Abstract
Targeted DNA double-strand breaks introduced by rare-cleaving designer endonucleases can be harnessed for gene disruption applications by engaging mutagenic nonhomologous end-joining DNA repair pathways. However, endonuclease-mediated DNA breaks are often subject to precise repair, which limits the efficiency of targeted genome editing. To address this issue, we coupled designer endonucleases to DNA end-processing enzymes to drive mutagenic break resolution, achieving up to 25-fold enhancements in gene disruption rates.
The introduction of a targeted DNA break by designer endonucleases is a powerful technology that leverages evolutionarily conserved repair pathways to conduct genome engineering at the site of the lesion1. For example, the resolution of a double-strand DNA break by ‘error-prone’ nonhomologous end joining (NHEJ) can be harnessed to create a genetic knockout, as the NHEJ process can result in insertions or deletions at the site of the break2. This approach has been successfully applied in a variety of cell types and organisms, including rats, zebrafish, fruit flies, pigs and humans.
Recent work has demonstrated that NHEJ is mediated by several subpathways, each of which has distinct mutational consequences2,3. The classical NHEJ (cNHEJ) pathway requires the KU/DNA-PKcs/Lig4/ XRCC4 complex, and it ligates broken DNA ends back together with minimal processing2. In the absence or failure of the cNHEJ pathway, alternative NHEJ (altNHEJ) pathways are able to act on DNA ends. altNHEJ pathways typically utilize sequence microhomologies and are considerably more mutagenic and translocation prone2. DNA breaks created by the current designer endonuclease platforms (such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and homing endonucleases (HEs)) all leave compatible overhang breaks that do not require processing before ligation and are thus excellent substrates for precise repair by the cNHEJ pathway4. Precise break repair with sustained nuclease expression leads to a cycle of cleavage and repair unless the break is resolved mutagenically, often via an altNHEJ pathway4,5. Thus, many of the DNA breaks created by designer endonucleases are unproductive for genome engineering.
To increase the frequency of targeted disruption, we sought a means to drive mutagenic end processing before the precise rejoining of endonuclease-induced breaks by the cNHEJ pathway. As cotransfection of the 3′ repair exonuclease 2 (Trex2) with the canonical homing endonuclease I-SceI has previously been shown to limit precise rejoining of I-SceI-induced breaks5, we hypothesized that coupling rarecleaving endonucleases with DNA end-processing enzymes would be a general approach to increasing targeted gene disruption.
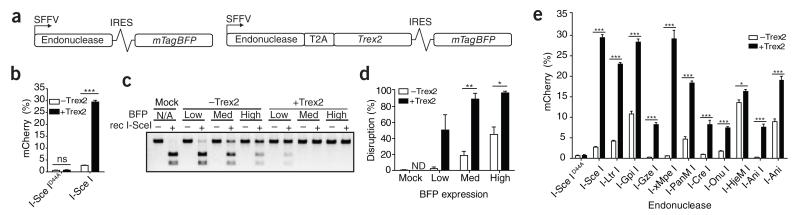
We assessed the effect of Trex2 on the mutagenic repair of doublestrand breaks generated by I-SceI. We developed expression vectors that drive coupled expression of both an endonuclease and Trex2 from a single promoter via a T2A ‘skip’ peptide motif6. We also included mTagBFP coexpression by an internal ribosomal entry site for tracking transfection efficiency (Fig. 1a). To measure the rate of targeted disruption, we transfected these constructs into a human HEK293T cell line containing a traffic-light reporter (TLR) harboring the cognate I-SceI target7 (Supplementary Fig. 1). This reporter generates mCherry-positive cells in response to a frame shift into the +3 reading frame at the target site, indicating targeted disruption. Although neither I-SceID44A (catalytically inactive) nor I-SceID44A coupled to Trex2 induced any measurable gene disruption, I-SceI coupled to Trex2 via T2A linkage produced a substantial increase in mCherry-positive cells compared to I-SceI alone (Fig. 1b; see Supplementary Fig. 2a for representative flow plots). Additionally, expression analysis of our BFP tracking fluorophore revealed that near-maximal gene disruption rates (~ 33% in the TLR) occurred at even very low endonuclease expression levels (Supplementary Fig. 2b).
figure 1.
Coupling endonucleases to exonucleases increases gene disruption. (a) Schematic of Trex2 exonuclease expression vectors. SFFV, spleen focus-forming virus promoter/enhancer; IRES, internal ribosomal entry site. (b) Quantification of mCherry expression in BFP-positive HEK293T cells transfected with the indicated vectors and analyzed 72 h after transfection (n = 3). (c) Sce-digestion NHEJ assay to measure total gene disruption. Cells were sorted based on the gates indicated in supplementary figure 2b. Rec, recombinant. (d) Quantification of band intensity (undigested out of total) of three independent experiments of the Sce digestion assay as performed in c. ND, not determined. (e) Quantification of mCherry expression in BFP-positive HEK293T cells harboring the respective TLR target for each of the indicated homing endonucleases (n = 3) and analyzed 72 h after transfection with the indicated vectors. Error bars, s.e.m. P values (*P < 0.05, **P < 0.005 and ***P < 0.0005) were calculated using the Student’s two-tailed unpaired t-test to compare the samples indicated in this and all subsequent figures.
To characterize in more depth the influence of Trex2 coexpression on double-strand break processing, we first sorted I-SceI.i.BFP- and I-SceI.T2A.Trex2.i.BFP-transfected cells based on varying BFP expression levels and PCR-amplified the area flanking the I-SceI target from each of the populations. We then digested the PCR product with recombinant I-SceI to look for a resistant band, indicative of a mutagenic event at the locus that destroyed the I-SceI target site. At low endonuclease expression levels, we observed a 25-fold increase in total gene disruption between I-SceI and I-SceI coupled to Trex2 (2.2% to 50.2%, respectively), and nearly 100% of targets were disrupted in the medium and high expression gates of I-SceI.T2A.Trex2 (90.3% and 97.1%, respectively) (Fig. 1c,d). These experiments indicate that whereas I-SceI exhibits a dose-dependent increase in gene disruption, I-SceI coupled to Trex2 quickly becomes saturated. Sequence analysis of the I-SceI target site in cells highly expressing I-SceI confirmed that 100% of cells were modified in the I-SceI.T2A.Trex2.i.BFP-treated cells (data not shown). Furthermore, sequencing of PCR amplicons encompassing the I-SceI cleavage site showed a trend toward smaller deletion events in the Trex2-treated cells (Supplementary Fig. 2c). In a kinetic analysis, although all constructs exhibited similar expression patterns, we found that Trex2 expression coincided with the appearance of disruption events at earlier time points (Supplementary Fig. 3). We also evaluated Trex2 activity at I-SceI-induced breaks in primary murine embryonic fibroblasts (MEFs) from a mouse model with an I-SceI site knocked into the interleukin-2 receptor subunit γ (IL2RG) locus (‘Sce-SCID’ mouse; G.C., J. Jarjour, D.J.R. and A.M.S., unpublished data). These experiments demonstrated that Sce-Trex2 expression produced a sixfold increase in disruption at the common γ-chain locus over that of I-SceI alone (I-SceI = 15.8%, I-SceI.T2A. Trex2 = 88.7%) (Supplementary Fig. 4). As IL2RG is only expressed in a subset of differentiated hematopoietic cells8, these experiments further demonstrate that Trex2 can facilitate high-frequency disruption at unexpressed loci. Finally, we evaluated the general applicability of Trex2-enhanced gene disruption for HE-induced breaks by generating HEK293T cell lines containing TLRs harboring the cognate target sites for a panel of different HEs (Supplementary Table 1). Transfection of each cell line with its respective enzyme with and without Trex2 demonstrated a consistent increase in disruption rates associated with Trex2 coupling, even with endonucleases that exhibit nearly undetectable activity when expressed alone (Fig. 1e).
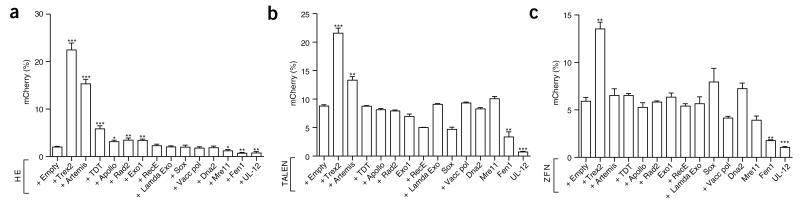
The practical utility of manipulating DNA break repair via coexpression of an endonuclease with a DNA end-processing enzyme would be enhanced if the strategy were generalizable. We therefore created a candidate library of 13 end-processing enzymes derived from mammalian, bacterial or viral species (Supplementary Table 2). We screened this library by expressing each enzyme in association with either the I-SceI HE, a VegF-targeted ZFN (VF2468)9 or a CCR5-targeted TALEN10 using HEK293T TLR lines containing the respective nuclease target sites (Fig. 2). Five of these enzymes (Artemis, TDT, Apollo, Rad2 and Exo1) increased the gene disruption efficiency of I-SceI (Fig. 2a), and none of the enzymes exhibited a substantial effect on homology-directed repair (Supplementary Fig. 5). We found that cotransfection of Trex2 with either the CCR5-targeted TALEN or VegF-targeted ZFN also resulted in increases in gene disruption, albeit to a lesser extent than when coupled to HEs (Fig. 2b,c). Coexpression of the CCR5 TALEN with the putative 5′ exonuclease, Artemis, promoted similar changes. Sequencing analysis of mutations demonstrated that Trex2 affects the resolution of ZFN- or TALEN-induced DNA breaks in a manner similar to the way it influences HE-induced breaks: there is an overall increase in total disruption rate, and a bias toward smaller deletion events (Supplementary Fig. 6).
figure 2.
DNA end-processing enzymes library screen. (a–c) Quantification of mCherry expression in BFP-positive HEK293T cells harboring the Sce-TLR (a; n = 5 or 6), the TLR with the CCR5 TALEN target site (b, n = 3) or the TLR with the VF2468 ZFN target site (c, n = 3). All quantifications were done at 72 h after transfection with the indicated vectors. Endonuclease expression was in all cases tracked with BFP. Asterisks indicate a significant difference between samples expressing an enzyme from the library with designer endonuclease and endonuclease with empty vector alone. Error bars, s.e.m.
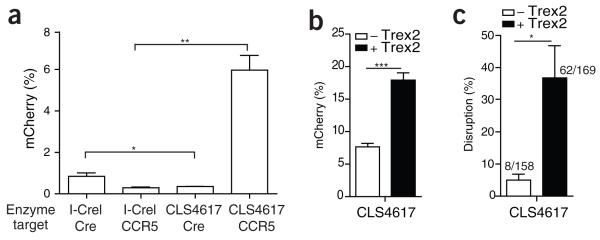
Finally, we evaluated this strategy in a therapeutically relevant context by engineering a homing endonuclease to cut a sequence in the gene encoding the HIV co-receptor CCR5 and delivering the endonuclease-exonuclease cassette using a lentiviral vector (LV). Targeted disruption of CCR5 has recently been established as a therapeutic modality for HIV and is being pursued in clinical trials11. This novel endonuclease was generated using combinatorial assembly of an archive of locally engineered I-CreI derivatives recognizing sequences differing from the wild-type I-CreI target by a few nucleotides12. This process yielded an enzyme (hereafter CLS4617) with 21 amino acid changes relative to the parental monomeric I-CreI recognizing a DNA target encoding the third transmembrane domain of CCR5 (Supplementary Fig. 7).
We first tested the activity and specificity of CLS4617 relative to its parental I-CreI using TLR cell lines that contained either the wild-type I-CreI target or the CCR5 target, and then we tested these lines with CLS4617 in the presence and absence of Trex2 (Figs. 3a,b and Supplementary Fig. 8a,b). The disruption capacity of CLS4617 expressed from LVs optimized for primary cells was substantially increased by the addition of Trex2. We then isolated and transduced CD34+ human hematopoietic stem cells derived from cord blood using LVs. We sequenced the endogenous CCR5 locus from flow-sorted BFP+ cells 72 h after transduction (Supplementary Fig. 8c) and scored the sequences for gene disruption (Fig. 3c). In these experiments, we observed a sevenfold increase in gene disruption of endogenous CCR5 with CLS4617 coupled to Trex2 over CLS4617 alone (5% and 37%, respectively).
figure 3.
Trex2 increases knockout of endogenous CCR5 with an engineered homing endonuclease. (a) Quantification of mCherry expression in BFP-positive HEK293T cells containing the TLR with the I-CreI cognate and CLS4617 targets 72 h after transfection with the indicated enzyme (n = 3). (b) Quantification of mCherry expression in BFP-positive HEK293T TLR cells with the CLS4617 target 72 h after transfection with the indicated vectors (n = 3). (c) Quantification of total disruption rate, analyzed by sequencing the CCR5 TLR target of BFP-sorted CD34+ cells transduced with the indicated lentiviral vectors at a multiplicity of infection of 20 (n = 3). Numbers above bars indicate total number of sequences (mutated/total). Error bars, s.e.m..
For all of the above experiments, we observed no overt toxicity as a result of overexpressing Trex2 in conjunction with any endonuclease, nor did we observe any effects of Trex2 on cell cycle (data not shown) or growth kinetics (Supplementary Fig. 9a,b) in the presence or absence of active endonucleases. We also observed no untoward effects of Trex2 on cells exposed to different types of model DNA-damaging agents (Supplementary Fig. 9c-h), which suggests that cells maintain high-fidelity repair at DNA lesions occurring independently of the endonuclease. Although it is expected that coupling Trex2 would result in increased mutagenesis at off-target sites, we observed minimal activity both with and without Trex2 at two previously characterized off-targets for the VegF ZFN13 despite clearly detectable activity at the endogenous locus (Supplementary Fig. 10). Further, we and others6 have found that the expression of Trex2 decreases the frequency of distal end joining (data not shown), which suggests that it may also decrease propensity for deleterious translocations.
In summary, we show that coupling DNA end-processing enzymes with the three main designer endonuclease platforms is a means to improve rates of targeted gene disruption in a variety of cell types and species, without notable associated cytotoxicity. Endo-exo coupling offers several advantages for investigators interested in using nuclease-induced genome engineering: a reduced need for promoters and expression systems that drive high-level persistent expression; elimination of ‘persistent’ breaks owing to reduced cleavage cycling, with an associated potential safety benefit of limiting break resolution by translocations and other deleterious repair events6; and enhanced potential for making multiple changes in a single round of mutagenesis via multi-allelic knockouts and multiplexing.
ONLINE METHODS
Construct assembly
The library of DNA processing enzymes was cloned into the pExodus vector with genes synthesized by Genscript as cDNA codon-optimized for human expression. pCVL Sce TLR, and subsequent reporters with different targets, were derived fromAddgene plasmid 31482, and constructs using the RRL backbone were derived from Addgene plasmid 12252. All other constructs were cloned into the RRL or CVL lentiviral backbones8 using standard molecular biology techniques. See Supplementary Note 1 for detailed sequence information and plasmid maps. Plasmids are available upon request.
Cell line derivation
HEK cell lines harboring the TLR were generated as previously reported8. Mice were maintained in a specific pathogen-free facility and were handled in accordance with protocols approved by the Institutional Animal Care and Use Committee of Seattle Children’s Research Institute. MEFs were isolated from ‘Sce-SCID’ embryos at 12-14 d gestation. Briefly, individual embryos were removed from the uterus and washed with PBS. The head and red tissue were removed from the embryo, and the remaining tissue was minced. The tissue was incubated with trypsin-EDTA for 10 min at 37 °C, followed by centrifugation at 10,000g for 5 min. The pellet was resuspended in MEF medium and plated at 37 °C. For isolation of CD34+ cells, human umbilical cord blood was obtained from the Puget Sound Blood Center cord-blood donor program, and CD34+ cells were isolated using Miltenyi CD34+ beads (Miltenyi Biotec) according to the manufacturer’s instructions. Purity of CD34+Lineage− cells was >90% in all experiments.
Cell culture
HEK and MEF cells were cultured in glutaminefree Dulbecco’s Modified Eagle’s Medium supplemented with 2 mM l-glutamine, 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. CD34+ cells were maintained and expanded in StemSpan (Stemcell Technologies) and supplemented with human cytokines (100 ng/mL SCF, TPO and FLT3-l; PeproTech) and 1 μM StemRegenin1 (Cellagen Technology). CD34+ medium with cytokines was exchanged every 2-3 d during culture. The levels of CD34 (CD34-PE, Becton Dickinson) and lineage markers (Lineage Cocktail-FITC, Becton Dickinson) were assessed by FACs every 2-3 d during culture, and all experiments were performed on cells with greater than 80% CD34+Lineage− cells.
Transfection
1.0 × 105 HEK293T cells were plated 24 h before transfection in 24-well plates. 0.5 μg DNA was used for each expression vector and was transfected using Fugene6 or XtremeGene9 (Roche) according to the manufacturer’s protocol. Cells were analyzed on a cytometer 72 h after transfection unless otherwise indicated.
Lentivirus generation
Recombinant lentivirus (LV) was produced by transient cotransfection of HEK293T cells as previously described14. Briefly, cells were transfected in 10-cm dishes using PEI (Polysciences, Inc.) with 6 μg vector, 1.5 μg pMD2G envelope plasmid (VSV-G) and 3 μg psPAX2 per plate. Virus was titered by Lenti-x p24 rapid ELISA kit (Clontech). MOI was calculated for fluorescently tagged virus experiments by dividing the amount of infectious units added by the number of cells plated. Infectious units were calculated by transducing 2.0 × 105 HEK293T cells with increasing amounts of viral stocks, analyzing them on a flow cytometer 72 h after transduction and applying the following formula for volumes of virus that yielded between 5% and 20% fluorescent-positive cells: ((2.0 × 105 × percent fluorescent-positive cells)/100)/ volume viral stock added).
Transduction
For Sce-SCID MEF experiments, 1.0 × 105 cells were seeded in a 24-well plate 24 h before transduction. Cells were transduced with indicated amounts of LV and 4 μg/mL polybrene. Cells were passaged 24 h later and analyzed 72 h after transduction. For CD34+ cells, cells were thawed and expanded in media and cytokines for 1 week before transduction. 5.0 × 106 cells were transduced with MND.4617.T2A.BFP and MND.4617.T2A.Trex2.T2A. BFP lentiviral vectors at an MOI of 20 for 8 h in 4 μg/mL polybrene. Cells were spun down, and new culture media was added. 72 h after transduction, BFP+ cells were sorted and washed in PBS, and pellets were frozen before genomic DNA isolation.
Flow cytometry
Cells were analyzed on a BD LSRII or BD FACSAria III. The mCherry fluorophore was excited using a 561-nm laser and acquired with a 610/20 filter. The mTagBFP fluorophore was excited on a 405-nm laser with a 450/50 filter. Data was analyzed using FlowJo software. In experiments defining expression via gating analysis (for example: low, medium and high), gates are set by visual inspection of the mean fluorescent intensity of the fluorophore and are applied consistently across control and experimental groups for each replicate.
Sce-digestion NHEJ assay
Genomic DNA was isolated from bulk or sorted cells as indicated using Qiagen’s DNeasy kit, and TLR target sites were PCR amplified as previously described8. 100 ng of each PCR product was digested in vitro with recombinant I-SceI (New England Biolabs) for 6 h at 37 °C. DNA was separated using a 1% agarose gel stained with ethidium bromide. Percent disruption was calculated by quantifying band intensity using ImageJ software and dividing the intensity of the undigested band by the total.
NHEJ sequencing
Genomic DNA was isolated from cells using Qiagen DNeasy kit (Qiagen) and the HE target region was amplified as previously described8. PCR products were cloned using a CloneJET PCR cloning kit (Fermentas) according to the manufacturer’s protocol, followed by transformation into chemically competent DH5α Escherichia coli bacteria. Bacterial colonies were directly sequenced using a standard colony-sequencing protocol. Sequences were analyzed using the Contig Express software provided in Invitrogen’s Vector NTI software suite.
CCR5 homing endonuclease engineering
The CLS4617 enzyme specific for CCR5 was designed as previously described12. Briefly, an archive of locally engineered I-CreI derivatives recognizing sequences differing from the I-CreI wild-type target by a few nucleotides were combinatorially assembled to generate a new endonucleases with the desired specificity.
TALEN engineering
A Xanthomonas pthXo1 Golden Gate destination ORF and an RVD plasmid library were kind gifts from the Voytas laboratory. The CCR5-specific TALEN pair was assembled with a Golden Gate cloning strategy as described15 into a pthXO1 scaffold truncated at positions N154 and C63 as previously described10.
Toxicity experiments
1.0 × 106 Sce-SCID MEFs were seeded in a 10-cm dish 24 h before transduction. 500 μL of 10× LV (pCVL.SFFV. sceD44A.IRES.BFP or T2A.TREX2.IRES.BFP) was added to the culture with 4 μg/mL polybrene. 24 h after transduction, cells were passaged to 15-cm plates. 72 h after transduction, 1.0 × 105 Sce-SCID MEFs were seeded in a 12-well plate with 1 mL media and treated as indicated with mitomycin C (Sigma Aldrich), xamptothecin (Sigma-Aldrich) or ionizing radiation. 48 h after exposure, cells were incubated in 0.5 μg/mL PI as above and analyzed by flow cytometry. For CD34+ cells, 72 h after transduction with Trex2-expressing LV, 2.0 × 105 CD34+ HSCs were seeded in a 96-well plate in 200 μL of media, DNA damaging agents were added to the media and plates were analyzed as above.
Statistical analysis
Error bars represent s.e.m. and P values (with * representing P < 0.05, ** P < 0.005 and *** P < 0.0005) were calculated using the Student’s two-tailed unpaired t-test to compare samples.
Supplementary Material
ACKNOWLEDGMENTS
M.T.C. was supported in part by the US Public Health Service and National Research Service Award (T32 GM07270) from the US National Institute of General Medical Sciences. Additional funding was from the US National Institutes of Health (RL1CA133832, UL1DE019582, R01-HL075453, PL1-HL092557, RL1-HL092553, RL1-HL92554 and U19-AI96111) and Seattle Children’s Center for Immunity and Immunotherapies. We would like to thank J. Stark (Beckman Research Institute and City of Hope) and D. Stetson (University of Washington) for initial Trex2 expression plasmids and D. Voytas (University of Minnesota) for TALEN cloning reagents. Zinc-finger nuclease expression plasmids were provided by C. Ramirez and K. Joung (Harvard University and Massachusetts General Hospital). J. Jarjour (Seattle Children’s Research Institute, presently Pregenen Inc.) designed the Sce-SCID mouse, and Sce-SCID MEFs were provided by A. Astrakhan and G. Metzler (University of Washington). We thank all members of the Northwest Genome Editing Consortium for their many insightful discussions.
Footnotes
Accession codes. pCVL Sce TLR, and subsequent reporters with different targets, were derived from Addgene plasmid 31482, and constructs using the RRL backbone were derived from Addgene plasmid 12252.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.T.C. designed and conceived experiments. M.T.C., K.S.G., R.K., B.S., G.C., M.B. and T.M. performed experiments. A.R.L., S.K.B., K.J., M.B., B.Y.R., H.-P.K., A.G. and F.P. provided reagents. M.T.C. wrote the paper and K.S.G., D.J.R. and A.M.S. edited the paper. D.J.R. and A.M.S provided technical support and conceptual advice.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McVey M, Lee SE. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mladenov E, Iliakis G. Mutat. Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Shrivastav M, De Haro LP, Nickoloff JA. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 6.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szymczak AL, Vignali DAA. Expert Opin. Biol. Ther. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- 8.Certo MT, et al. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Nat. Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JC, et al. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 11.Cannon P, June C. Curr. Opin. HIV AIDS. 2011;6:74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J, et al. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sather BD, et al. Mol. Ther. 2011;19:515–525. doi: 10.1038/mt.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cermak T, et al. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.