Abstract
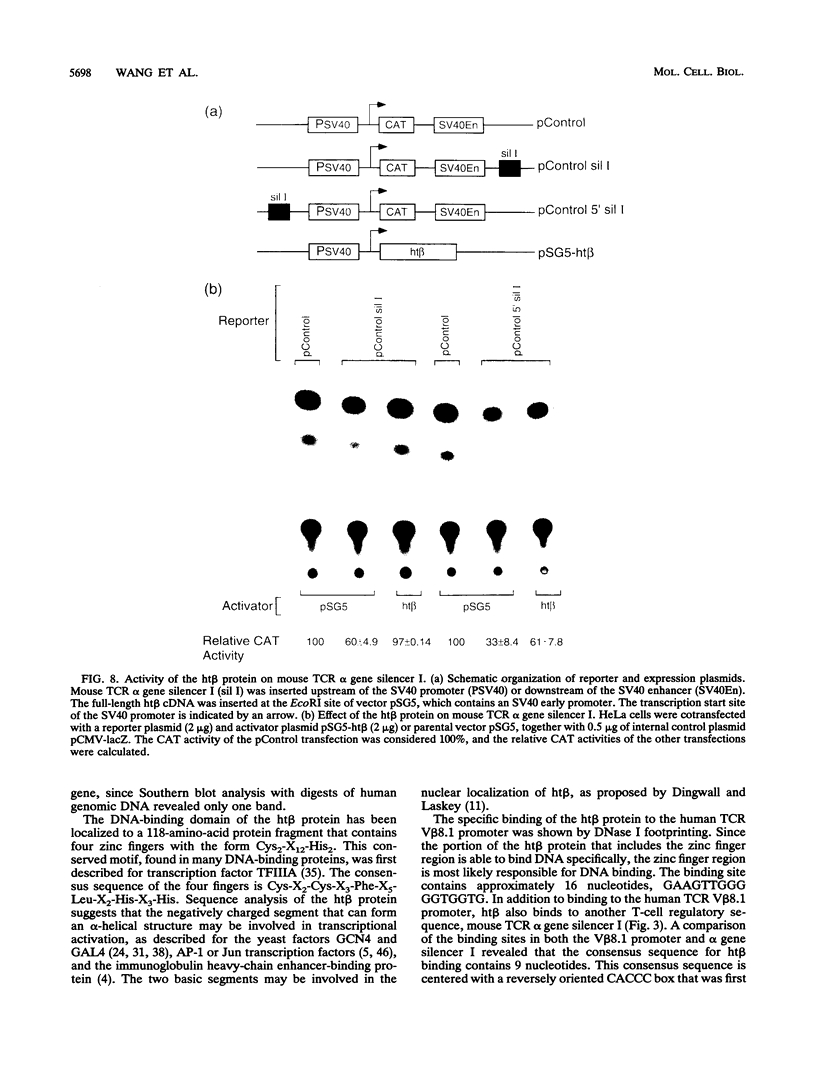
A gene encoding a novel CACCC box-binding protein that binds to the promoter region of the human T-cell receptor (TCR) V beta 8.1 gene and the mouse TCR alpha gene silencer has been cloned. This gene, termed ht beta, contains four zinc fingers of the class Cys2-X12-His2 that may be responsible for DNA binding and a highly negatively charged region that defines a putative transcriptional activation domain. Analysis of the expression of ht beta mRNA revealed similar expression levels and patterns in various cell lines. The bacterially expressed ht beta protein can bind to the CACCC box in both the human TCR V beta 8.1 gene promoter and the mouse TCR alpha gene silencer. The CACCC box is essential for efficient transcription of the V beta 8.1 promoter. Cotransfection with an ht beta expression plasmid and a reporter vector indicated that ht beta can activate human TCR V beta 8.1 gene transcription. ht beta also is able to counteract the silencing effect of the mouse TCR alpha gene silencer. The CACCC box has been found in almost all V beta 8.1 gene subfamily members and in both TCR alpha and beta gene enhancers in humans and mice. These results suggest that the CACCC box-binding protein may have an important regulatory function for TCR gene expression in alpha beta T cells versus gamma delta T cells.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S. J., Chou H. S., Loh D. Y. A conserved sequence in the T-cell receptor beta-chain promoter region. Proc Natl Acad Sci U S A. 1988 May;85(10):3551–3554. doi: 10.1073/pnas.85.10.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Bohmann D., Bos T. J., Admon A., Nishimura T., Vogt P. K., Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987 Dec 4;238(4832):1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Iwashima M., Kaplan K. B., Elliott J. F., Davis M. M. A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. 1987 Jun 25-Jul 1Nature. 327(6124):677–682. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Iwashima M., Wettstein D. A., Kaplan K. B., Elliott J. F., Born W., Davis M. M. T-cell receptor delta gene rearrangements in early thymocytes. Nature. 1987 Dec 24;330(6150):722–727. doi: 10.1038/330722a0. [DOI] [PubMed] [Google Scholar]
- Chien Y., Becker D. M., Lindsten T., Okamura M., Cohen D. I., Davis M. M. A third type of murine T-cell receptor gene. Nature. 1984 Nov 1;312(5989):31–35. doi: 10.1038/312031a0. [DOI] [PubMed] [Google Scholar]
- Diamond D. J., Nelson F. B., Reinherz E. L. Lineage-specific expression of a T cell receptor variable gene promoter controlled by upstream sequences. J Exp Med. 1989 Apr 1;169(4):1213–1231. doi: 10.1084/jem.169.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- Fan C. M., Maniatis T. A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev. 1990 Jan;4(1):29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- Fujita T., Shibuya H., Ohashi T., Yamanishi K., Taniguchi T. Regulation of human interleukin-2 gene: functional DNA sequences in the 5' flanking region for the gene expression in activated T lymphocytes. Cell. 1986 Aug 1;46(3):401–405. doi: 10.1016/0092-8674(86)90660-4. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglioni B., Comi P., Ronchi A., Mantovani R., Ottolenghi S. The same nuclear proteins bind the proximal CACCC box of the human beta-globin promoter and a similar sequence in the enhancer. Biochem Biophys Res Commun. 1989 Oct 16;164(1):149–155. doi: 10.1016/0006-291x(89)91695-1. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk L. R., Leiden J. M. Identification and functional characterization of the human T-cell receptor beta gene transcriptional enhancer: common nuclear proteins interact with the transcriptional regulatory elements of the T-cell receptor alpha and beta genes. Mol Cell Biol. 1990 Oct;10(10):5486–5495. doi: 10.1128/mcb.10.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan M. V., Pothier F., Leclerc S., Palaniswami R., Xie B. Human glucocorticoid receptor gene promotor-homologous down regulation. J Steroid Biochem Mol Biol. 1991;40(1-3):317–323. doi: 10.1016/0960-0760(91)90197-d. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Nielsen E. A., Kavaler J., Cohen D. I., Davis M. M. Sequence relationships between putative T-cell receptor polypeptides and immunoglobulins. Nature. 1984 Mar 8;308(5955):153–158. doi: 10.1038/308153a0. [DOI] [PubMed] [Google Scholar]
- Ho I. C., Bhat N. K., Gottschalk L. R., Lindsten T., Thompson C. B., Papas T. S., Leiden J. M. Sequence-specific binding of human Ets-1 to the T cell receptor alpha gene enhancer. Science. 1990 Nov 9;250(4982):814–818. doi: 10.1126/science.2237431. [DOI] [PubMed] [Google Scholar]
- Ho I. C., Yang L. H., Morle G., Leiden J. M. A T-cell-specific transcriptional enhancer element 3' of C alpha in the human T-cell receptor alpha locus. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6714–6718. doi: 10.1073/pnas.86.17.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Kappes D. J., Browne C. P., Tonegawa S. Identification of a T-cell-specific enhancer at the locus encoding T-cell antigen receptor gamma chain. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2204–2208. doi: 10.1073/pnas.88.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert H., Assert R., Benecke B. J. A single base pair deletion from the inactive octamer-like motif of the 7S K distal sequence element brings full functionality in vivo. J Biol Chem. 1991 Dec 15;266(35):23872–23877. [PubMed] [Google Scholar]
- Kranz D. M., Saito H., Heller M., Takagaki Y., Haas W., Eisen H. N., Tonegawa S. Limited diversity of the rearranged T-cell gamma gene. 1985 Feb 28-Mar 6Nature. 313(6005):752–755. doi: 10.1038/313752a0. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Kulozik A. E., Bellan-Koch A., Bail S., Kohne E., Kleihauer E. Thalassemia intermedia: moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood. 1991 May 1;77(9):2054–2058. [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Maeda M., Oshiman K., Tamura S., Kaya S., Mahmood S., Reuben M. A., Lasater L. S., Sachs G., Futai M. The rat H+/K(+)-ATPase beta subunit gene and recognition of its control region by gastric DNA binding protein. J Biol Chem. 1991 Nov 15;266(32):21584–21588. [PubMed] [Google Scholar]
- McDougall S., Peterson C. L., Calame K. A transcriptional enhancer 3' of C beta 2 in the T cell receptor beta locus. Science. 1988 Jul 8;241(4862):205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmacek M. S., Vora A. J., Shen T., Barr E., Jung F., Leiden J. M. Identification and characterization of a cardiac-specific transcriptional regulatory element in the slow/cardiac troponin C gene. Mol Cell Biol. 1992 May;12(5):1967–1976. doi: 10.1128/mcb.12.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C., Pitiot G., Plumb M., Lowe S., de Verneuil H., Grandchamp B. Characterization of hypersensitive sites, protein-binding motifs, and regulatory elements in both promoters of the mouse porphobilinogen deaminase gene. J Biol Chem. 1991 Jun 5;266(16):10562–10569. [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Redondo J. M., Hata S., Brocklehurst C., Krangel M. S. A T cell-specific transcriptional enhancer within the human T cell receptor delta locus. Science. 1990 Mar 9;247(4947):1225–1229. doi: 10.1126/science.2156339. [DOI] [PubMed] [Google Scholar]
- Royer H. D., Reinherz E. L. Multiple nuclear proteins bind upstream sequences in the promotor region of a T-cell receptor beta-chain variable-region gene: evidence for tissue specificity. Proc Natl Acad Sci U S A. 1987 Jan;84(1):232–236. doi: 10.1073/pnas.84.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Kranz D. M., Takagaki Y., Hayday A. C., Eisen H. N., Tonegawa S. A third rearranged and expressed gene in a clone of cytotoxic T lymphocytes. Nature. 1984 Nov 1;312(5989):36–40. doi: 10.1038/312036a0. [DOI] [PubMed] [Google Scholar]
- Saito H., Kranz D. M., Takagaki Y., Hayday A. C., Eisen H. N., Tonegawa S. Complete primary structure of a heterodimeric T-cell receptor deduced from cDNA sequences. 1984 Jun 28-Jul 4Nature. 309(5971):757–762. doi: 10.1038/309757a0. [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. Identification of a novel lymphoid specific octamer binding protein (OTF-2B) by proteolytic clipping bandshift assay (PCBA). EMBO J. 1988 Dec 20;7(13):4221–4229. doi: 10.1002/j.1460-2075.1988.tb03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle R., Muller M., Otsuka-Murakami H., Renkawitz R. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature. 1988 Mar 3;332(6159):87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- Siu G., Strauss E. C., Lai E., Hood L. E. Analysis of a human V beta gene subfamily. J Exp Med. 1986 Nov 1;164(5):1600–1614. doi: 10.1084/jem.164.5.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988 Apr 14;332(6165):649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Treisman J., Desplan C. The products of the Drosophila gap genes hunchback and Krüppel bind to the hunchback promoters. Nature. 1989 Sep 28;341(6240):335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Tsai S. F., Strauss E., Orkin S. H. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991 Jun;5(6):919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., LaMarco K. L., Johnson P. F., Landschulz W. H., McKnight S. L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988 Jul;2(7):801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- Waterman M. L., Jones K. A. Purification of TCF-1 alpha, a T-cell-specific transcription factor that activates the T-cell receptor C alpha gene enhancer in a context-dependent manner. New Biol. 1990 Jul;2(7):621–636. [PubMed] [Google Scholar]
- Winoto A., Baltimore D. A novel, inducible and T cell-specific enhancer located at the 3' end of the T cell receptor alpha locus. EMBO J. 1989 Mar;8(3):729–733. doi: 10.1002/j.1460-2075.1989.tb03432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. Alpha beta lineage-specific expression of the alpha T cell receptor gene by nearby silencers. Cell. 1989 Nov 17;59(4):649–655. doi: 10.1016/0092-8674(89)90010-x. [DOI] [PubMed] [Google Scholar]
- Winoto A., Mjolsness S., Hood L. Genomic organization of the genes encoding mouse T-cell receptor alpha-chain. 1985 Aug 29-Sep 4Nature. 316(6031):832–836. doi: 10.1038/316832a0. [DOI] [PubMed] [Google Scholar]
- Xu M. Z., Stavnezer J. Regulation of transcription of immunoglobulin germ-line gamma 1 RNA: analysis of the promoter/enhancer. EMBO J. 1992 Jan;11(1):145–155. doi: 10.1002/j.1460-2075.1992.tb05037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi Y., Yoshikai Y., Leggett K., Clark S. P., Aleksander I., Mak T. W. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984 Mar 8;308(5955):145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]