual contributions to this manuscript, Dr. Sarah H. Allgeier for assistance with the Scanning Electron Microscopy and Mr. Ron Schimmel, Ms. Amber Lashua and Mr. Kurt Throckmorton for technical assistance.
Funding
This research was supported by NIH DK083425 (to C.M.V.), NSF DGE-0718123 (to K.P.K.) and NIH DK082888 (to X.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abler LL, Keil KP, Mehta V, Joshi PS, Schmitz CT, Vezina CM. A high-resolution molecular atlas of the fetal mouse lower urogenital tract. Dev Dyn. 2011a;240:2364–2377. doi: 10.1002/dvdy.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abler LL, Mehta V, Keil KP, Joshi PS, Flucus CL, Hardin HA, Schmitz CT, Vezina CM. A high throughput in situ hybridization method to characterize mRNA expression patterns in the fetal mouse lower urogenital tract. J Vis Exp. 2011b;54:e2912. doi: 10.3791/2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abler LL, Mehta V, Keil KP, Joshi PS, Schmitz CT, Vezina CM. Accession ID 14274: Axin2 expression in the E14.5 Mouse Lower Urinary Tract. 2012. [Google Scholar]
- Allgeier SH, Lin TM, Moore RW, Vezina CM, Abler LL, Peterson RE. Androgenic regulation of ventral epithelial bud number and pattern in mouse urogenital sinus. Dev Dyn. 2010;239:373–385. doi: 10.1002/dvdy.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cremediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Chang CH, Jiang TX, Lin CM, Burrus LW, Chuong CM, Widelitz R. Distinct Wnt members regulate the hierarchical morphogenesis of skin regions (spinal tract) and individual feathers. Mech Dev. 2004;121:157–171. doi: 10.1016/j.mod.2003.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TC, Wallace MC, Merlie JP, Olson EN. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 1993;261:215–218. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- Cook C, Vezina CM, Allgeier SH, Shaw A, Yu M, Peterson RE, Bushman W. Noggin is required for normal lobe patterning and ductal budding in the mouse prostate. Dev Biol. 2007;312:217–230. doi: 10.1016/j.ydbio.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha GR, Battle E, Young P, Brody J, Donjacour A, Hayashi N, Kinbara H. Role of epithelial-mesenchymal interactions in the differentiation and spatial organization of visceral smooth muscle. Epithelial Cell Biol. 1992;1:76–83. [PubMed] [Google Scholar]
- Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. The endocrinology and developmental biology of the prostate. Endocr Rev. 1987;8:338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- Faraldo MM, Taddei-De La Hosseraye I, Teuliere J, Deugnier MA, Moumen M, Thiery JP, Glukhova MA. Mammary gland development: Role of basal myoepithelial cells. J Soc Biol. 2006;200:193–198. doi: 10.1051/jbio:2006021. [DOI] [PubMed] [Google Scholar]
- Feng JQ, Chen D, Cooney AJ, Tsai MJ, Harris MA, Tsai SY, Feng M, Mundy GR, Harris SE. The mouse bone morphogenetic protein-4 gene. Analysis of promoter utilization in fetal rat calvarial osteoblasts and regulation by COUP-TFI orphan receptor. J Biol Chem. 1995;270:28364–28373. doi: 10.1074/jbc.270.47.28364. [DOI] [PubMed] [Google Scholar]
- Fox J, Weisberg S. An [R] Companion to Applied Regression. Second ed. Thousand Oaks, CA: Sage; 2011. [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- Grishina IB, Kim SY, Ferrara C, Makarenkova HP, Walden PD. BMP7 inhibits branching morphogenesis in the prostate gland and interferes with Notch signaling. Dev Biol. 2005;288:334–347. doi: 10.1016/j.ydbio.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR. Interactions between adult human prostatic epithelium and rat urogenital sinus mesenchyme in a tissue recombination model. Differentiation. 1998;63:131–140. doi: 10.1046/j.1432-0436.1998.6330131.x. [DOI] [PubMed] [Google Scholar]
- Joesting MS, Cheever TR, Volzing KG, Yamaguchi TP, Wolf V, Naf D, Rubin JS, Marker PC. Secreted frizzled related protein 1 is a paracrine modulator of epithelial branching morphogenesis, proliferation, and secretory gene expression in the prostate. Dev Biol. 2008;317:161–173. doi: 10.1016/j.ydbio.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil KP, Mehta V, Abler LL, Joshi PS, Schmitz CT, CM, V Visualization and quantification of mouse prostate development by in situ hybridization. Differentiation. 2012;84:232–239. doi: 10.1016/j.diff.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131:4955–4964. doi: 10.1242/dev.01384. [DOI] [PubMed] [Google Scholar]
- Lamm ML, Podlasek CA, Barnett DH, Lee J, Clemens JQ, Hebner CM, Bushman W. Mesenchymal factor bone morphogenetic protein 4 restricts ductal budding and branching morphogenesis in the developing prostate. Dev Biol. 2001;232:301–314. doi: 10.1006/dbio.2001.0187. [DOI] [PubMed] [Google Scholar]
- Lasnitzki I, Mizuno T. Prostatic induction: interaction of epithelium and mesenchyme from normal wild-type mice and androgen-insensitive mice with testicular feminization. J Endocrinol. 1980;85:423–428. doi: 10.1677/joe.0.0850423. [DOI] [PubMed] [Google Scholar]
- Lin TM, Rasmussen NT, Moore RW, Albrecht RM, Peterson RE. Region-specific inhibition of prostatic epithelial bud formation in the urogenital sinus of C57BL/6 mice exposedin utero to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicological Sciences. 2003;76:171–181. doi: 10.1093/toxsci/kfg218. [DOI] [PubMed] [Google Scholar]
- Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, Taketo MM, Morrisey EE, Atit R, Dlugosz AA, Millar SE. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Thirumangalathu S, Gallant NM, Yang SH, Stoick-Cooper CL, Reddy ST, Andl T, Taketo MM, Dlugosz AA, Moon RT, Barlow LA, Millar SE. Wnt-beta-catenin signaling initiates taste papilla development. Nat Genet. 2007;39:106–112. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- Mehta V, Abler LL, Keil KP, Schmitz CT, Joshi PS, Vezina CM. Atlas of Wnt and R-spondin gene expression in the developing male mouse lower urogenital tract. Dev Dyn. 2011;240:2548–2560. doi: 10.1002/dvdy.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noramly S, Freeman A, Morgan BA. beta-catenin signaling can initiate feather bud development. Development. 1999;126:3509–3521. doi: 10.1242/dev.126.16.3509. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Seifert AW, Harfe BD, Cohn MJ. Cell lineage analysis demonstrates an endodermal origin of the distal urethra and perineum. Dev Biol. 2008;318:143–152. doi: 10.1016/j.ydbio.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P, Park D, Pond AC, Seethammagari M, Chiou SH, Cho K, Carstens JL, Decker WK, McCrea PD, Ittmann MM, Rosen JM, Spencer DM. Activation of Wnt signaling by chemically induced dimerization of LRP5 disrupts cellular homeostasis. PLoS One. 2012;7:e30814. doi: 10.1371/journal.pone.0030814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P, Seethammagari MR, Valdez JM, Xin L, Spencer DM. Wnt and Notch pathways have interrelated opposing roles on prostate progenitor cell proliferation and differentiation. Stem Cells. 2011;29:678–688. doi: 10.1002/stem.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoretti S, Pires MM, Lindauer M, Horner JW, Grisanzio C, Dhar S, Majumder P, McKeon F, Kantoff PW, Sellers WR, Loda M. p63 regulates commitment to the prostate cell lineage. Proc Natl Acad Sci U S A. 2005;102:11355–11360. doi: 10.1073/pnas.0500165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BW, Hurley PJ, Huang Z, Ross AE, Miller R, Marchionni L, Berman DM, Schaeffer EM. Wnt signaling though beta-catenin is required for prostate lineage specification. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Timms BG, Mohs TJ, Didio LJ. Ductal budding and branching patterns in the developing prostate. J Urol. 1994;151:1427–1432. doi: 10.1016/s0022-5347(17)35273-4. [DOI] [PubMed] [Google Scholar]
- Vezina CM, Allgeier SH, Fritz WA, Moore RW, Strerath M, Bushman W, Peterson RE. Retinoic acid induces prostatic bud formation. Dev Dyn. 2008;237:1321–1333. doi: 10.1002/dvdy.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BE, Wang XD, Ernst JA, Polakis P, Gao WQ. Regulation of epithelial branching morphogenesis and cancer cell growth of the prostate by Wnt signaling. PLoS ONE. 2008;3:e2186. doi: 10.1371/journal.pone.0002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Lu J, Chuong CM. beta-catenin in epithelial morphogenesis: conversion of part of avian foot scales into feather buds with a mutated beta-catenin. Dev Biol. 2000;219:98–114. doi: 10.1006/dbio.1999.9580. [DOI] [PubMed] [Google Scholar]
- Wu X, Daniels G, Shapiro E, Xu K, Huang H, Li Y, Logan S, Greco MA, Peng Y, Monaco ME, Melamed J, Lepor H, Grishina I, Lee P. LEF1 identifies androgen-independent epithelium in the developing prostate. Mol Endocrinol. 2011;25:1018–1026. doi: 10.1210/me.2010-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang Y, Jiang M, Bierie B, Roy-Burman P, Shen MM, Taketo MM, Wills M, Matusik RJ. Activation of beta-Catenin in mouse prostate causes HGPIN and continuous prostate growth after castration. Prostate. 2009;69:249–262. doi: 10.1002/pros.20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TJ, Hoffman BG, Ruiz de Algara T, Helgason CD. SAGE reveals expression of Wnt signalling pathway members during mouse prostate development. Gene Expr Patterns. 2006;6:310–324. doi: 10.1016/j.modgep.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Andl T, Yang SH, Teta M, Liu F, Seykora JT, Tobias JW, Piccolo S, Schmidt-Ullrich R, Nagy A, Taketo MM, Dlugosz AA, Millar SE. Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development. 2008;135:2161–2172. doi: 10.1242/dev.017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–B) In vivo cre activation prior to and during prostatic bud formation in mice carrying the Shhcre allele was demonstrated by revealing β-galactosidase activity (blue) in E12.5 whole-mount cloaca and in E18.5 whole-mount Shhcre;R26R UGS. To demonstrate epithelial specificity of β- galactosidase activity (inset images), whole-mount stained tissues were cut into sections and counterstained with DAPI to label all cells. Note that β-galactosidase activity was confined to all E12.5 cloacal epithelial cells and E18.5 UGE. (C) Timing and distribution of in vivo Shhcre activation is summarized with respect to other known events important for lower urogenital tract development. (D–F) In vivo cre activation prior to and during prostatic bud formation in mice carrying the ShhcreERT2 allele was demonstrated by treating pregnant dams with tamoxifen (Tam) and progesterone as described and visualizing β-galactosidase activity in whole-mount ShhcreERT2;R26R UGS at E14.5, 15.5 and E18.5. Whole-mount stained tissues were cut into sections (inset image) and counterstained with DAPI. Black line marks the mesenchymal-epithelial interface. Note that β-galactosidase activity was detected in some but not all E18.5 UGE. (G) Timing and distribution of in vivo ShhcreERT2 activation is summarized with respect to other known events important for lower urogenital tract development. (H) In vitro cre activation during prostatic bud formation in cultured UGSs carrying the ShhcreERT2;R26R allele was demonstrated by growing E14.5 male UGSs in media containing DHT (10 nM) and 4-hydroxytamoxifen (4OHTam, 1 µm) and revealing β-galactosidase activity after 4 d in culture. Results represent at least 3 UGSs per genotype and per experimental condition. Abbreviations are bl: bladder, cl: cloaca, sv: seminal vesicle; wd: Wolffian duct. Arrowheads indicate prostatic buds. Scale bars are 500 µM.
E18.5 male Shh+/+ (control) and ShhcreERT2/+ mouse UGSs were stained by ISH to visualize the prostatic bud marker Nkx3-1 (purple). Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. Abbreviations are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers were determined as mean ± SE of five independent samples per group from at least three litters.
E14.5, E15.5 and E16.5 wild type control mouse near mid-sagittal UGS sections were immunofluorescently labeled to visualize CDH1 and UGS basal epithelial markers KRT14 and TRP63. Cell nuclei stained with DAPI. Results in each panel represent three males. Abbreviations are bl: bladder, sv: seminal vesicle, wd: Wolffian duct.
Pregnant wild type control dams were administered a solution containing tamoxifen (25 mg/kg maternal dose) and progesterone (18.75 mg/kg maternal dose) or corn oil (vehicle control, 2.5 ml/kg maternal dose) via once daily intraperitoneal injections on E13.5 and E14.5. (A–B) E18.5 whole-mount male UGSs from treated dams were stained by ISH to visualize Nkx3-1 (purple). Staining patterns represents five males. Blue arrowheads indicate ventral prostatic buds, red arrowheads are anterior prostatic buds and green arrowheads are dorsolateral prostatic buds. Abbreviations are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers are mean ± SE of five independent samples per group from at least three litters. An asterisk indicates a significant difference (p <0.05) from vehicle control.
E18.5 male Shh+/+;Ctnnb1tm1Mmt/tm1Mmt (control) and ShhcreERT2/+;Ctnnb1tm1Mmt/tm1Mmt (Ctnnb1iGOF) UGS sections. (A–B) Sections were immunofluorescently labeled to detect the smooth muscle marker ACTA2. Nuclei stained with DAPI (blue). (C–D) UGS sections were stained by ISH to visualize mRNA expression (purple) of the urethral mesenchymal pad marker Fgf10 and counterstained by IHC to visualize UGE marked by CDH1. Arrowheads indicate prostatic buds. Staining patterns in each panel represent three males. Abbreviations are bl: bladder, sv: seminal vesicle.
E14.5 male C57BL/6J wild type control mouse UGSs were incubated for 4 d in organ culture medium containing 10 nM 5α dihydrotesterosterone and vehicle (0.1% ethanol) or 4-hydroxytamoxifen (1 µM). Media and supplements were replenished after the second day. (A–B) UGSs were stained by ISH to visualize Nkx3-1 mRNA (purple). Staining patterns in each panel represent five males. Abbreviations used are bl: bladder, sv: seminal vesicle. (C) Nkx3-1+ bud numbers are the mean ± SE of five independent samples per group from at least three litters.