Abstract
The two LIM domain-containing proteins from plants (LIMs) typically exhibit a dual cytoplasmic–nuclear distribution, suggesting that, in addition to their previously described roles in actin cytoskeleton organization, they participate in nuclear processes. Using a south-western blot-based screen aimed at identifying factors that bind to plant histone gene promoters, we isolated a positive clone containing the tobacco LIM protein WLIM2 (NtWLIM2) cDNA. Using both green fluorescent protein (GFP) fusion- and immunology-based strategies, we provide clear evidence that NtWLIM2 localizes to the actin cytoskeleton, the nucleus, and the nucleolus. Interestingly, the disruption of the actin cytoskeleton by latrunculin B significantly increases NtWLIM2 nuclear fraction, pinpointing a possible novel cytoskeletal–nuclear crosstalk. Biochemical and electron microscopy experiments reveal the ability of NtWLIM2 to directly bind to actin filaments and to crosslink the latter into thick actin bundles. Electrophoretic mobility shift assays show that NtWLIM2 specifically binds to the conserved octameric cis-elements (Oct) of the Arabidopsis histone H4A748 gene promoter and that this binding largely relies on both LIM domains. Importantly, reporter-based experiments conducted in Arabidopsis and tobacco protoplasts confirm the ability of NtWLIM2 to bind to and activate the H4A748 gene promoter in live cells. Expression studies indicate the constitutive presence of NtWLIM2 mRNA and NtWLIM2 protein during tobacco BY-2 cell proliferation and cell cycle progression, suggesting a role of NtWLIM2 in the activation of basal histone gene expression. Interestingly, both live cell and in vitro data support NtWLIM2 di/oligomerization. We propose that NtWLIM2 functions as an actin-stabilizing protein, which, upon cytoskeleton remodeling, shuttles to the nucleus in order to modify gene expression.
Key words: actin, BY-2, cytoskeleton, DNA-binding, histone genes, LIM, Nicotiana tabacum, promoter regulation, trans-acting factors
INTRODUCTION
Histones are well-conserved proteins that are highly abundant in eukaryotic cell nuclei. They are classified into five subtypes: the four core histones H2A, H2B, H3, and H4, which package and arrange chromosomal DNA into nucleosomes; and the linker histone H1 which is associated with the DNA stretch interconnecting the nucleosomes (Luger et al., 1997; Robinson and Rhodes, 2006).
In plants, histone-mediated changes in chromatin structure and DNA organization affect various processes such as root growth, flowering time, as well as gametophyte or embryo formation (Berr et al., 2011). Similarly to histone biosynthesis in yeast and animals, the coordinated and mostly S phase-specific activation of plant histone expression is for the most part regulated by transcriptional and posttranscriptional processes (Atanassova et al., 1992; Terada et al., 1993; Reichheld et al., 1995; Ohtsubo et al., 1997; Reichheld et al., 1998; Taoka et al., 1999). The preferential activity of histone gene promoters in dividing cells suggests transcriptional regulation as a predominant control mechanism for plant histone gene expression (Atanassova et al., 1992; Brignon and Chaubet, 1993; Terada et al., 1995; Chaubet et al., 1996; Atanassova et al., 1998; Taoka et al., 1999; Minami et al., 2000). Accordingly, analyses of plant histone promoters in transient (Nakayama et al., 1992; Lepetit et al., 1993; Sakamoto et al., 1996; Taoka et al., 1998) or stable approaches (Terada et al., 1995; Chaubet et al., 1996; Ohtsubo et al., 1997) have revealed the presence of positive and negative cis-acting elements of transcription.
Two cis-elements are reported to be common to all known replication-dependent plant histone genes: the moderately conserved nonameric motif (NON, CATCCAACG), which has been originally identified as a positive cis-element of wheat H3 and maize H4 genes (Nakayama et al., 1992; Lepetit et al., 1993); and the CCGTC (CG) element, which has been described as a trans-factor-binding site of the Arabidopsis H4, as well as of maize H3 and H4 gene promoters (Brignon and Chaubet, 1993). Interestingly, NON and CG motifs, which are known to participate in tissue-specific histone gene transcription, such as in meristems (Brignon and Chaubet, 1993; Chaubet et al., 1996), are frequently present in a pairwise fashion and, accordingly, may function interactively (Brignon and Chaubet, 1993; Chaubet and Gigot, 1998; Meshi et al., 2000). Another nonameric motif, the AGATCGACG stretch of the Arabidopsis histone H4A748 gene promoter, which is known to be essential for proliferation-specific histone promoter activity, has been identified by in vivo footprinting and was recognized as a positive regulator of histone gene expression in meristematic tissues (Brignon and Chaubet, 1993; Chaubet et al., 1996; Shen and Gigot, 1997; Meshi et al., 2000). The well-conserved CAT-named cis-element (GCCAAT) was identified in a maize H3 promoter as a nuclear factor-binding site by in vivo footprinting experiments (Brignon and Chaubet, 1993). Interestingly, the same experimental approach revealed a reversed CAT motif in the H4A748 gene promoter, where it acts as a strong positive cis-element (Chaubet et al., 1996).
One highly conserved cis-element, namely the octamer CGCGGATC (Oct), is common to most plant histone gene promoters. This octamer appears in at least one copy in the proximal part of promoters and can be present in a direct or reverse orientation. In several promoters, these octamer copies appear as an imperfect or degenerate motif (dOct) (Chaubet et al., 1996; Robertson et al., 1997; Taoka et al., 1999; Meshi et al., 2000; Okada et al., 2005). Analyses in transient and stable expression assays have identified the octamer as a proliferation-coupled and S phase-specific cis-element (Chaubet et al., 1996; Ohtsubo et al., 1997; Taoka et al., 1999), which also confers tissue-specific expression, such as in newly developing meristems (Brignon and Chaubet, 1993; Terada et al., 1995; Chaubet et al., 1996; Taoka et al., 1999; Minami et al., 2000). However, octameric cis-elements are also known to be involved in basal, replication-independent histone gene transcription, such as in adult tissues and quiescent cells (Brignon and Chaubet, 1993; Chaubet et al., 1996; Robertson et al., 1996). Octamers often act in combination with another module, thereby forming three types (types I, II, and III) of Oct-containing composite elements (OCEs), which all function as separable S phase-specific elements (Yang et al., 1995; Ohtsubo et al., 1997; Taoka et al., 1998, 1999; Meshi et al., 2000). The type I element (CCACGTCANCGATCCGCG) consists of a reverse-oriented Oct paired with another conserved histone promoter motif, the hexameric ACGTCA sequence (HEX). The HEX element of type I OCEs has also been identified as a separate motif in wheat H3 and H4 promoters as a recognition site of wheat DNA-binding factors (Tabata et al., 1989, 1991; Minami et al., 2000). Mutational knockout or deletion of this hexamer reduces promoter strength and leads to a loss of cell-cycle-specific expression (Terada et al., 1995; Taoka et al., 1999). In type II OCEs, the Oct motif is paired with a TCA module to form a conserved 11-bp element (TCACGCGGATC), whereas the type III element (GATCCGCG-N14-ACCAATCA) is composed of a reverse-oriented Oct and an 8-bp sequence (ACCAATCA, referred to as CCAAT-box), which are separated by a 14-bp spacer (Taoka et al., 1999; Meshi et al., 2000). Interestingly, the 8-bp CCAAT-box, which functions in a variety of animal histone promoters and is targeted by distinct DNA-binding proteins (Maity and de Crombrugghe, 1998; Mantovani, 1998), seems unrelated to the above-mentioned CAT motifs (GCCAAT) from Arabidopsis and maize (Meshi et al., 2000). In plants, the CCAAT-box has been described as a positive cis-element of the wheat histone H1 promoter TH315 (Taoka et al., 1998).
In order to identify DNA-protein interactions within plant histone promoters, various biochemical assays, including in vivo footprinting, gel shift, and UV crosslinking experiments, have been conducted. Such assays revealed the existence of NON-binding proteins in maize H3 and H4 promoters, as well as in a tobacco H3 promoter (Brignon and Chaubet, 1993; Reichheld et al., 1998). Additional in vivo footprinting experiments revealed the presence of Oct-binding proteins for maize H3 and H4, tobacco H3, and Arabidopsis H4 promoter sequences (Brignon and Chaubet, 1993; Chaubet et al., 1996; Shen and Gigot, 1997; Reichheld et al., 1998). Gel shift assays unraveled cell cycle-specific Oct- and HEX-binding activities for nuclear extracts from wheat and tobacco, respectively (Shen and Gigot, 1997; Minami et al., 2000). However, our knowledge about proteins that interact with plant histone cis-elements is still rather limited (Mikami et al., 1987; Tabata et al., 1989, 1991; Kawaoka and Ebinuma, 2001; Kaothien et al., 2002).
The two LIM domain-containing proteins from plants (plant LIMs) are short (~200-aa) proteins, which are structurally related to the vertebrate cysteine-rich proteins (CRPs) and characterized by two tandemly arranged LIM domains (Baltz et al., 1992; Weiskirchen and Gunther, 2003; Kadrmas and Beckerle, 2004; Arnaud et al., 2007). The term ‘LIM’ has derived from the first letter of the proteins LIN-11 (Freyd et al., 1990), Isl1 (Karlsson et al., 1990), and MEC-3 (Way and Chalfie, 1989) from which the LIM domain was historically identified. In animals, the LIM domain is found in numerous cytoplasmic and/or nuclear proteins, and is usually considered to function as a protein-binding interface (Kadrmas and Beckerle, 2004). Like the vertebrate CRPs, plant LIMs display a dual nuclear–cytoplasmic distribution and, for both protein families, several isoforms have lately been shown to fulfill cytoplasmic functions as actin-binding and -bundling proteins (Grubinger and Gimona, 2004; Tran et al., 2005; Thomas et al., 2006; Wang et al., 2008; Jang and Greenwood, 2009; Papuga et al., 2010). In comparison to the family of vertebrate CRPs, the size of the LIM family in plants is rather limited and, in dependence on their expression patterns, plant LIM family members have been classified into two groups: the widely expressed LIM proteins (WLIMs), which are expressed in most sporophytic tissues and display no or only little expression in pollen; and the PLIM proteins, which show predominant and abundant expression in pollen grains (Eliasson et al., 2000; Arnaud et al., 2007; Papuga et al., 2010).
Implication of plant LIMs in nuclear processes has initially been suggested by the in vitro binding of the sunflower pollen protein PLIM-1 to DNA and by the trans-acting properties of tobacco protein WLIM1 (Baltz et al., 1996; Kawaoka et al., 2000; Kawaoka and Ebinuma, 2001; Kaothien et al., 2002). We have identified tobacco LIM protein WLIM2 (NtWLIM2) in a south-western blot-based screen for proteins binding to plant histone gene promoters. Using both green fluorescent protein (GFP) fusion- and immunochemistry-based strategies, we provide clear evidence that NtWLIM2 exhibits a typical dual nuclear and cytoskeletal localization. Bimolecular fluorescence complementation analyses indicate that NtWLIM2 homodimerizes in live cells, which is consistent with in vitro biochemical data. We demonstrate the ability of NtWLIM2 to directly bind to actin filaments and to induce the formation of actin bundles in vitro. Furthermore, electrophoretic mobility shift assays show that NtWLIM2 specifically targets DNA sequences, namely the Oct motifs present in the histone H4A748 gene promoter of Arabidopsis. Transient protoplast assays reveal that NtWLIM2 activates the transcription of a histone H4A748 promoter-driven reporter gene. We discuss potential functions of NtWLIM2, which, in addition to acting as an actin-binding and -bundling protein in the cytoplasm, may play a role in the nucleus as a trans-acting factor that, through its interaction with octameric cis-elements, participates in the activation of basal histone gene expression.
RESULTS
Isolation of NtWLIM2 cDNA
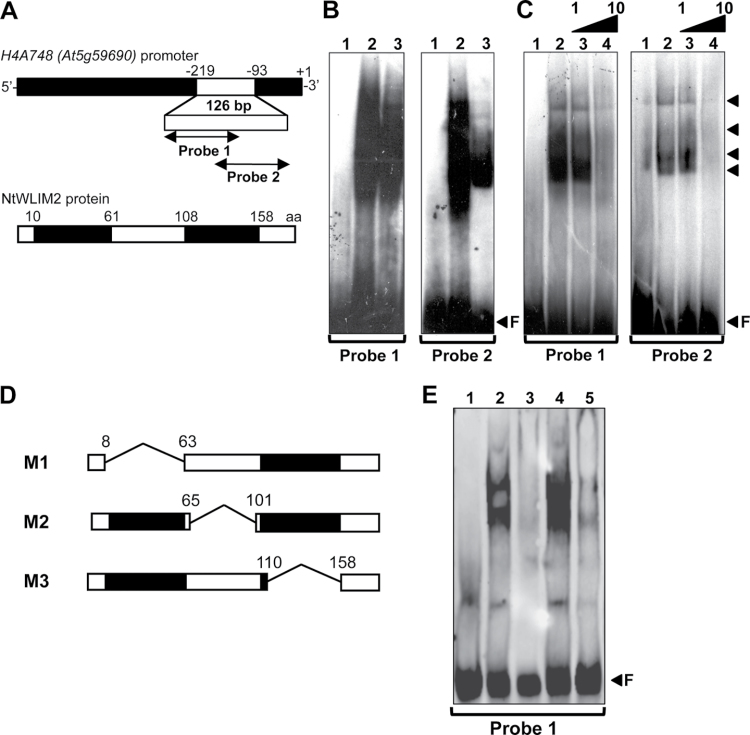
To identify proteins capable of binding to conserved cis-elements of plant histone gene promoters, we performed a south-western blot-based screen of a tobacco BY2 cDNA expression library (Shen and Gigot, 1999). As a probe, we used a double-stranded 126-bp-long oligonucleotide of the Arabidopsis histone H4A748 gene promoter (At5g59690). This 126-bp fragment (position –93 to –219 relative to transcription start) was previously shown to bear several highly conserved plant histone promoter cis-elements that preferentially direct proliferation-coupled and S phase-specific expression (Atanassova et al., 1992; Chaubet et al., 1996). One of the sequenced positive clones contained a 966-bp-long cDNA of the tobacco WLIM2 gene (NtWLIM2; NCBI Accession No. Y11002 and AF184886). The NtWLIM2 cDNA encodes a protein of 190amino acids, which shares sequence homologies (52% identity and 66% similarity; see Supplemental Figure 1A) with the tobacco WLIM1 protein (NtWLIM1; NCBI Accession No. AF184109) (Eliasson et al., 2000), which has been previously described as a DNA-binding and promoter-activating protein (Kawaoka et al., 2000; Kaothien et al., 2002). Like NtWLIM1, NtWLIM2 is structurally related to the vertebrate CRP family with a short N-terminus, two LIM domains separated by a long (40–50 residues) spacer (interLIM domain) and a C-terminal domain, which exhibits the highest degree of variability (Supplemental Figure 1B).
NtWLIM2 Binds Via Its LIM Domains to the Histone H4A748 Promoter In Vitro
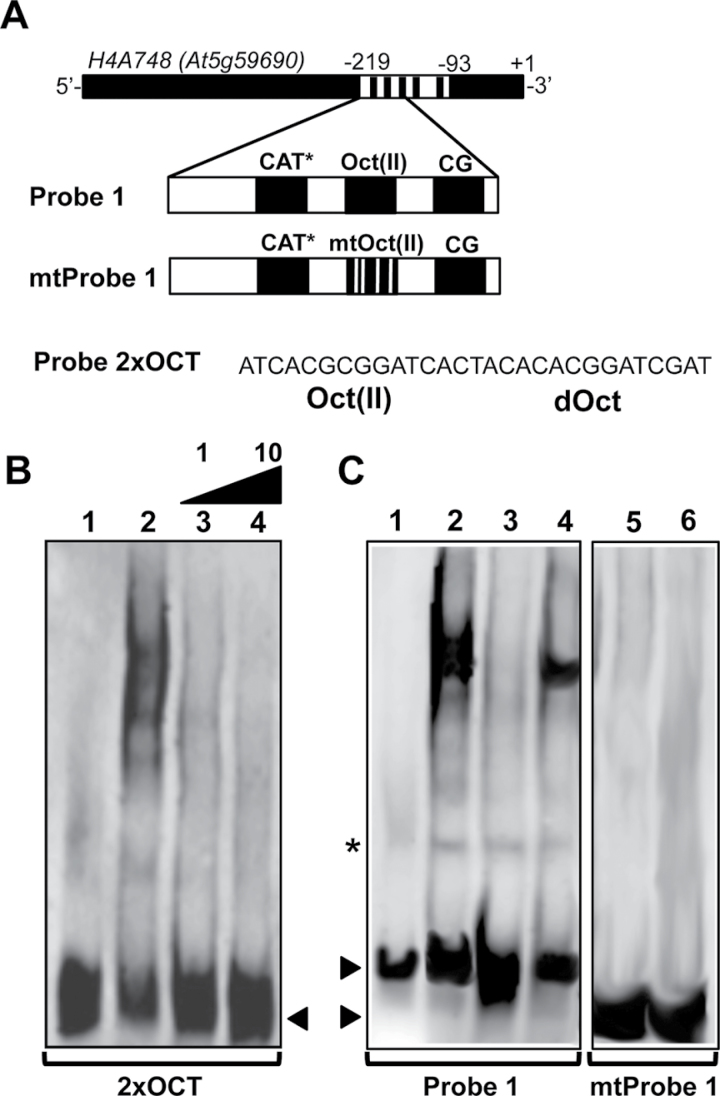
In order to confirm the binding of NtWLIM2 to the Arabidopsis histone H4A748 gene promoter, we expressed and purified a recombinant NtWLIM2 protein, which we tested for its DNA-binding ability by electrophoretic mobility shift assay (EMSA). In parallel, we tested the DNA-binding capability of nuclear protein extracts from tobacco BY-2 cells. As probes, we used two γ32P-labeled double-stranded oligonucleotides which together cover the 126-bp-long fragment of the Arabidopsis histone H4A748 gene promoter (position –93 to –219) used in the primary screen (Figure 1A). Both probes bear several highly conserved and frequently described plant histone promoter cis-elements (Atanassova et al., 1992; Chaubet et al., 1996). To not disrupt any of these cis-elements, the probes were designed to overlap by 17bp. EMSA experiments performed with Probe 1 (Figure 1B, left panel) and Probe 2 (Figure 1B, right panel) showed that migration of both probes was strongly retarded in the presence of either nuclear BY-2 protein extract (Figure 1B, lanes 2) or recombinant NtWLIM2 (lane 3), when compared to probe migration in the absence of any protein (lane 1). The large smear observed in the presence of nuclear BY-2 cell extract hinted at a multitude of presumably different proteins interacting with both histone promoter probes. Several DNA-binding protein complexes were detected in the presence of recombinant NtWLIM2 protein (Figure 1C, right panel, arrows lanes 2 and 3), suggesting that the protein can form oligomers. To verify the binding specificity of the recombinant NtWLIM2 protein to Probes 1 and 2, competition assays were conducted by respectively adding non-labeled Probe 1 (Figure 1C, left panel) or Probe 2 (Figure 1C, right panel) to the binding assay. Whereas the presence of equimolar amounts of unlabeled specific competitor Probes 1 and 2 hardly weakened the retarded bands caused by NtWLIM2 (Figure 1C, lane 3), the use of a 10-fold molar excess visibly diminished the shifted signal (Figure 1C, lane 4). This result confirmed the ability of NtWLIM2 to directly interact with the proximal region of the histone H4A748 promoter covered by Probes 1 and 2.
Figure 1.
In Vitro DNA-Binding Ability of NtWLIM2 in Electrophoretic Mobility Shift Assays (EMSAs).(A) Scheme of the Arabidopsis histone H4A748 gene promoter and the derived double-stranded probes. Nucleotide positions are given relative to the transcription start (+1). To avoid disruption of any promoter cis-element, Probe 1 (–219 to –147) and Probe 2 (–161 to –93) were chosen to overlap by 17bp. Schematic representation of the recombinant NtWLIM2 wild-type protein used in the following EMSAs (B, C).(B) EMSA to test the DNA-binding activity of NtWLIM2. One ng of radiolabeled Probe 1 (left panel) or Probe 2 (right panel) was incubated with either DNA-binding buffer (lane 1), nuclear BY-2 protein extract (1μg, lane 2), or purified recombinant NtWLIM2 (3μg, lane 3).(C) Competitive EMSA to check the DNA-binding affinity of NtWLIM2. One ng of radiolabeled Probe 1 (left panel) or Probe 2 (right panel) was incubated with either DNA-binding buffer (lane 1) or purified recombinant NtWLIM2 protein (3μg, lanes 2–4). Lanes 3 and 4 correspond to binding reactions supplemented with equimolar and a 10-fold molar excess of probe, respectively.(D) In addition to the wild-type NtWLIM2, deletion mutants lacking either the LIM1 domain (M1), the interLIM region (M2), or the LIM2 domain (M3) were expressed as 6xHis fusions, affinity-purified, and tested in EMSAs. (E) EMSA with DIG-labeled probes to determine the DNA-binding domains of NtWLIM2. One ng of labeled Probe 1 was incubated with either DNA-binding buffer (lane 1) or 3μg of different recombinant proteins (lanes 2–5): wild-type NtWLIM2 (lane 2) or one of the deletion mutants M1, M2, and M3 (lane 3, 4, and 5, respectively). Arrows mark the migrated free probe (F) and the DNA–protein complexes formed.
To identify the NtWLIM2 domain(s) implicated in DNA-binding, three deletion mutants of wild-type NtWLIM2 were generated: M1 and M3 were deleted for the LIM1 and LIM2 domain, respectively, and M2 was deprived of the interLIM domain (Figure 1D). EMSAs with the truncated NtWLIM2 proteins revealed that the deletion of either LIM1 or LIM2 domain (Figure 1E, lane 3 and 5, respectively) led to the abolishment of the shifted signal obtained with full-length NtWLIM2 (Figure 1E, lane 2). In contrast, truncation of the interLIM domain had no effect on the binding of NtWLIM2 to the labeled probe (Figure 1E, lane 4).
Together, the here-presented EMSA data indicate that NtWLIM2 binds directly and specifically to the Arabidopsis H4A748 promoter and that this binding involves both LIM domains of NtWLIM2.
NtWLIM2 Localizes to Both the Cytoplasm and the Nucleus, where It Forms Dimers
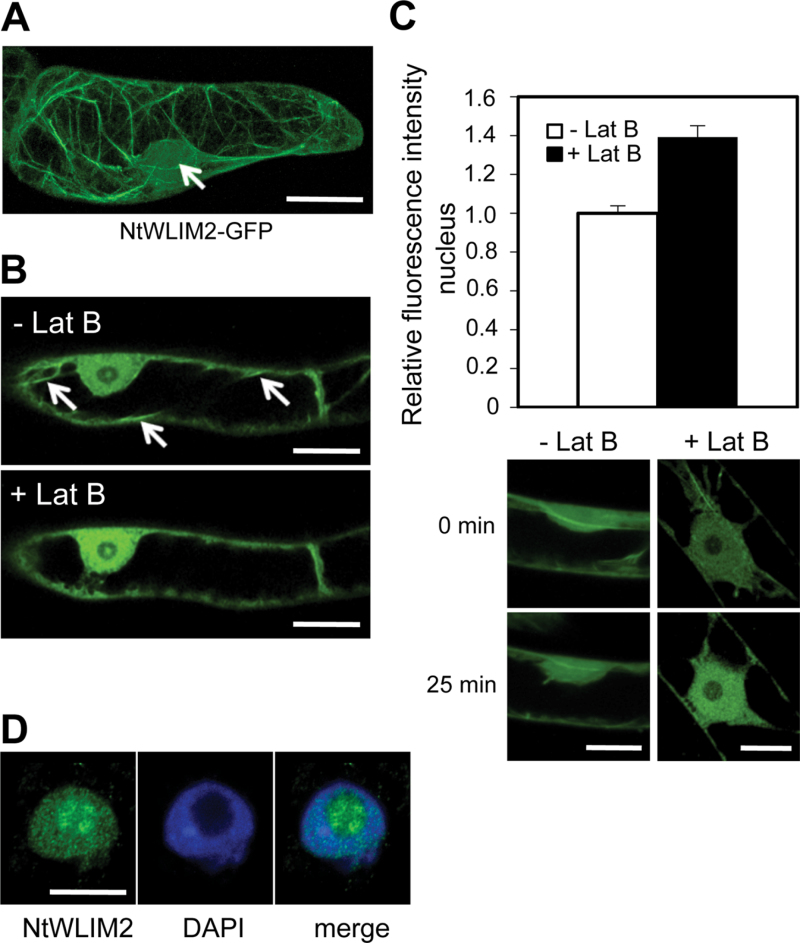
The DNA-binding ability of NtWLIM2 implies a nuclear localization of this tobacco LIM protein. To test this hypothesis, a stable transgenic BY-2 cell line expressing a NtWLIM2–GFP fusion protein was generated and submitted to confocal microscopy investigations. In the cytoplasm, NtWLIM2–GFP decorated a filamentous network (Figure 2A), which was confirmed as the actin cytoskeleton by rhodamine–phalloidine co-labeling (Supplemental Figure 2). In addition to its cytoplasmic localization, NtWLIM2–GFP substantially accumulated within the nucleoplasm (Figure 2A, arrow). An identical subcellular distribution in BY-2 cells was observed for an N-terminal fusion of NtWLIM2 with GFP (GFP–NtWLIM2, data not shown).
Figure 2.
NtWLIM2 Subcellular Localization in BY-2 Cells. (A) Typical distribution of NtWLIM2–GFP in stably transformed BY-2 cell lines. Beside a clear nuclear localization (arrow), NtWLIM2–GFP interacted with filamentous cytoplasmic structures, later identified as the actin cytoskeleton (Supplemental Figure 2). (B) Modification of NtWLIM2–GFP subcellular distribution following Lat B treatment. Transgenic NtWLIM2–GFP cells were treated with 2.5μM Lat B and GFP fluorescence distribution was monitored over 25min. A notable increase of nuclear signal is observed upon Lat B treatment. Arrows point to prominent cables (upper panel), which disappeared after Lat B treatment (lower panel).(C) Quantification of GFP fluorescence in the nucleus of BY-2 cells after 25-min mock or Lat B treatment (white column and black column, respectively). GFP fluorescence intensity in the nucleus after 25min of mock or Lat B treatment was determined in a defined area using ImageJ software and normalized to the GFP fluorescence measured in the same area at the start of the time course. Error bars denote the 95% confidence interval obtained for these GFP fluorescence ratios (n=23 for Lat B-treated cells, n=7 for mock-treated cells, p-value obtained by Student’s t-test=1.27e–12). Image panel: typical GFP fluorescence intensity in the nucleus of BY-2 cells before (upper panels) and after treatment with mock buffer (control, left panels) or Lat B (right panels). Scale bars: 5μm.(D) Immunodetection of endogenous NtWLIM2 (left) by anti-WLIM2 antibody in isolated BY-2 nuclei and nucleoli (see Supplemental Figure 3 for the specificity of this antibody). DAPI counterstain (middle panel) enabled localization of WLIM2 (left panel) relative to the nuclear shape in the merged picture (right panel). Scale bar: 5μm.
To test a possible and direct link between the cytoskeletal and nuclear fractions of NtWLIM2, the effects of the actin filament disrupting drug latrunculin B (Lat B) were analyzed. Data indicate that a 25-min incubation with Lat B provokes a marked increase of the nuclear GFP fluorescence (Figure 2B). Quantification of the nuclear fluorescence revealed that Lat B treatment induced a 40 % increase of the NtWLIM2–GFP nuclear fraction (Figure 2C). In contrast, the nuclear amount of NtWLIM2–GFP in control, mock-treated cells remained unchanged over time. Although a detailed analysis of the mechanism underlying the nuclear shuttling of NtWLIM2 is beyond the scope of this work, the above data suggest that NtWLIM2 is part of a signaling pathway connecting the actin cytoskeleton and the nucleus.
To gain a better insight into the nuclear localization of the endogenous NtWLIM2, we performed immunolocalization assays on paraformaldehyde-fixed, isolated nuclei from wild-type BY-2 cells using a polyclonal anti-WLIM2 antibody, whose specificity for NtWLIM2 was controlled by immunoblot analyses (Supplemental Figure 3). Confocal analyses confirmed and extended the above GFP fusion-based data with a clear accumulation of endogenous NtWLIM2 in both the nucleus and nucleoli of BY-2 cells (Figure 2D).
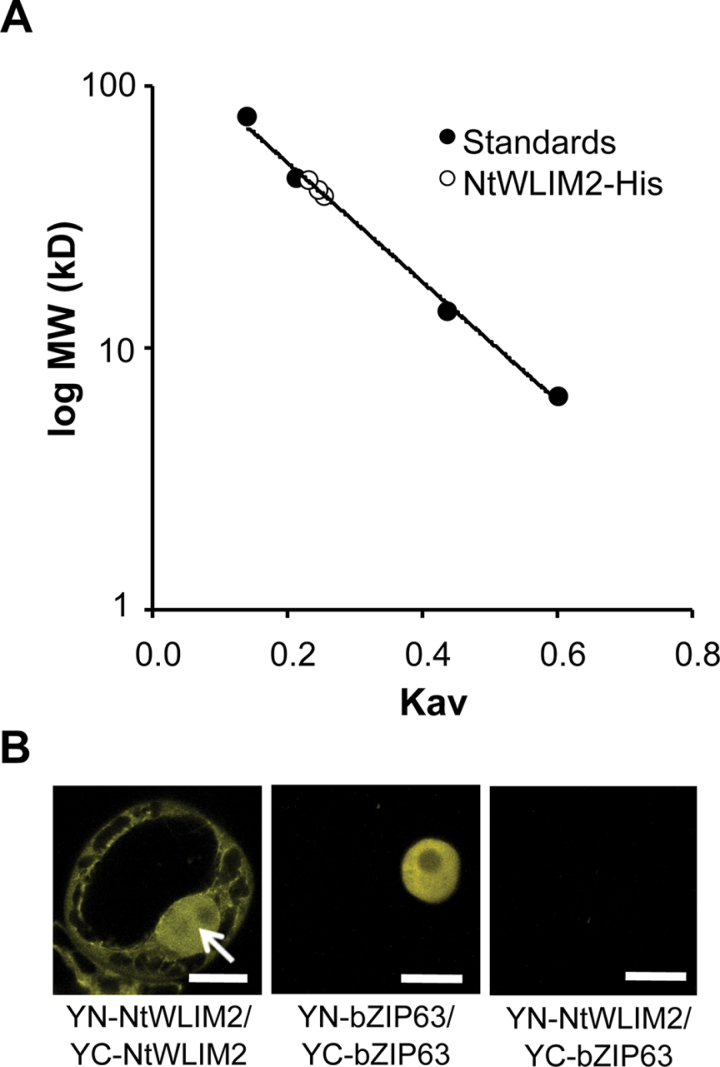
Interestingly, preparative purification of recombinant NtWLIM2 protein via size exclusion chromatography yielded the elution of NtWLIM2 as a 40-kD-sized protein (Figure 3A), whereas, when deduced from its amino acid sequence, NtWLIM2 mass is predicted to be 21kD. This experi mentally observed size duplication indicates dimerization of NtWLIM2 and thus corroborates the conclusions drawn from the observation of multiple supershifted bands in the preceding EMSA data (see Figure 1C). To follow up on this question in the cellular context, we performed bimolecular fluorescence complementation analyses (BiFC) in living BY-2 cells. Co-expression of two NtWLIM2 peptides fused to the N-terminal and the C-terminal part of the yellow fluorescent protein, respectively (YN-NtWLIM2 and YC-NtWLIM2), led to the formation of BiFC complexes, confirming the formation of NtWLIM2 homodimers in the nucleus (Figure 3B, left panel). Co-expression of YN- and YC-constructs of the homodimer-forming Arabidopsis transcription factor bZIP63 (At5g28770) (Walter et al., 2004; Waadt et al., 2008), which were used as positive control, yielded a YFP-fluorescence intensity comparable to that obtained with the NtWLIM2 constructs (Figure 3B, middle panel). As negative controls, we combined either the YN-NtWLIM2 plasmid with the YC-fusion of bZIP63 or the YN-bZIP63 plasmid with the YC-fusion of NtWLIM2. Both combinations yielded no or only very weak fluorescence (Figure 3B, right panel and data not shown, respectively).
Figure 3.
NtWLIM2 Dimerizes In Vitro and In Vivo. (A) Molecular weight determination of an affinity-purified NtWLIM2–His recombinant protein. Molecular weight was determined by size exclusion chromatography using a Tricorn Superdex 75 10/300 GL column (GE Healthcare) calibrated with aprotinin (6.5kD), ribonuclease (13.7kD), ovalbumin (44kD), conalbumin (75kD), and Blue dextran (2000kD). The partition coefficient Kav was calculated for the standard proteins and plotted against the log of the molecular weight (filled circles). The Kav values of NtWLIM2 obtained from three independent experiments were plotted as open circles on the standard curve and molecular weight of 40kD was deduced. (B) Bimolecular fluorescence complementation assays in the nucleus (arrow) of living BY-2 cells co-expressing pSPYNE(R)173–NtWLIM2 and pSPYCE(MR)–NtWLIM2. The yellow fluorescent signal due to the formation of functional YFP indicates NtWLIM2 homodimerization (left panel). Combination of pSPYNE(R)–bZIP63 and pSPYCE(MR)–bZIP63 served as a positive control (middle panel). Co-expression of either pSPYCE(MR)–bZIP63 with pSPYNE(R)173–NtWLIM2 (right panel) or pSPYCE(MR)–NtWLIM2 with pSPYNE–bZIP63 (data not shown) yielded no or only background fluorescence (right panel). Scale bars: 5μm.
NtWLIM2 Directly Binds To and Bundles Actin Filaments In Vitro
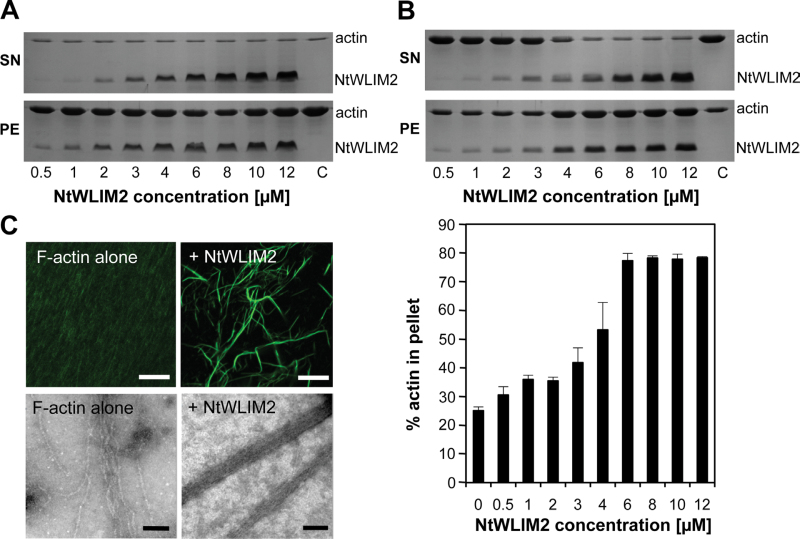
The co-localization of NtWLIM2–GFP with the actin cytoskeleton in the cytoplasm of BY-2 cells prompted us to investigate whether NtWLIM2 displays the same actin regulatory activities as other, previously described plant LIM proteins (Thomas et al., 2006, 2007; Wang et al., 2008; Papuga et al., 2010). First, the capability of NtWLIM2 to bind to F-actin in a direct manner was tested in high-speed cosedimentation assays (Figure 4A). Increasing amounts of recombinant NtWLIM2 (0.5–12µM) were incubated for 1h with polymerized F-actin (4µM), the mixture was centrifuged, and the resulting pellet and supernatant fractions were analyzed by SDS–PAGE. Control experiments showed that NtWLIM2 did not sediment significantly when centrifuged in the absence of F-actin (data not shown). When centrifuged in the presence of F-actin, NtWLIM2 was found enriched in the pellet fraction, indicating its ability to directly interact with actin filaments. An apparent equilibrium dissociation constant (K d±SD) of 1.83±0.18µM was calculated from three independent experiments and, noticeably, this K d value was similar to that determined for its homolog NtWLIM1 (1.7±0.2µM) (Thomas et al., 2007).
Figure 4.
In Vitro Characterization of Actin Regulatory Activities of NtWLIM2. (A) High-speed cosedimentation assays. Increasing concentrations of NtWLIM2 (0.5–12μM) were cosedimented (100000g, 45min) with AFs (4μM). The resulting pellet and supernatant fractions were subjected to SDS–PAGE and Coomassie Blue stain (see representative gel). (B) Low-speed cosedimentation assays. F-actin (4μM) was cosedimented (12500g, 45min) in the presence of increasing amounts of NtWLIM2 (0.5–12μM). After centrifugation, pellet and supernatant fractions were analyzed by SDS–PAGE and subsequent Coomassie Brilliant Blue stain. A representative gel is shown in the upper panel. Gels were scanned and the percentages of actin present in the pellet and in the supernatant were quantified. Results are expressed as the percentage of actin that sediments as a function of the concentration of NtWLIM2 (lower panel). In the absence of NtWLIM2, 25%±1.3% of actin is found in the pellet. Values represent the mean of three independent experiments. Error bars represent standard deviations. SN, supernatant fraction; PE, pellet fraction. (C) Direct visualization of NtWLIM2-induced actin bundles by fluorescence light microscopy (upper panel) and electron microscopy (lower panel). Alexa 488-labeled AFs (4μM) were polymerized alone (left) or in the presence of NtWLIM2 (8μM; right) and observed under a confocal microscope. Bars: 10μm. Electron micrographs show the negatively stained preparations of AFs alone (8μM; left) and in the presence of NtWLIM2 (4μM; right). Bars: 100nm.
To investigate whether NtWLIM2 functions as an F-actin crosslinking protein, we performed low-speed cosedimentation assays. Four micromolar AFs were copolymerized in the presence of increasing NtWLIM2 concentrations (0.5–12µM) and submitted to a centrifugation at 12500g, which only pellets higher-order actin structures. In the absence of NtWLIM2, most of the actin was detected in the supernatant fraction (Figure 4B, upper panel). By contrast, NtWLIM2 induced actin sedimentation in a concentration-dependent manner, indicating that it crosslinks AFs into higher-order structures. Three independent low-speed cosedimentation experiments were used to quantify the respective amounts of actin in the supernatant and pellet fractions. The results, which are expressed as the percentage of actin in the pellet fraction (Figure 4B, lower panel), not only confirm the progressive sedimentation of actin shown by the representative SDS–PAGE (Figure 4B, upper panel), but also reveal that saturation occurs at an NtWLIM2 concentration of between 6 and 8µM.
To visualize the higher-order actin structures triggered by NtWLIM2, we used fluorescence light microscopy and negative-staining electron microscopy (Figure 4C, upper and lower panels, respectively). Both strategies unambiguously demonstrated that NtWLIM2 crosslinks AFs into long and thick actin bundles.
In conclusion, NtWLIM2, like other plant LIM proteins from lily, Arabidopsis, or tobacco, functions as an actin-binding protein, which triggers the formation of actin bundles in an autonomous manner.
NtWLIM2 Specifically Activates pH4A748-Reporter Expression in Protoplasts
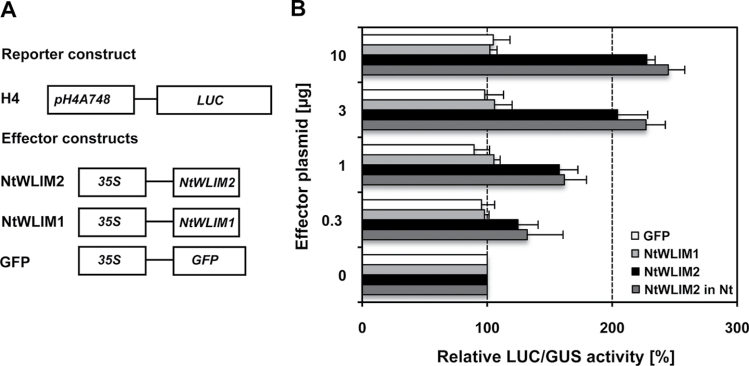
To elucidate the role of NtWLIM2 in targeting the histone H4A748 promoter in vivo, we cloned appropriate effector and reporter constructs for transient expression analyses (Figure 5A). The reporter construct contained the firefly luciferase (LUC) gene under the control of a 604-bp-long fragment of the Arabidopsis histone H4A748 gene promoter (pH4A748; NCBI Accession No. M17132, position +1 to –529 relative to transcription start), which comprised TATA-box, transcription start, and the (74-bp-long) complete 5’-UTR (Chabouté et al., 1987). Effector plasmids allowed constitutive expression of NtWLIM2 or NtWLIM1 (control) under the control of the CaMV 35S promoter. Regulation of reporter gene expression by tobacco WLIM effectors was analyzed in a transient protoplast system (Yoo et al., 2007; Moes et al., 2008). Either Arabidopsis thaliana or Nicotiana tabacum protoplasts were transfected with both the promoter-LUC reporter and effector plasmids. In addition, protoplasts were co-transformed with a 35S driven β-glucuronidase (GUS) reporter gene for standardization of LUC expression. Transient expression of LUC and GUS in Arabidopsis or tobacco protoplasts was measured 20h after co-transfection of reporter, effector, and control plasmids, and the measured LUC activity was normalized relative to the constitutive GUS activity. As shown in Figure 5B, co-expression of the H4-controlled LUC reporter with the NtWLIM2 effector construct led to a clear activation of the pH4A748-driven LUC expression. Indeed, addition of 0.3–10µg of NtWLIM2 expressing plasmid resulted in an up to 2.5-fold amplification of pH4A748-controlled LUC expression in Arabidopsis protoplasts, when compared to the LUC activity registered in the presence of 0.3–10µg of a GFP-expressing plasmid, the latter being used as an unspecific effector plasmid (Figure 5B, black and white bars, respectively). Similar pH4A748-controlled LUC activities were obtained for tobacco protoplasts (Figure 5B, dark gray bars). Additionally, constitutive expression of a GFP–NtWLIM2 expressing effector plasmid led to comparable levels of LUC activity (data not shown). Interestingly, the NtWLIM2-dependent, 2.5-fold increase in pH4A748-controlled LUC expression is comparable to the trans-activating activity previously reported for NtWLIM1, which increased the activity of its specific target promoter by three- to fourfold in similar transient assays (Kawaoka and Ebinuma, 2001). Interestingly, in both protoplast systems, the 3.3-fold increase of effector plasmid from 3 to 10µg led to relatively slight raises in LUC activity. Whereas the 3.3-fold increases in effector plasmid from 0.3 to 1µg and from 1 to 3µg amplified LUC activity by ~ 25%, only a 10% increase of LUC activity could be observed when the amount of effector plasmid was raised from 3 to 10µg. This result might be due to a dose-dependent saturation of the reporter–effector system. Remarkably, and similarly to what was observed with the GFP control effector, no activation of pH4A748 could be observed in the presence of increasing amounts of an NtWLIM1-expressing effector plasmid (Figure 5B, light gray and white bars, respectively). To amplify the data about LIM-dependent Arabidopsis H4A748 promoter regulation, we have documented the DNA-binding and promoter trans-activation abilities of the Arabidopsis homologs of NtWLIM2 and NtWLIM1, namely AtWLIM2a (At2g39900) and AtWLIM1 (At1g10200), respectively (Arnaud et al., 2007). LUC reporter activity tests performed with AtWLIM effectors in the transient Arabidopsis protoplast system confirmed the results observed with NtWLIM effectors (Supplemental Figure 4A and 4B): whereas constitutive expression of AtWLIM2a led to a more than twofold increase in pH4A748-driven reporter expression, no or only very weak activation of pH4A748-driven LUC expression was observed in the presence of AtWLIM1 effector. In agreement with the in cellula experiments, EMSA analysis shows a clear in vitro binding ability of AtWLIM2a to the Arabidopsis H4A748 promoter (Supplemental Figure 4C). Thus, the results obtained with AtWLIM2a in both EMSA and protoplast assays support our results obtained with NtWLIM2. Together, all these data indicate that NtWLIM2 acts as a specific activator of histone H4A748 promoter-driven gene expression.
Figure 5.
Effect of NtWLIM2 Expression on Arabidopsis H4A748 Gene Promoter Activation in Arabidopsis and Tobacco Protoplasts. (A) Reporter and effector constructs used in the study. In the H4 reporter construct, firefly luciferase (LUC) expression was controlled by a 604-bp-long fragment of the Arabidopsis histone H4A748 gene promoter. Effector constructs allowed cDNA expression of either full-length NtWLIM2, NtWLIM1, or GFP under control of the CaMV 35S promoter (35S). (B) NtWLIM2 activates pH4A748-controlled LUC expression in protoplasts. Effector and reporter constructs were co-transfected into Arabidopsis or tobacco protoplasts. LUC activity in protoplast extracts was measured 20h after transfection and normalized against GUS activity. The signal of reporter activity in the presence of each effector is given relative to the signal measured with the same concentrations of an empty vector, which is set to 100% (NtWLIM2, NtWLIM1, GFP: black, light gray, white bars in Arabidopsis protoplasts). Additional experiments were conducted for the NtWLIM2 effector construct in tobacco protoplasts (dark gray bars). Data represent mean values of several independent transfections (n≥3) and error bars indicate standard deviations.
NtWLIM2 Interacts with the Oct Cis-Element In Vitro
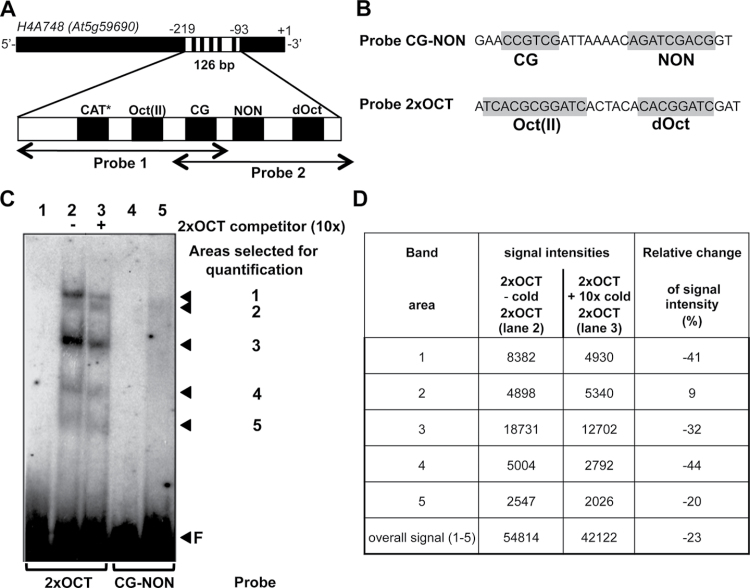
The above results indicate a direct interaction of NtWLIM2 with one or several of the conserved plant histone promoter cis-elements located within the 126-bp-long fragment of the Arabidopsis histone H4A748 promoter, which is covered by Probes 1 and 2. Probe 1 includes three of these current promoter motifs (Figure 6A). It contains the reversed GCCACT-like (CAT) motif (Chaubet et al., 1996; Meshi et al., 2000), as well as the highly conserved octamer (Oct) motif CGCGGATC, which is paired with a TCA module to form a type-II OCE, named Oct(II) (Taoka et al., 1999; Meshi et al., 2000). The third conserved histone promoter motif of Probe 1 is the hexameric CG-element (CCGTCG) (Chaubet et al., 1996; Meshi et al., 2000). Probe 2 contains the same CG motif as Probe 1 and, in addition, it holds the Arabidopsis-specific NON element (AGATCGACG), as well as the less conserved degenerated octamer (dOct; CACGGATC) (Chaubet et al., 1996; Meshi et al., 2000), the latter being located downstream of the NON element (Figure 6A). As the CG motif is the unique cis-element located in the overlapping region of Probes 1 and 2, the interaction of NtWLIM2 with both probes might reflect its ability to recognize the CCGTCG sequence. Alternatively, NtWLIM2 might interact with the two nearly identical Oct(II) and dOct motifs present in Probes 1 and 2, respectively. As a last scenario, NtWLIM2 might recognize unrelated elements specific to each of the probes. To discriminate between these possibilities and further characterize the NtWLIM2–histone promoter complex, additional gel retardation analyses were conducted, using two novel, shorter DNA probes containing different H4A748 promoter cis-elements (Figure 6B). The first probe contained the above-mentioned hexameric CG motif and the NON sequence of Arabidopsis (Probe CG–NON, Figure 6B), since CG and nonameric elements frequently appear in a pairwise fashion and are suggested to function interactively (Brignon and Chaubet, 1993; Chaubet and Gigot, 1998). The second probe included the Oct(II) and dOct cis-elements of the H4A748 gene (Probe 2xOCT, Figure 6B).
Figure 6.
NtWLIM2 Interaction with Conserved, Plant Histone Promoter cis-Elements. (A) Position of conserved plant histone promoter cis-elements within Arabidopsis H4A748 Probes 1 and 2. Cis-elements located on the reverse strand are marked by an asterisk. CAT, GCAAT-like element; Oct(II), type-II octamer-containing composite element TCACGCGGATC; CG, CCGTC motif; dOct, type II-like, degenerated octamer CACGGATC; NON, nonameric motif AGATCGACG. (B) Conserved plant histone promoter motifs present in Probe 2xOCT and Probe CG–NON (highlighted in gray). (C) EMSA reveals complexes formed between the recombinant NtWLIM2 protein and Arabidopsis pH4A748 promoter cis-elements. One ng of either radiolabeled Probe 2xOCT (lanes 1–3) or CG–NON (lanes 4–5) was incubated with protein storage buffer (lanes 1 and 4, respectively) or 6xHis-tagged affinity-purified NtWLIM2 protein (3μg, lanes 2, 3, and 5). A 10-fold molar excess (lane 3) of cold, double-stranded oligonucleotide 2xOCT was added as a specific competitor to the binding reactions. Arrows mark the migrated free probe (F) and the DNA–protein complexes, whose intensities were quantified by Image J (see (D)). (D) Intensity of shifted bands as determined by ImageJ. Signal intensities for every band are given in the presence and absence of cold 2xOCT probe (band area 1–5). In addition, the sum of the signal over all bands is indicated (overall signal (1–5)). To express the changes in signal intensity produced by the addition of 2xOCT competitor, the ratio of the signal intensity values is calculated for every band and expressed in percent.
Figure 6C shows the result obtained from gel retardation assays performed with 2xOCT probe (lanes 1–3) and Probe CG–NON (lanes 4–5). Compared to the migration of the 2xOCT probe in the absence of NtWLIM2 protein (lane 1), a clearly shifted signal composed of five distinct bands could be observed when NtWLIM2 was added to the DNA-binding reaction (lane 2). The addition of specific 2xOCT competitor to a 10-fold molar excess reduced the formation of the NtWLIM2–DNA complex (lane 3). In contrast to the 2xOCT probe, the CG–NON probe did not yield any band shift (lanes 4–5). The competitive experiments with labeled 2xOCT probe and a 10-fold excess of cold 2xOCT probe appeared to be less efficient than the competitive EMSAs previously obtained for Probes 1 and 2 (Figure 1C). To better characterize the differences in band shift intensities obtained by the addition of cold 2xOCT, the signal intensity of all five shifted bands was estimated using ImageJ software (Figure 6D). In the presence of 2xOCT as a competitor, four out of five previously described bands showed a 20%–40% reduction in their signal intensity, thereby supporting an effective competition. Surprisingly, the signal of band 2 was not competed out by cold 2xOCT, which may indicate an unspecific interaction between the labeled 2xOCT DNA and NtWLIM2.
To confirm the specific recognition of H4A748 octameric cis-elements by NtWLIM2, additional competitive EMSAs were carried out (Figure 7). In the first experiment, we used 2xOCT as a labeled probe and Probe 1 as an unlabeled competitor (Figure 7B). The successful competition of labeled 2xOCT carried out with either an equimolar amount or a 10-fold molar excess of unlabeled Probe 1 corroborated the formation of specific complexes between the H4A748 octamer motifs and NtWLIM2 (Figure 7B). To further analyze the specificity of NtWLIM2 for octameric elements, we performed two additional EMSA experiments, the first with labeled Probe 1 and either Probe 1 or mtProbe 1, a mutated form of Probe 1, as an unlabeled competitor (Figure 7C, left panel). The mtProbe1 DNA comprised base exchanges within the Oct(II) element, which are known to reduce the promoter activity of wheat H3 and Arabidopsis H4 genes (Terada et al., 1995; Chaubet et al., 1996). In this competition EMSA, the band shift obtained with recombinant NtWLIM2 was efficiently out-competed by unlabeled Probe 1 (Figure 7C, lanes 2 and 3, respectively). Interestingly, this was not the case when unlabeled mtProbe1 was used as a competitor (Figure 7C, lane 4). If labeled mtProbe 1 was incubated with either protein storage buffer (Figure 7C, lane 5) or recombinant protein (3µg, Figure 7C, lane 6), no band shift was obtained for NtWLIM2. Together, the above EMSA data demonstrate that NtWLIM2 specifically binds to Oct-related cis-elements and does not interact with CG motifs.
Figure 7.
Determination of NtWLIM2 Binding Specificity by Competitive EMSAs. (A) Scheme of the Arabidopsis histone H4A748 gene promoter and the derived double-stranded Probe 1, mtProbe 1, and 2xOCT used as labeled probes and competitors in the following EMSAs. The mtProbe 1 comprises base exchanges within the conserved Oct(II) element (5’-TCACGCGGATC-3’ changed to 5’-TCACGaaGcTt-3’), which are known to reduce plant histone H3 and H4 promoter activities (Terada et al., 1995; Chaubet et al., 1996). (B) Competition EMSA carried out with 1ng of DIG-labeled 2xOCT probe, which was incubated with either protein storage buffer (lane 1) or purified recombinant NtWLIM2 protein (3μg, lanes 2–4). The shift of the labeled oligo was competed out by the addition of either an equimolar amount or a 10-fold molar excess of unlabeled Probe 1 (lanes 3 and 4, respectively). Arrows mark the migrated free probe. (C) EMSA to analyze the binding specificity of NtWLIM2 to the H4A748 Oct(II) element. A competitive assay (left panel) was performed with 1ng of DIG-labeled Probe 1 (lanes 1–4), which was incubated with either protein storage buffer (lane 1) or recombinant NtWLIM2 protein (3μg, lanes 2–4). A 10-fold molar excess of either cold Probe 1 (lane 3) or cold mtProbe 1 (lane 4) was added as a specific competitor to the binding reactions. In a binding assay (right panel), 1ng of DIG-labeled mtProbe1 (lanes 5–6) was incubated with either protein storage buffer (lane 5) or with 3μg of recombinant NtWLIM2 protein (lane 6). Arrows mark the migrated free probes. An unspecific band shift is indicated by an asterisk.
In Vivo Binding of NtWLIM2 to pH4A748 Involves Octameric Cis-Elements
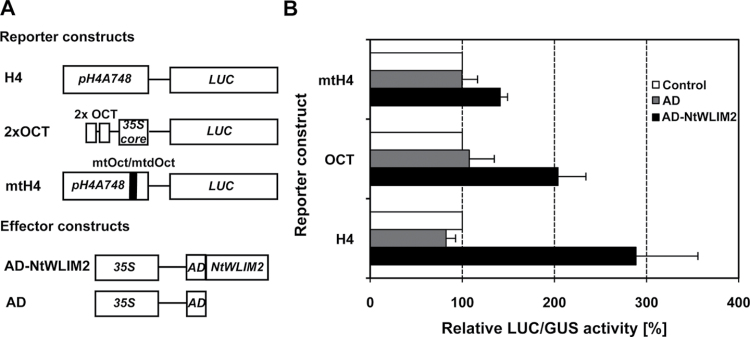
To test the functionality of NtWLIM2 binding to the octameric cis-elements in living cells, we analyzed the trans-activating ability of a p35S-driven AD–NtWLIM2 effector construct, which contained the NtWLIM2 cDNA fused to the transcriptional activation domain (AD) of herpes simplex virus protein VP16 (Sadowski et al., 1988). The VP16 AD represents a widely used tool to characterize transcription factors in transient Arabidopsis protoplast assays, such as the DNA-binding and transcription-regulating NAC proteins from soybean (Hao et al., 2010). The effect of AD–NtWLIM2 was tested on the LUC activity of various reporter constructs in Arabidopsis protoplasts (Figure 8A). Protoplasts that express pH4A748-controlled LUC in the presence of the AD–NtWLIM2 effector gave almost three times higher LUC activities than protoplasts that have been transformed with either an unfused AD effector plasmid or a GFP-expressing control construct (Figure 8B). The presence of AD–NtWLIM2 in protoplasts expressing the 2xOCT-controlled LUC reporter induced a twofold increase in the LUC activity, supporting that Oct-elements may play a central role in the DNA-binding of NtWLIM2 (Figure 8B). We therefore also tested a mutagenized form of the Arabidopsis histone H4A748 gene promoter (mtH4), containing base substitutions in both Oct(II) and dOct motifs, which were previously shown to reduce promoter activity of the wheat H3 and the Arabidopsis H4 genes (Terada et al., 1995; Chaubet et al., 1996). Accordingly, the mtH4 promoter is only weakly activated in the presence of AD–NtWLIM2 effector, showing an only 1.3-fold increase in LUC activity (Figure 8B). This again supports the importance of the Oct-elements for NtWLIM2-binding. Similar results were obtained when the LUC activity of the three reporter constructs (H4, mtH4, and 2xOCT) was measured in the presence of constitutively expressed NtWLIM2–AD effector (data not shown). These results indicate that the in vivo interaction of NtWLIM2 with the histone H4A748 gene promoter requires functional octamer elements.
Figure 8.
Trans-Activation of pH4A748-, Mutated pH4A748-, and Oct Motif-Controlled Reporters by Trans-Activation Domain-Fused NtWLIM2. (A) Reporter and effector constructs used in this study. H4 reporter was as previously described (Figure 4). The 2xOCT reporter plasmid contained two tandemly oriented copies of the EMSA oligonucleotide 2xOCT fused upstream to the –46CaMV 35S core promoter (35S core) that controlled LUC expression. In mtH4, LUC was under the control of a mutated version of pH4A748 that comprised point mutations within the octamer and degenerated octamer elements. The effector construct AD–NtWLIM2 consisted in a fusion of the trans-activation domain (AD) of herpes simplex virus protein (Sadowski et al., 1988) to NtWLIM2. An unfused AD construct was used as a negative control effector. (B) Level of trans-activation of pH4A748-, mutated pH4A748-, and 2xOCT motif-controlled reporters by AD-fused NtWLIM2 in Arabidopsis protoplasts. The LUC activity measured in the presence of 1μg of effector plasmid is given relative to the LUC activity measured in the presence of 1μg of GFP-expressing vector, the latter being set to 100% (white bars, control). The two used effector constructs allowed ectopic expression of either AD–NtWLIM2 (black bars) or unfused AD (dark gray bars; AD). Data represent mean values of three independent transfections and error bars indicate standard deviations.
NtWLIM2 Gene Transcription and Protein Levels Are Independent of Cell Division and Proliferation
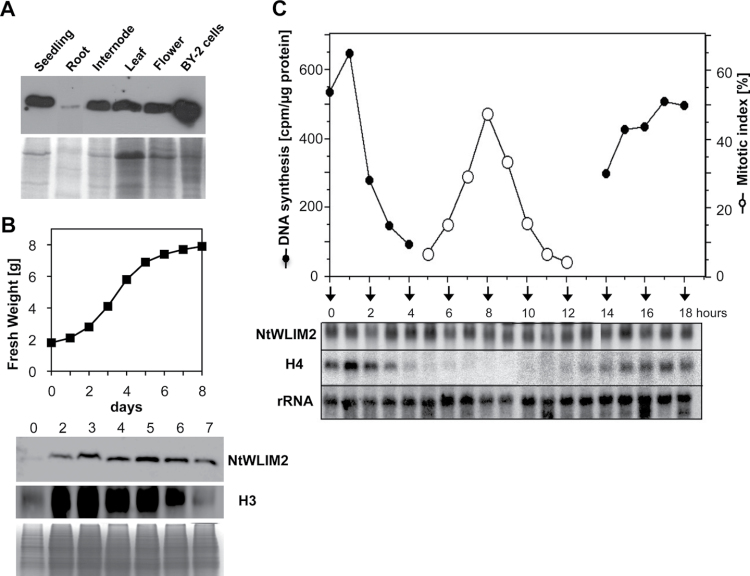
Previous histochemical analyses of transgenic pH4A748–GUS tobacco and Arabidopsis plants have shown dominant reporter expression in proliferating tissues such as root and shoot meristems, young leaves, and exponentially growing suspension cultures, but also a basal constitutive expression in adult plants, such as in internodes, leaves, flowers, and roots (Atanassova et al., 1992; Chaubet et al., 1996). To see whether the NtWLIM2 protein content correlates with the expression of its target gene H4A748, total protein extracts were prepared from 3-week-old seedlings, various organs of 8-week-old plants, and 3-day-old BY-2 suspension cultures, and analyzed by Western blotting using our specific anti-WLIM2 antibody. NtWLIM2 was abundant in seedlings, internodes, leaves, and flowers, whereas it was only weakly present in roots (Figure 9A). Thus, in non-proliferative Arabidopsis tissues, the distribution of NtWLIM2 protein paralleled the areas of basal histone H4A748 promoter activity. Interestingly, NtWLIM2 was most abundant in BY-2 suspension cultures. The strong NtWLIM2 protein accumulation in BY-2 cells and the fact that plant histones are mostly abundant in highly dividing cells induced us to have a closer look at protein expression through growth cycle and cell cycle phases. We traced protein levels through the growth cycle of a freshly subcultured BY-2 cell suspension culture. To mark the different growth phases of the BY-2 cell culture, we monitored the levels of histone H3 protein, the latter showing a progressive increase during the exponential growth phase and a substantial decay during the stationary phase, namely when the BY-2 cell proliferation rate is reduced (Figure 9B). Regarding NtWLIM2, relatively low protein amounts were detected at the time of subculture (Figure 9B, Day 0), but then NtWLIM2 accumulated to a steady-state level that remained constant over the exponential and stationary growth phases (Figure 9B, Days 2–5 and Days 6–7, respectively). Such a constitutive, growth-phase-independent expression pattern clearly differed from the proliferation-dependent abundance of histone H3 protein.
Figure 9.
Expression Profile of NtWLIM2. (A) Immunodetection of endogenous NtWLIM2 in tobacco protein extracts. Total protein (10μg) was extracted from 3-week-old whole seedlings and 8-week-old plant organs of Nicotiana tabacum, as well as from suspension-cultured BY-2 cells. Proteins were separated by SDS–PAGE and transferred onto a PVDF membrane. NtWLIM2 was detected at the expected size of 22kD using a polyclonal anti-NtWLIM2 antibody. Coomassie-stained gels are shown as loading control. (B) Time course of NtWLIM2 and histone H3 protein expression through the growth cycle of a BY-2 suspension culture. Fresh weight of BY-2 cells was determined from 10-ml samples after culture medium removal. Ten μg of total BY-2 protein was loaded on an SDS–PAGE and blotted onto a PVDF membrane. Immunodetection of NtWLIM2 was performed as described in Figure 1A. Histone H3 protein was detected at the predicted size of 18kD using a monoclonal anti-histone H3 antibody. (C) Time course of NtWLIM2 and histone H4A748 expression during BY-2 cell cycle. Samples of aphidicolin-synchronized BY-2 cells were taken at different time points and analyzed for DNA synthesis and mitotic index. Total RNA (25μg) was extracted at the indicated time points and analyzed via Northern blot using NtWLIM2- and histone H4A748-specific probes. An 18S rRNA specific probe was used to ascertain equal loading of RNA samples.
Furthermore, we used an aphidicolin-synchronized BY-2 cell suspension culture to address the transcriptional activation of NtWLIM2 through the cell cycle (Reichheld et al., 1995; Shen and Gigot, 1999). Cells were synchronously released into the S phase and analyzed until the next S phase took place. Cell cycle progression was monitored by 3H-thymidine incorporation into newly replicated DNA and the percentage of mitotic cells was determined by microscopic analysis following Hoechst 33342-staining (Figure 9C, black and white circles, respectively). A high percentage (up to 50%) of the BY-2 cells proceeded synchronously through different phases of the cell cycle. Total RNA was isolated from the synchronized cells corresponding to different cell cycle time points and was used in Northern blot analyses (Figure 9C). In these cells, histone H4 transcription occurred in parallel to DNA replication, with maximal mRNA levels at S phase of the first cycle (1h), minimal mRNA levels during G2 and M phase (5–12h), and a gradual increase in mRNA levels during G1 and S phase of the second cycle (13–18h). The NtWLIM2 gene, however, exhibits constitutive expression throughout the whole cell cycle, as indicated by constantly high mRNA levels.
In summary, the NtWLIM2 gene transcription and protein levels were independent of cell division and proliferation and, thus, did not correlate with transcript and protein levels of histones in BY-2 cells.
DISCUSSION
In this study, we have characterized NtWLIM2 as a multifunctional protein that localizes to the cytoplasm, the nucleus, and the nucleolus of BY-2 cells. Besides its cytoplasmic activities as an actin-binding and -bundling protein, NtWLIM2 functions as a trans-activator of plant histone gene expression in the nucleus, able to bind to the conserved OCE Oct(II) (TCACGCGGATC) and the degenerated solo-octamer dOct motif (CACGGATC).
Noticeably, octamer motifs are highly conserved among plant histone gene promoters and found in various species, such as Arabidopsis, maize, wheat, or tobacco (Brignon and Chaubet, 1993; Chaubet et al., 1996; Huh et al., 1997; Atanassova et al., 1998; Reichheld et al., 1998). The role of octamers (Oct) and their composite elements (OCEs) in the regulation of plant histone gene expression is a matter of debate. Mutational analyses of Arabidopsis pH4A748::GUS lines have underscored the importance of both H4A748 octamer elements Oct(II) and dOct for basal transcription (Chaubet et al., 1996), whereas transient and stable expression assays have identified the octamer as a proliferation-coupled and S phase-specific cis-element (Chaubet et al., 1996; Ohtsubo et al., 1997; Taoka et al., 1999). Remarkably, octamer motifs are unable to confer detectable histone gene expression levels on their own (Taoka et al., 1999; Meshi et al., 2000). Indeed, gain-of-function experiments in tobacco cell lines and plants showed that the Oct motifs of all three OCE types can confer S phase-specific expression, but only if both constituents of the OCE are intact (Taoka et al., 1999). This implies that Oct-binding proteins interact with other DNA-binding factors that specifically recognize the second OCE-constituent, namely the HEX, TCA, or CCAAT-box to form various multiprotein–DNA complexes. Furthermore, the activity of OCEs can be modulated by additional cis-elements and their respective trans-acting factors, the latter interacting with the OCE binding proteins (Taoka et al., 1999). Accordingly, it has been postulated that the Arabidopsis H4A748 octamers interplay with adjacent up- and downstream cis-elements, such as the NON and CG motifs, to mobilize proliferation-specific transcription factors for example (Chaubet et al., 1996). The residual NtWLIM2-mediated trans-activation observed for an Oct-mutagenized form of the Arabidopsis histone H4A748 promoter (Figure 6B) supports the idea that promoter activation by NtWLIM2 does not exclusively depend on octamer motifs. Interestingly, the monitoring of NtWLIM2 transcript and protein levels over the BY-2 cell cycle provides evidence for a constitutive presence of NtWLIM2, which therefore does not correlate with the cell cycle-regulated pattern of histone expression. Although octamer-binding proteins have been barely characterized so far, gel shift assays have disclosed, on the one hand, proteins with optimal octamer-binding activity during the S phase and, on the other hand, proteins with constant octamer-binding activity throughout the cell cycle. Indeed, in the case of the wheat histone H3 promoter, three octamer-binding regulatory factors (OBRFs) were identified, among which OBRF-1 appeared predominantly during S phase, whereas OBRF-2 and -3 were constantly bound to the Oct motif (Minami et al., 2000). Similarly to OBRF-2 and -3, nuclear proteins from tobacco, which most likely include NtWLIM2, were shown to interact throughout the cell cycle with a probe containing the Arabidopsis H4A748 Oct(II) motif (Shen and Gigot, 1997). Moreover, in vivo footprints, which were observed for two octamer elements in an uninduced maize histone promoter, intensified upon induction of proliferation-dependent transcription (Brignon and Chaubet, 1993).
Together, the above data support a model in which plant histone octamers and their interacting proteins play a key role in both basal and cell-division-induced histone gene expression, the first being assured by constitutively present transcription factors like the Oct-binding proteins and the second being fulfilled by other, proliferation-specific trans-acting proteins. Regarding the octamer motifs of the Arabidopsis histone H4A748 gene promoter, it has been suggested that proliferation-specific factors would be directed to their respective positive cis-elements, namely the nonamer and CCGTCG motifs, through their interaction with the constitutively present Oct-binding proteins (Brignon and Chaubet, 1993; Chaubet et al., 1996). In view of this model and of the constitutive NtWLIM2 expression, one may consider the Oct-binding NtWLIM2 protein as one of the general, replication-independent transcriptional modulators, which, apart from assuring basal histone gene transcription, would interact with replication-specific transcription factors to form a multiprotein complex coordinating S phase-specific histone gene expression. Correspondingly, the role of a basic transcription factor has already been attributed to the NtWLIM2-related protein NtWLIM1, as the latter has been suggested to function as a weak activator in the maintenance of basal phenylpropanoid biosynthesis gene expression (Kawaoka and Ebinuma, 2001). Nevertheless, the activity of replication-independently expressed NtWLIM2 could still be regulated by cell cycle-regulated post-transcriptional mechanism(s), such as phosphorylation or interaction with co-factors, and thereby vary in a cell cycle-dependent manner. Accordingly, Reichheld et al. (1998) proposed a model in which histone gene activation at G1/S transition results from the structural or chemical modification of protein(s) constantly bound to the histone promoter, rather than from novel interactions emerging between constitutive and S phase-specific regulators. Indeed, protein–DNA complex formation at the histone H3 nonameric cis-element from tobacco was shown to depend on phosphorylation. However, DNA-binding activity of Oct-interacting proteins has been reported to be independent of their phosphorylation status (Shen and Gigot, 1997).
Interestingly, three structural counterparts of NtWLIM2, namely the mammalian Cysteine-rich proteins CRP1, CRP2, and CRP3, are described as compounds of transcriptional complexes that link different transcription factors and integrate the activities of multiple nuclear proteins to coordinate gene expression (Chang et al., 2003, 2007; Zheng and Zhao, 2007; Boateng et al., 2009; Gunkel et al., 2009). CRP3, also known as muscle LIM protein (MLP), is proposed to act as a cofactor in transcription complexes of muscle basic helix-loop-helix (bHLH) proteins, thereby enhancing the DNA-binding activity of myogenic transcription factors (Kong et al., 1997). In addition, CRP1 and CRP2 are suggested to function as transcriptional co-adaptors which assemble the nuclear factors GATA4/5/6 and serum response factor (SRF) into a multiprotein–DNA complex that mediates strong activation of smooth muscle cell-specific genes (Chang et al., 2003, 2007). Remarkably, the Arabidopsis histone H4A748 gene promoter carries an oligonucleotide stretch (ACTAATATGA) showing similarity to the binding site of human SRF (‘CArG’ box; CCTAATATGG) and being located adjacently to the Oct(II) element (Shen and Gigot, 1997). However, mammalian CRPs were not reported to bind directly to DNA so far, suggesting that the molecular mechanisms underlying the transcriptional regulatory activity of CRPs and NtWLIM2 at least partially differ.
Similarly to the previously described plant LIM proteins (Thomas et al., 2006; Wang et al., 2008; Papuga et al., 2010) and to the three mammalian CRPs (Louis et al., 1997; Weiskirchen and Gunther, 2003), NtWLIM2 also localizes to the cytoplasm, where it concentrates on the filamentous actin cytoskeleton. Interestingly, the nuclear fraction remarkably and rapidly increases after treatment with the F-actin disrupting drug Lat B and the subsequent release of free NtWLIM2 in the cytoplasm. This observation raises the possibility that the compartmentalization of NtWLIM2 is, at least partly, modulated by the polymerization status of the actin cytoskeleton. Since the latter most likely changes in a significant manner during cell proliferation and division, future studies should establish how the nuclear fraction of NtWLIM2 evolves during these processes and whether it correlates with the changes of histone gene expression levels.
Recent findings about the subcellular distribution of cardiac-enriched CRP3 have highlighted the importance of LIM protein distribution between the cytoskeletal and nuclear compartments (Boateng et al., 2007, 2009). CRP3 has been shown to play important roles in both the remodeling and maintenance of myocyte cytoarchitecture, as well as in the regulation of nuclear gene expression and the activation of ribosomal protein synthesis in the nucleolus (Arber et al., 1997; Boateng et al., 2007, 2009). In addition, CRP3 has been found to translocate from the cytoplasm to the nucleus and nucleolus in response to mechanical cues (Boateng et al., 2007, 2009). By homology to the mechanosensing role proposed for CRP3 in cardiac myocytes (Gunkel et al., 2009), NtWLIM2 could function as a plant sensor protein, able to transmit signals from the actin cytoskeleton to the nucleus and nucleolus.
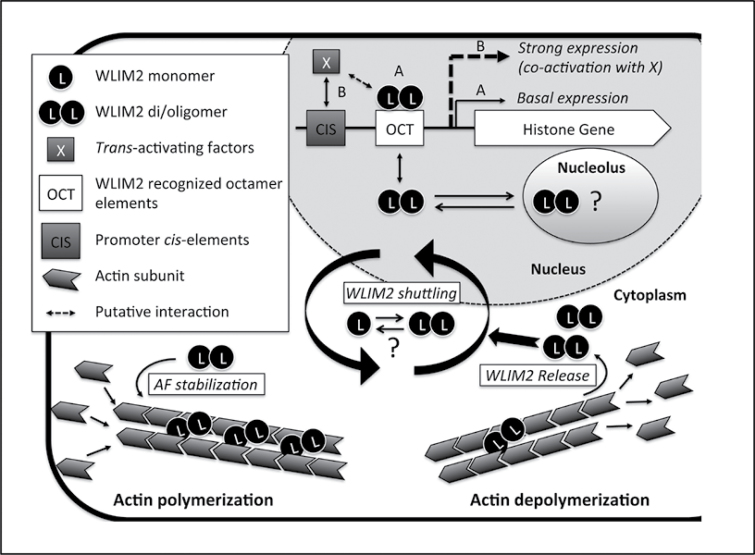
Figure 10 summarizes the functions of NtWLIM2 suggested by the present work and proposes an actin-based regulatory mechanism of NtWLIM2 cytoplasmic–nuclear shuttling. In the cytoplasm, NtWLIM2 would increase actin cytoskeleton stability by crosslinking actin filaments into bundles, and thereby contribute to maintain the integrity of intracellular structures, such as transvacuolar strands, and long-distance tracks for myosin-dependent transport (Thomas et al., 2009). In the nucleus, NtWLIM2 would activate basal histone gene transcription through direct binding to Oct(II) and dOct histone promoter elements. Although the overall NtWLIM2 transcript and protein content is relatively stable over cell division and proliferation (Figure 9), one cannot exclude that NtWLIM2 also triggers specific changes in histone gene transcription. Indeed, its activity could be regulated by posttranslational modifications or by interplay with other, such as cell cycle-regulated co-factors. Alternatively, upon conditions favoring a more dynamic, less polymerized state of the actin cytoskeleton, such as during mitosis or rapid cell growth phases, NtWLIM2 would be released from depolymerizing filaments and accumulate in the nucleus where its activity would, in turn, increase.
Figure 10.
Model of NtWLIM2 Functions and Cytoplasmic–Nuclear Shuttling. In the cytoplasm, NtWLIM2 (WLIM2) directly interacts with actin filaments, promotes the formation of actin bundles, and thereby contributes to stabilize the actin cytoskeleton. In the nucleus, WLIM2 interacts with Oct(II) and dOct histone promoter elements (OCT) and activates basal histone gene transcription (pathway A). Modulation of histone transcription levels would require additional trans-activating factors (X) able to target their corresponding cis-elements (CIS) in response to specific conditions (pathway B). In addition, WLIM2 might be further activated by interacting with co-factors (dashed double arrow) and/or by posttranslational modifications. Finally, both GFP-fused and endogenous forms of WLIM2 localize in the nucleolus, suggesting that WLIM2 participates in rDNA and/or ribosomal gene regulation as well. There are several lines of evidence that NtWLIM2 di/oligomerizes in the different subcellular compartments, although the significance of this process remains to be addressed. The exact mechanism underlying WLIM2 cytoplasmic–nuclear shuttling also requires further investigation. However, our data support that the intracellular distribution of NtWLIM2 is influenced by the cellular actin polymerization status. Upon actin filament depolymerization (e.g. by latrunculin B), WLIM2 is released in the cytoplasm and subsequently accumulates in the nucleus. Based on the present study and recent data obtained for the mammalian counterparts of plant LIMs, namely the cysteine-rich proteins (Kihara et al., 2011), we propose that the two LIM domain-containing proteins function as cytoskeletal stabilizers able to sense and transmit signals from actin filaments to the nucleus, where they modulate gene expression.
Interestingly, the oligomerization of CRP3 appears to be a prerequisite for its translocation to the nucleus (Boateng et al., 2007). The here-demonstrated formation of NtWLIM2 dimers suggests that dimerization or oligomerization is a property retained by all CRPs and plant CRP-like proteins. However, in contrast to CRP3, which is exclusively monomeric in the nucleus, NtWLIM2 does not exhibit a clear compartment-dependent change in oligomerization status, since it di/oligomerizes both in the cytoplasm and the nucleus (Figure 3B and Hoffmann et al., unpublished data). Apart from its impact on histone gene expression in the nucleus, NtWLIM2 could act on nucleolar processes, such as rDNA or ribosomal protein gene transcription, as has been suggested for CRP3 (Boateng et al., 2007), and thereby, in a wider context, affect cell division and proliferation.
METHODS
Cell Culture Maintenance and Synchronization
Stable transgenic BY-2 lines containing the pTA7002–NtWLIM2–GFP construct (Aoyama and Chua, 1997) were obtained by Agrobacterium tumefaciens-mediated transformation (Criqui et al., 2000). Expression of NtWLIM2–GFP in the transgenic cell lines was induced by overnight incubation on BY-2 medium supplied with 10µM Dexamethasone. Selection of cell lines was based on hygromycine-resistence and epifluorescence screening of individual transformed calli.
Suspension-cultured tobacco BY-2 (Nicotiana tabacum L. cv. Bright Yellow) cells were grown in the dark at 27°C on a rotary shaker and weekly subcultured by 1:10 dilution. BY-2 cell synchronization, tracking of DNA synthesis, and mitotic index determination were based on previous protocols (Reichheld et al., 1995; Shen and Gigot, 1997). Briefly, stationary phase cells were diluted (1:8) into fresh medium and treated for 24h with aphidicolin (5mgl–1, Sigma-Aldrich). After removal of aphidicolin, cells entered S phase and proceeded through the cell cycle in a synchronous manner. For DNA synthesis, 1ml of cell suspension (≈2×106cells) was taken at different time points after aphidicolin treatment and incubated with 3H-TTP according to preceding descriptions (Reichheld et al., 1995; Shen and Gigot, 1997). Subsequently, incorporation of 3H-TTP into the newly synthesized DNA was measured (Lepetit et al., 1992). Estimation of the mitotic index was done by UV light microscopy analysis of 200 cells stained with Hoechst 33342 (Sigma-Aldrich).
RNA Gel Blot Analysis
Total RNA (25µg) was extracted from samples of synchronous BY-2 suspension cells and analyzed by electrophoresis on formaldehyde/agarose gels and blotted onto Hybond-N nylon membranes (Amersham). Hybridization with gene-specific, 32P-labeled probes was performed, as previously described, under standard high-stringency conditions (Shen and Gigot, 1999). For the NtWLIM2-probe, primers amplified full cds (5’-ATGTCTTTATTGGGACACAACAG-3’ and 5’-AAGAATCTGGAACGGTTGCAG-3’). The histone H4A748-probe, which corresponded to the coding regions of the Arabidopsis H4A748 cds, has been previously described (Reichheld et al., 1995; Shen and Gigot, 1999). Transcript levels were quantified by a PhosphorImager (Molecular Dynamics).
Construction of a Tobacco cDNA Expression Library
The cDNA expression library was constructed according to previous protocols using the ZAP-cDNA® Library Construction Kit (Stratagene) (Shen and Gigot, 1999). Briefly, 5µg of poly(A)-RNA was isolated from S phase cells of a synchronized BY-2 suspension culture and used for cDNA synthesis. The cDNA was unidirectionally ligated into ZAP Express™ vector via EcoRI and XhoI restriction sites. In vitro packaging and transformation of E. coli yielded a total of 107 plaque-forming units for the primary library.
Expression and Purification of Recombinant Tobacco WLIM2 Protein
Subcloning of tobacco WLIM2 coding sequence into the pQE60 vector, as well as the expression of 6xHis-tagged NtWLIM2 (NtWLIM2–His) in M15[pREP4] bacteria and its imidazole-based purification using a Ni-NTA resin were performed according to the manufacturer’s instructions (Qiagen). For EMSA analysis, the purified protein was dialyzed against DNA-binding buffer (30mM Tris, 0.2mM EDTA, 2mM DTT, 250mM NaCl, 50µM ZnCl2, 1mM MgCl2, 25mM imidazole, pH8.0) and stored at 4°C. Protein quality check and quantification were performed as reported for Arabidopsis LIM proteins (Papuga et al., 2010).
Size Exclusion Chromatography and Molecular Weight Determination
Protein eluted from a Ni-NTA matrix was loaded on a Superdex 75 10/300 GL column (GE Healthcare) equilibrated with Tris/NaCl buffer (50mM Tris, 200mM NaCl, 5mM β-Mercaptoethanol). The column was calibrated using proteins of the gel filtration calibration kit LMW (GE Healthcare) under the same conditions. Kav was calculated using the equitation Kav=(Ve–Vo)/(Vt–Vo). Fractions were collected and analyzed by SDS–PAGE followed by Coomassie Blue staining.
Preparation of Nuclear Extracts
Tobacco BY-2 cells were harvested by vacuum filtration and frozen in liquid nitrogen. Preparation of nuclei and nuclear extracts from frozen cells was performed at 4°C using the CelLytic™ PN Kit according to the manufacturer’s protocol (Sigma-Aldrich). For EMSA analysis, nuclear proteins were buffer-exchanged (30mM Tris, 0.2mM EDTA, 2mM DTT, 250mM NaCl, 50µM ZnCl2, 1mM MgCl2, 25mM imidazole, pH8.0) using Micro Bio-Spin™ Chromatography Columns (BioRad). Generally, 0.5ml of nuclear protein extract containing up to 1.5mgml–1 of protein were obtained from 5g of cells. Aliquots of nuclear protein extracts were snap-frozen in liquid nitrogen and stored at –80°C.
Electrophoretic Mobility Shift Assay (EMSA)
For radioactive EMSAs, nuclear extracts (1µg) or recombinant NtWLIM2 protein (3µg) were incubated for 20min on ice in DNA-binding buffer (10mM Tris-HCl, 5% glycerol, 50mM NaCl, 0.5mM EDTA, 0.5mM DTT, 1mM MgCl2, 2mgml–1 poly [d(I-C)], 2ngml–1 BSA, 80ngml–1 spermidine, pH8.5) supplemented with 1ng of γ32P-labeled double-stranded oligonucleotides and 1:100 (v/v)-diluted Protease Inhibitor Cocktail for Plants (P9599, Sigma-Aldrich). DNA sense strand oligonucleotides of EMSA probes were as follows: ‘Probe 1’ (histone H4A748 promoter) 5’-CGCCACATGTAAA ACAAAAGACGATTCTTAGTGGCTATCACTGCCATCA CGCGGATCACTAATATGAACCGTCG-3’and‘Probe2’(histoneH4A748promoter):5’-CGGTTCATATTAG TGATCCGCGTGATGGCAGTGATAGCCACTAAGAATCGTCTTTT GTTTTACATGTGGCG-3’,probes‘OCT’:5’- ATCACGCGGATCACTACAACACGGATC-3’and‘CG–NON’: 5’-GAACCGTCGATTAAA ACAGATCGACGG-3’. Annealing and labeling procedures were adapted from previous protocols (Morceau et al., 2006). EMSA samples were loaded on a 5% native acrylamide gel and separated by electrophoresis (3h, 16mA) in 0.5TBE buffer. Gels were dried and autoradiographed for 2h using the Cyclone Storage Phosphoimager System (Packard). Non-radioactive EMSA assays were performed using a DIG Gel Shift kit (Roche, Mannheim, Germany) following the manufacturer’s instructions.
High- and Low-Speed Cosedimentation Assays
To assess the actin-binding and crosslinking activities of NtWLIM2, high-speed and low-speed cosedimentation assays were used as previously described (Thomas et al., 2006, 2007). Briefly, rabbit muscle actin (Cytoskeleton) was diluted at 2mgml–1 in A-buffer (5mM Tris-HCl, pH8.0, 0.2mM CaCl2, 0.2mM Na2ATP, and 0.5mM DTT). Polymerization was induced by the addition of actin polymerization inducer (final concentration of 2mM MgCl2, 1mM ATP, 50mM KCl). Prior to cosedimentation experiments, recombinant NtWLIM2 was pre-clarified at 50000g (45min). For high-speed cosedimentation assays, actin (4µM) was copolymerized with different amounts of NtWLIM2 (0.5–12mM) for 1h at 25°C. Samples were then centrifuged at 100000g for 45min at 4°C to pellet AFs. The presence of NtWLIM2 in the supernatants (F-actin unbound fraction) and pellets (F-actin bound fraction) was analyzed by SDS–PAGE and Coomassie Brilliant Blue staining. The determination of the dissociation constant (Kd) was performed as previously described (Thomas et al., 2006, 2007). Briefly, the amount of NtWLIM2 in the pellet and supernatant fractions of three independent high-speed cosedimentation assays was quantified using ImageJ software (National Institutes of Health) and the mean Kd value for NtWLIM2 was calculated by fitting the data of bound NtWLIM2 protein versus free NtWLIM2 protein to a hyperbolic function with Sigmaplot V10 software (Systat Software).
In low-speed cosedimentation assays, actin was copolymerized with NtWLIM2 as described above and subsequently centrifuged at 12500g for 45min at 4°C and analyzed by SDS–PAGE as previously described. After quantification using ImageJ software, results were expressed as percentage of actin in the pellet as a function of NtWLIM2 protein concentration. The presence of higher-order actin structures in the samples was checked as previously described by direct visualization using fluorescence or electron microscopy (Thomas et al., 2007).
Plasmids for Transient Expression
The pH4A748::LUC (H4) and pH4A748mt::LUC (mtH4) reporter plasmids were derived from the pSK-based pRAB18::LUC construct (Moes et al., 2008), the latter containing the LUC gene from pGEM®–luc (Promega). For pH4A748::LUC, a 604-bp promoter fragment of the Arabidopsis histone H4A748 gene (At5g59690) was PCR amplified from Arabidopsis (Col-0) genomic DNA using the primers 5’-tccccgcggacacaaactttgtaaatttgg-3’ (fwd) and 5’-cgcggatcctcagcgaagatttagtttatctcg-3’ (rev). These primers introduced single BamHI and SacII sites into the promoter fragment and allowed the fusion of pH4A748 to the LUC gene using the SacII and BamHI sites of pSK. The mutated version of pH4A748 (mtH4) comprised base exchanges within both the conserved octamer (Oct(II)) and degenerated octamer (dOct) motifs (Oct(II): 5’-CGCGGATC-3’ changed to 5’-CGaaGcTt-3’; dOct: 5’-CACGGATC-3’ changed to 5’-gtCGacTC-3’) that are known to reduce plant histone H3 and H4 promoter activities (Terada et al., 1995; Chaubet et al., 1996). The mtH4 fragment with SacII and BamHI sites was obtained by DNA synthesis (DNA2.0). The 2xOCT reporter contained two copies of the oligonucleotide 5’-CTAGTCACGCGGATCACTACACACGGATCGAT-3’ comprising the Oct(II) (TCACGCGGATC) and the dOct (CACGGATC) elements. These elements were cloned in tandem orientation upstream of a minimal –46CaMV 35S core promoter (Odell et al., 1985), the latter driving LUC reporter gene expression (Himmelbach et al., 2002). The effector plasmids used for transient expression in protoplasts are all derivatives of the pMAV–YN vector (Stolpe et al., 2005), in which PCR products of NtWLIM1 and NtWLIM2 cDNA were inserted via BamHI–SacI, thereby replacing the N-terminal YFP fragment. The trans-activation domain (AD) of herpes simplex virus protein VP16 was synthesized with BamHI sites (DNA2.0) and cloned into pMAV (DNA2.0).
Protoplast Analysis
Isolation of protoplasts from rosette leaves of 3-week-old Arabidopsis and tobacco plants, as well as polyethylene glycol-mediated protoplast transfection, were performed as previously described (Yoo et al., 2007). Approximately 5×104 protoplasts were transfected with 10µg of reporter and 0.1–10µg of effector plasmid. For internal standardization, 4µg of p35S::GUS plasmid was included in each transfection. LUC activity was assayed in a luminometer (Berthold Technologies) with a 90-s integration period of light emission and was given relative to the GUS activity (relative light units, RLU). GUS quantification was performed as previously described (Breyne et al., 1993). Each protoplast assay was repeated at least twice.
Generation of a Polyclonal Anti-NtWLIM2 Antibody in Mice
To generate the anti-NtWLIM2 antibody, BALB-c mice were immunized by intraperitoneal injection of recombinant affinity-purified NtWLIM2–His protein. For each of the three injections, 30µg of NtWLIM2–His was re-suspended in PBS and diluted 1:1 with either complete or incomplete Freund’s adjuvant (first and second to third injection, respectively). Injections were performed with a 3-week interval. Three days prior to collection of the mouse blood, a final boost was performed with 30µg of NtWLIM2–His re-suspended in PBS. The collected blood was centrifuged for 45min at 4°C and 14000rpm, and the polyclonal serum was then affinity-purified from an immunoblot loaded with recombinant NtWLIM2 protein as previously described (Harlow and Lane, 1988). The serum specificity for NtWLIM2 was verified by Western blot (Supplemental Figure 3). The monoclonal anti-histone H3 antibody was obtained from abcam (ab24834).
Immunochemical Analyses
For immunolocalization of NtWLIM2 in BY-2 cells, the paraformaldehyde-based fixation of poly-L-lysine-immobilized cells was performed according to previously established protocols (Thomas et al., 2006). After a 1-h blocking step in PBS supplemented with 5% BSA, 5% normal goat serum, and 0.1% cold water fish skin gelatin (Aurion), cells were incubated overnight at 4°C with the polyclonal mouse anti-NtWLIM2 antibody (dilution 1:3). After four PBS washing steps, cells were incubated for 2h in the dark and at RT with a secondary goat anti-mouse IgG1-Alexa Fluor® 568-antibody (Invitrogen), diluted 1:200 in PBS. For nuclear counterstain, cells were incubated in PBS supplemented with 1µM DAPI. Finally, cells were washed four times with PBS and mounted with Citifluor™ anti-fading solution before confocal microscopy observation.
Confocal Microscopy and Imaging
Fluorescence of GFP or YFP fusion proteins in BY-2 cells was monitored as previously described using a Zeiss LSM510 META confocal laser scanning microscope (Papuga et al., 2010). For Lat B-induced NtWLIM2–GFP relocalization experiments, BY-2 cells were immobilized on a poly-L-lysine (0.01%) coated cover slip treated with 2.5µM Lat B and imaged using the ‘Multi Time Lapse’ module of Zeiss LSM software. Single planes and stacks of neighboring 1-µm sections were recorded at 25-min intervals after Lat B addition. To ensure comparability between single cells, all images for the Lat B and control time courses have been taken under identical experimental conditions (time after subculture, time of Dexamethasone induction, treatment with Lat B) and by preserving the same microscope settings. Co-labeling of NtWLIM2–GFP expressing BY-2 cells with rhodamine–phalloidine was performed as previously described (Papuga et al., 2010).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the Ministry of Culture, Higher Education and Research. S.G. was supported by a PhD fellowship from the Ministry of Culture, Higher Education and Research, Luxembourg. Financial support from the National Research Fund, Luxembourg (FNR), is gratefully acknowledged (TR-PDR BFR07-100, C10/BM/784171-HUMCRP and FNR/12/AM4/31).

Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Jörg Kudla (University of Münster, Germany) for providing us with the BiFC plasmids. We also thank Dr Annette Kuehn and Tanja Scheuermann (Laboratory of Immunogenetics and Allergology, CRP-Santé, Luxembourg) for the support regarding size exclusion chromatography and we are grateful to Dr Petr Nazarov (Microarray Center, CRP-Santé, Luxembourg) for giving advice concerning statistical analyses. No conflict of interest declared.
REFERENCES
- Aoyama T, Chua N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612 [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter J.J, Ross V, Jr, Hongo M, Sansig G, Borg J, Perriard J.C, Chien K.R, Caroni P. (1997). MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 88, 393–403 [DOI] [PubMed] [Google Scholar]
- Arnaud D, Déjardin A, Leplé J.-C, Lesage-Descauses M.-C, Pilate G. (2007). Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14, 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova R, Chaubet N, Gigot C. (1992). A 126bp fragment of a plant histone gene promoter confers preferential expression in meristems of transgenic Arabidopsis. Plant J. 2, 291–300 [DOI] [PubMed] [Google Scholar]
- Atanassova R, Flénet M, Gigot C, Chaubet N. (1998). Functional analysis of the promoter region of a maize (Zea mays L.) H3 histone gene in transgenic Arabidopsis thaliana. Plant Mol. Biol. . 37, 275–285 [DOI] [PubMed] [Google Scholar]
- Baltz R, Evrard J.-L, Bourdon V, Steinmetz A. (1996). The pollen-specific LIM protein PLIM-1 from sunflower binds nucleic acids in vitro. Sex. Plant Rep. 9, 264–268 [Google Scholar]
- Baltz R, Evrard J.-L., Domon C, Steinmetz A. (1992). A LIM motif is present in a pollen-specific protein. Plant Cell. 4, 1465–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Shafiq S, Shen W.-H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta—Gene Regulatory Mechanisms. 1809, 567–576 [DOI] [PubMed] [Google Scholar]
- Boateng S.Y, Belin R.J., Geenen D.L, Margulies K.B, Martin J.L, Hoshijima M, de Tombe P.P, Russell B. (2007). Cardiac dysfunction and heart failure are associated with abnormalities in the subcellular distribution and amounts of oligomeric muscle LIM protein. Am. J. Physiol. Heart Circ. Physiol. 292, H259–H269 [DOI] [PubMed] [Google Scholar]
- Boateng S.Y, Senyo S.E, Qi L, Goldspink P.H, Russell B. (2009). Myocyte remodeling in response to hypertrophic stimuli requires nucleocytoplasmic shuttling of muscle LIM protein. J. Mol. Cell. Cardiol. 47, 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P., De Loose M, Dedonder A, Van Montagu M, Depicker A. (1993). Quantitative kinetic analysis of beta-glucuronidase activities using a computer-directed microtiter plate reader. Plant Mol. Biol. Rep. 11, 21–31 [Google Scholar]
- Brignon P, Chaubet N. (1993). Constitutive and cell-division-inducible protein–DNA interactions in two maize histone gene promoters. Plant J. 4, 445–457 [DOI] [PubMed] [Google Scholar]
- Chabouté M.-E, Chaubet N, Philipps G, Ehling M, Gigot C. (1987). Genomic organization and nucleotide sequences of two histone H3 and two histone H4 genes of Arabidopsis thaliana. Plant Mol. Biol. Rep. 8, 179–191 [DOI] [PubMed] [Google Scholar]
- Chang D.F, Belaguli N.S, Chang J, Schwartz R.J. (2007). LIM-only protein, CRP2, switched on smooth muscle gene activity in adult cardiac myocytes. Proc. Natl Acad. Sci. U S A. 104, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.F, et al. (2003). Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell. 4, 107–118 [DOI] [PubMed] [Google Scholar]
- Chaubet N, Gigot C. (1998). Histone gene expression. In Plant Cell Division Francis D, Dudits D, Inzé D, eds (London: Portland Press; ), pp. 269–283 [Google Scholar]
- Chaubet N, Flénet M, Clément B, Brignon P, Gigot C. (1996). Identification of cis-elements regulating the expression of an Arabidopsis histone H4 gene. Plant J. 10, 425–435 [DOI] [PubMed] [Google Scholar]
- Criqui M.C, Parmentier Y, Derevier A, Shen W.-H, Dong A., Genschik P. (2000). Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 24, 763–773 [DOI] [PubMed] [Google Scholar]
- Eliasson A, Gass N, Mundel C, Baltz R, Krauter R, Evrard J.L, Steinmetz A. (2000). Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol. Gen. Genet. 264, 257–267 [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim S.K, Horvitz H.R. (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 344, 876–879 [DOI] [PubMed] [Google Scholar]
- Grubinger M, Gimona M. (2004). CRP2 is an autonomous actin-binding protein. FEBS Lett. 557, 88–92 [DOI] [PubMed] [Google Scholar]
- Gunkel S, Knoll R, Heineke J, Hilfiker-Kleiner D. (2009). MLP: a stress sensor goes nuclear. J. Mol. Cell Cardiol. 47, 423–425 [DOI] [PubMed] [Google Scholar]
- Hao Y.-J, Song Q.-X, Chen H.-W, Zou H.-F, Wei W, Kang X.-S, Ma B, Zhang W.-K., Zhang J.-S, Chen S.-Y. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta. 232, 1033–1043 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. (1988). Antibodies: A Laboratory Manual (New York: Cold Spring Harbor Laboratory; ). [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh G.H, Nakayama T, Meshi T, Iwabuchi M. (1997). Structural characteristics of two wheat histone H2A genes encoding distinct types of variants and functional differences in their promoter activity. Plant Mol. Biol. 33, 791–802 [DOI] [PubMed] [Google Scholar]
- Jang H.S, Greenwood J.A. (2009). Glycine-rich region regulates cysteine-rich protein 1 binding to actin cytoskeleton. Biochem. Biophys. Res. Commun. 380, 484–488 [DOI] [PubMed] [Google Scholar]
- Kadrmas J.L, Beckerle M.C. (2004). The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- Kaothien P, Kawaoka A, Ebinuma H, Yoshida K, Shinmyo A. (2002). Ntlim1, a PAL-box binding factor, controls promoter activity of the horseradish wound-inducible peroxidase gene. Plant Mol. Biol. 49, 591–599 [DOI] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys–His domain. Nature. 344, 879–882 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Ebinuma H. (2001). Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry. 57, 1149–1157 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H. (2000). Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 22, 289–301 [DOI] [PubMed] [Google Scholar]
- Kihara T, Shinohara S, Fujikawa R, Sugimoto Y, Murata M, Miyake J. (2011). Regulation of cysteine-rich protein 2 localization by the development of actin fibers during smooth muscle cell differentiation. Biochem. Biophys. Res. Commun. 411, 96–101 [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick M.J, Kudla A.J, Konieczny S.F. (1997). Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol. Cell. Biol. 17, 4750–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit M., Ehling M, Atanassova R, Chaubet N, Gigot C. (1993). Replication-independent cis-acting element of a maize histone gene promoter. Plant Science. 89, 177–184 [Google Scholar]
- Lepetit M, Ehling M, Chaubet N, Gigot C. (1992). A plant histone gene promoter can direct both replication-dependent and -independent gene expression in transgenic plants. Mol. Gen. Genetics. 231, 276–285 [DOI] [PubMed] [Google Scholar]
- Louis H.A, Pino J.D, Schmeichel K.L, Pomies P, Beckerle M.C. (1997). Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. J. Biol. Chem. 272, 27484–27491 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader A.W, Richmond R.K, Sargent D.F, Richmond T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389, 251–260 [DOI] [PubMed] [Google Scholar]
- Maity S.N, de Crombrugghe B. (1998). Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends. Biochem. Sci. 23, 174–178 [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T, Taoka K.-i, Iwabuchi M. (2000). Regulation of histone gene expression during the cell cycle. Plant Mol. Biol. 43, 643–657 [DOI] [PubMed] [Google Scholar]
- Mikami K, Tabata T, Kawata T, Nakayama T, Iwabuchi M. (1987). Nuclear protein(s) binding to the conserved DNA hexameric sequence postulated to regulate transcription of wheat histone genes. FEBS Lett. 223, 273–278 [DOI] [PubMed] [Google Scholar]
- Minami M, Meshi T, Iwabuchi M. (2000). S Phase-specific DNA-binding proteins interacting with the Hex and Oct motifs in type I element of the wheat histone H3 promoter. Gene. 241, 333–339 [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. (2008). Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 54, 806–819 [DOI] [PubMed] [Google Scholar]
- Morceau F, Schnekenburger M, Blasius R, Buck I, Dicato M, Diederich M. (2006). Tumor necrosis factor alpha inhibits aclacinomycin A-induced erythroid differentiation of K562 cells via GATA-1. Cancer Lett. 240, 203–212 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Sakamoto A, Yang P, Minami M, Fujimoto Y, Ito T, Iwabuchi M. (1992). Highly conserved hexamer, octamer and nonamer motifs are positive cis-regulatory elements of the wheat histone H3 gene. FEBS Lett. 300, 167–170 [DOI] [PubMed] [Google Scholar]
- Odell J.T, Nagy F, Chua N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 313, 810–812 [DOI] [PubMed] [Google Scholar]
- Ohtsubo N, Nakayama T, Kaya H, Terada R, Shimamoto K, Meshi T, Iwabuchi M. (1997). Cooperation of two distinct cis-acting elements is necessary for the S phase-specific activation of the wheat histone H3 promoter. Plant J. 11, 1219–1225 [Google Scholar]
- Okada T, Endo M, Singh M.B, Bhalla P.L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44, 557–568 [DOI] [PubMed] [Google Scholar]
- Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A., Thomas C. (2010). Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell. 22, 3034–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.-P, Gigot C, Chaubet-Gigot N. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Res. 26, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.-P, Sonobe S., Clément B., Chaubet N, Gigot C. (1995). Cell cycle-regulated histone gene expression in synchronized plant cells. Plant J. 7, 245–252 [Google Scholar]
- Robertson A.J, Kapros T, Waterborg J.H. (1997). A cell cycle-regulated histone H3 gene of alfalfa with an atypical promoter structure. DNA Seq. 7, 209–216 [DOI] [PubMed] [Google Scholar]
- Robertson A.J, Kapros T., Dudits D, Waterborg J.H. (1996). Identification of three highly expressed replacement histone H3 genes of alfalfa. DNA Seq. 6, 137–146 [DOI] [PubMed] [Google Scholar]
- Robinson P.J.J, Rhodes D. (2006). Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Op. Struct. Biol. 16, 336–343 [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J, Triezenberg S, Ptashne M. (1988). GAL4-VP16 is an unusually potent transcriptional activator. Nature. 335, 563–564 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Omirulleh S, Nakayama T, Iwabuchi M. (1996). A zinc-finger-type transcription factor WZF-1 that binds to a novel cis-acting element of histone gene promoters represses its own promoter. Plant Cell Physiol. 37, 557–562 [DOI] [PubMed] [Google Scholar]
- Shen W.H, Gigot C. (1997). Protein complexes binding to cis elements of the plant histone gene promoters: multiplicity, phosphorylation and cell cycle alteration. Plant Mol. Biol. 33, 367–379 [DOI] [PubMed] [Google Scholar]
- Shen W.H, Gigot C. (1999). Characterization of Prt1, a gene encoding for one of the subunits of the translation initiation factor 3 (eIF3), from Nicotiana tabacum. Plant Science. 143, 45–54 [Google Scholar]
- Stolpe T, Süßlin C, Marrocco K, Nick P, Kretsch T, Kircher S. (2005). In planta analysis of protein–protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma. 226, 137–146 [DOI] [PubMed] [Google Scholar]
- Tabata T, Nakayama T, Mikami K, Iwabuchi M. (1991). HBP-1a and HBP-1b: leucine zipper-type transcription factors of wheat. EMBO J. 10, 1459–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Takase H, Takayama S, Mikami K, Nakatsuka A, Kawata T, Nakayama T, Iwabuchi M. (1989). A protein that binds to a cis-acting element of wheat histone genes has a leucine zipper motif. Science. 245, 965–967 [DOI] [PubMed] [Google Scholar]
- Taoka K.-I, Kaya H, Nakayama T, Araki T, Meshi T, Iwabuchi M. (1999). Identification of three kinds of mutually related composite elements conferring S phase-specific transcriptional activation. Plant J. 18, 611–623 [DOI] [PubMed] [Google Scholar]
- Taoka K.-I, Ohtsubo N, Fujimoto Y, Mikami K, Meshi T, Iwabuchi M. (1998). The modular structure and function of the wheat H1 promoter with S Phase-specific activity. Plant Cell Physiol. 39, 294–306 [DOI] [PubMed] [Google Scholar]
- Terada R, Nakayama T, Iwabuchi M, Shimamoto K. (1993). A wheat histone H3 promoter confers cell division-dependent and -independent expression of the gus A gene in transgenic rice plants. Plant J. 3, 241–252 [DOI] [PubMed] [Google Scholar]
- Terada R, Nakayama T, Iwabuchi M, Shimamoto K. (1995). A type I element composed of the hexamer (ACGTCA) and octamer (CGCGGATC) motifs plays a role(s) in meristematic expression of a wheat histone H3 gene in transgenic rice plants. Plant Mol. Biol. 27, 17–26 [DOI] [PubMed] [Google Scholar]
- Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell. 18, 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Hoffmann C, Gatti G, Steinmetz A. (2007). LIM proteins: a novel class of actin cytoskeleton organizers in plants. Plant Signaling Behav. 2, 99–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton. 66, 940–957 [DOI] [PubMed] [Google Scholar]
- Tran T.C, Singleton C, Fraley T.S, Greenwood J.A. (2005). Cysteine-rich protein 1 (CRP1) regulates actin filament bundling. BMC Cell Biol. 6, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt L.K, Lohse M, Hashimoto K, Bock R, Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H.J, Wan A.R, Jauh G.Y. (2008). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147, 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J.C, Chalfie M. (1989). The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 3, 1823–1833 [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Gunther K. (2003). The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. Bioessays. 25, 152–162 [DOI] [PubMed] [Google Scholar]
- Yang P, Taoka K, Nakayama T, Iwabuchi M. (1995). Structural and functional characterization of two wheat histone H2B promoters. Plant Mol. Biol. 28, 155–172 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D, Cho Y.-H, Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protocols. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhao Y. (2007). The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein protein interaction. Biol. Cell. 99, 489–502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.