Abstract
A small number of rare, recurrent genomic copy number variants (CNVs) are known to substantially increase susceptibility to schizophrenia. As a consequence of the low fecundity in people with schizophrenia and other neurodevelopmental phenotypes to which these CNVs contribute, CNVs with large effects on risk are likely to be rapidly removed from the population by natural selection. Accordingly, such CNVs must frequently occur as recurrent de novo mutations. In a sample of 662 schizophrenia proband–parent trios, we found that rare de novo CNV mutations were significantly more frequent in cases (5.1% all cases, 5.5% family history negative) compared with 2.2% among 2623 controls, confirming the involvement of de novo CNVs in the pathogenesis of schizophrenia. Eight de novo CNVs occurred at four known schizophrenia loci (3q29, 15q11.2, 15q13.3 and 16p11.2). De novo CNVs of known pathogenic significance in other genomic disorders were also observed, including deletion at the TAR (thrombocytopenia absent radius) region on 1q21.1 and duplication at the WBS (Williams–Beuren syndrome) region at 7q11.23. Multiple de novos spanned genes encoding members of the DLG (discs large) family of membrane-associated guanylate kinases (MAGUKs) that are components of the postsynaptic density (PSD). Two de novos also affected EHMT1, a histone methyl transferase known to directly regulate DLG family members. Using a systems biology approach and merging novel CNV and proteomics data sets, systematic analysis of synaptic protein complexes showed that, compared with control CNVs, case de novos were significantly enriched for the PSD proteome (P=1.72 × 10−6). This was largely explained by enrichment for members of the N-methyl-D-aspartate receptor (NMDAR) (P=4.24 × 10−6) and neuronal activity-regulated cytoskeleton-associated protein (ARC) (P=3.78 × 10−8) postsynaptic signalling complexes. In an analysis of 18 492 subjects (7907 cases and 10 585 controls), case CNVs were enriched for members of the NMDAR complex (P=0.0015) but not ARC (P=0.14). Our data indicate that defects in NMDAR postsynaptic signalling and, possibly, ARC complexes, which are known to be important in synaptic plasticity and cognition, play a significant role in the pathogenesis of schizophrenia.
Keywords: CNV, de novo, DLG, EHMT1, postsynaptic, schizophrenia
Introduction
Genome-wide association studies have found strong evidence for association between schizophrenia and a number of genetic variants, both common and rare.1 So far, the evidence for rare variants comes mainly from the analysis of deletions and duplications of segments of DNA known as copy number variants (CNVs). Cumulatively, as a general class, large (>100 kb) rare (<1%) CNVs occur more frequently in those with schizophrenia2, 3 than controls, and several individual CNV loci have been strongly implicated as risk factors for schizophrenia with high degrees of statistical confidence. These include deletions at 1q21.1, NRXN1, 3q29, 15q11.2, 15q13.3, 22q11.2 and duplications at VIPR2, 16p11.2, 16p13.1 and 15q11-q13.2, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Pleiotropic effects are common, the same CNV often conferring risk for a range of neurodevelopmental phenotypes including autism, mental retardation, attention deficit hyperactivity disorder and epilepsy, although interestingly, and in contrast to the findings with common risk alleles, there is little evidence that schizophrenia-associated CNVs confer risk for bipolar disorder.14
All of the currently known risk CNVs are rare (control frequencies typically <0.001) and confer substantial effects on risk (odds ratios 3–30). The known risk CNVs occur in 2–3% of cases, but it is likely that many other risk CNV loci remain to be identified. Most schizophrenia-associated CNVs span multiple genes, limiting our ability to make strong inferences regarding pathogenesis. Important exceptions are deletions of NRXN1, encoding the presynaptic neuronal cell adhesion molecule neurexin 1,11, 15 pointing to the importance of as yet unspecified abnormalities of synaptic function in the disorder. Also, obscuring mechanistic insights from the CNV data are that most reported CNVs occurring in cases are too rare to allow clear demonstration of association statistically. One way to circumvent this is to test whether particular functionally related groups or sets of genes are enriched among case CNVs, rather than trying to interpret the results from individual CNVs. A limitation of this approach is that the enrichment of CNVs seen in case–control studies is modest;2 indeed, one large study has reported no overall excess of CNVs in cases at all.4 This implies that among sets of CNVs drawn from cases, only a small proportion can be expected to be true risk factors for the disorder. Nevertheless, gene-set enrichment studies have supported conclusions drawn from consideration of genes affected by individual CNVs in schizophrenia16 by observing enrichment in schizophrenia of genes involved in a range of brain functions, for example, those encoding products involved in nitric oxide signalling, synaptic long-term potentiation and glutamate receptor signalling,17 or genes in a broad category corresponding to the gene ontology (GO) category ‘synaptic transmission'.18 However, it has been noted that the early gene-set studies did not allow for important confounders, in particular the large size of genes implicated in brain function, and that the conclusions that can be drawn are consequently unclear.19
Schizophrenia is associated with reduced fecundity, ∼40% that of the general population,20 or even lower according to the largest population-based study.21 It follows that schizophrenia-related mutations of large effect should be rare because of intense purifying selection, and those that occur in multiple unrelated individuals are likely to do so through independent de novo mutations.7, 22, 23, 24 One study on de novo CNV mutation in schizophrenia24 showed that the rate of de novo CNV mutation in probands with no family history was 8 times higher in cases than in controls. This marked elevation in the rate of de novo CNVs contrasts with the relatively modest elevation in the rate of CNVs seen in case–control studies,2, 4 and suggests that sets of de novos might be more informative for gene-set enrichment analyses.
Here, we report the largest analysis of de novo CNVs in schizophrenia to date. Our aims were to identify novel CNVs that increase risk of schizophrenia and to illuminate aspects of the pathophysiology of the disorder through gene-set enrichment analyses informed by recently curated proteomics data sets of synaptic protein complexes.
Materials and methods
Samples
Bulgaria
The sample for de novo CNV analysis comprised 662 Bulgarian parent–proband trios from 638 families. We did not exclude probands (N=61) with a history of psychosis in a parent as none of the risk CNVs identified to date are sufficiently penetrant to fully explain the disorder in carriers. All cases had been hospitalised and met DSM-IV (Diagnostic and Statistical Manual of Mental Disorders-fourth edition) criteria for schizophrenia or schizoaffective disorder based upon SCAN (Schedules for Clinical Assessment in Neuropsychiatry) interview by psychiatrists, and review of case notes. Cases were recruited from general adult psychiatric services and were typical of those attending those services. Although they did not have formal IQ assessments, all attended mainstream schools from which people with known mental retardation were excluded. All participants provided informed consent. Further details concerning ascertainment and diagnostic practices are provided in the Supplementary Material. All DNA samples were derived from peripheral venous blood.
Icelandic control de novos
deCODE Genetics provided data for 2623 complete parent–offspring trios from the Icelandic population.7 Probands known to be affected with neurodevelopmental/psychiatric disorders (schizophrenia, autism, attention deficit hyperactivity disorder, mental retardation and bipolar affective disorder) had been excluded.
Autism case and control de novos
Data on de novo rates in autism cases and their unaffected siblings are directly taken from the recent large study of Sanders et al.25 based upon the Illumina (San Diego, CA, USA) 1M high-density array.
Case–control data sets
We used four large publicly available data sets to which we also had access to the raw data. (1) The International Schizophrenia Consortium (ISC),2 which included 3391 cases and 3181 controls genotyped with Affymetrix 6.0 or 5.0 arrays (Affymetrix, Santa Clara, CA, USA). Note that 328 Bulgarian cases from that study are probands in our trios (although their parents were not genotyped for de novo calling in the ISC study). We excluded those subjects from the ISC data. The ISC also included 605 unrelated controls recruited by us in Bulgaria (details in ref. 2) and those publicly available data were included in the present study. (2) The Molecular Genetics of Schizophrenia (MGS) Consortium,4 which included 3192 cases and 3437 controls genotyped with Affymetrix 6.0 arrays. (3) A UK case–control study of 471 schizophrenia and 2792 controls genotyped using the Affymetrix (Affymetrix) GeneChip500K Mapping Array (see ref. 3 for details of the sample and CNV calling). (4) CNV data reported by Ikeda et al.26 comprising a Japanese sample of 519 cases and 513 controls. Including the data from the current study on transmissions and non-transmissions to affected offspring, and excluding the 328 overlapping Bulgarian cases, the combined case–control data sets contain a total of 7907 independent cases and 10 585 controls.
Genotyping and CNV analysis
Bulgarian samples
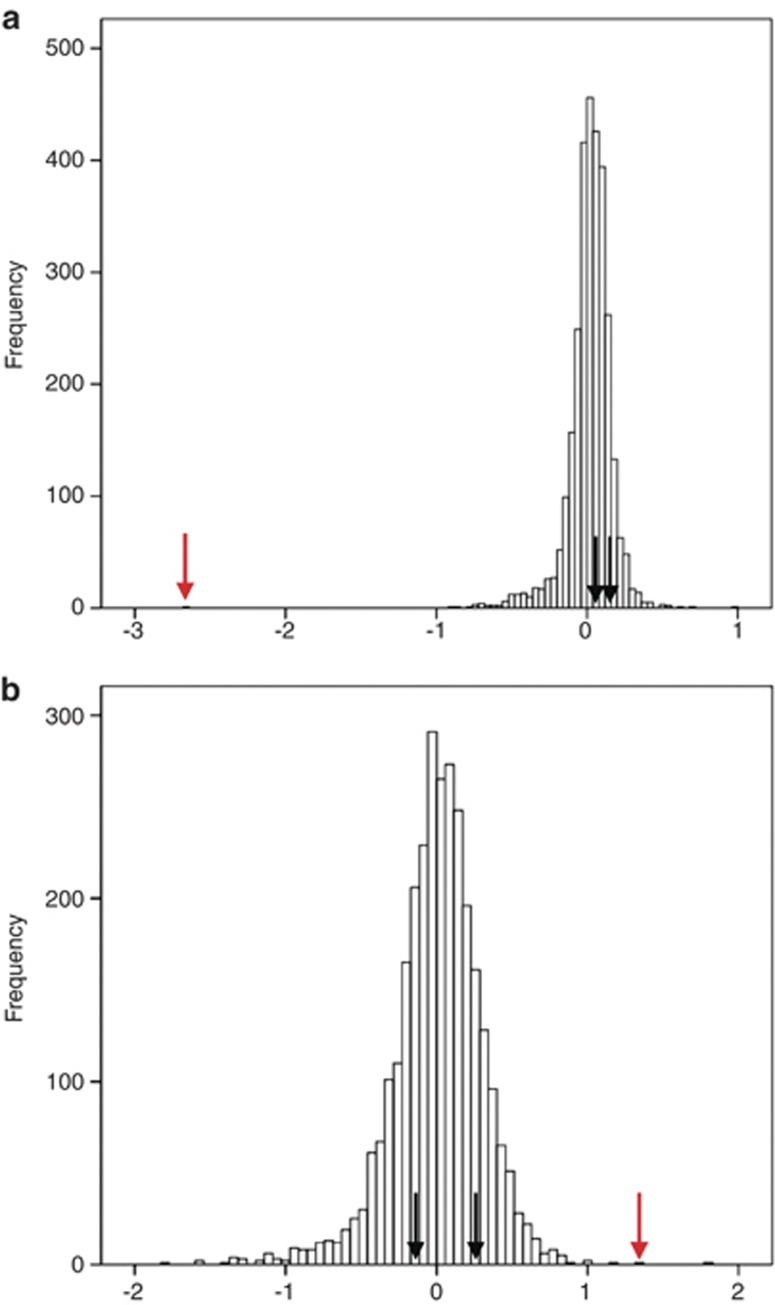
Full details are provided in the Supplementary Material (Sections 1–3). All participants were genotyped with Affymetrix 6.0 arrays (Affymetrix) at the Broad Institute of Harvard and Massachusetts Institute of Technology. As an initial screen, we used Genotyping Console 4.0 software (Affymetrix, Santa Clara, CA, USA) to call autosomal CNVs, restricting initial calls to ⩾10 kb and ⩾10 probes. We next excluded individuals with >50 CNV calls, as these were outliers from the distribution, followed by CNV loci with a frequency >1% in the whole sample. We then excluded putative CNVs <15 kb, covered by <15 probes, or where >50% of their length overlapped low copy repeats. Calls compatible with a de novo were made if a proband CNV was not spanned >50% of its length by a CNV in either parent. Probands who had large numbers of apparent de novos (>10) were excluded. After this initial screen with relaxed criteria to capture as many potential de novos as possible, we measured probe Log2 ratios derived from PennCNV.27 We then used a slight modification of the MeZOD algorithm12 (Supplementary Section 3) to visualise outlier signals in probands potentially indicative of de novos (Figure 1). Again, we used relaxed criteria, only excluding clear false positives (Supplementary Section 3). For those whose patterns were either highly suggestive of a de novo (N=40, Figure 1a) or were ambiguous (N=33, Figure 1b), proband and parent DNAs were examined on custom Agilent SurePrint G3 Human CGH Microarrays on which 50–200 probes were placed to cover each CNV (depending on CNV size). For quality control purposes, we also included probes on all putative de novos identified by the first-pass Genotyping Console analysis, but that were subsequently rejected as false positives by the MeZOD method.
Figure 1.
Histograms of distributions of z-scores. (a) A suggestive de novo and (b) an ambiguous de novo MeZOD call. Black arrows indicate the position of a parent, and red arrows of a child. The x axis shows the median z-scores for all individuals for a particular copy number variant (CNV) region.
To re-call CNVs in the Bulgarian controls, we used the same filtering criteria and accepted only those considered highly suggestive by the MeZOD. Although we did not validate these on Agilent arrays, our calls have a demonstrable low false positive rate (<1%, Supplementary Section 4). This is much less than the corresponding false positive rate for de novos whose rarity confers more unfavourable signal-to-noise characteristics.
Icelandic samples
These were genotyped using Illumina bead arrays (HumanHap317, HumanHap370 and HumanHap1M). BeadStudio (Illumina, San Diego, CA, USA; version 2.0) was used to call genotypes, normalise signal intensity data and establish the log R ratio and B allele frequency at every single-nucleotide polymorphism. Samples passing quality control were examined using PennCNV (10.1101/gr.6861907). Calls required 10 consecutive markers based upon the subset of markers present on all genotyping chips listed above (the HumanHap317 content). All putative de novo events were visually inspected using DosageMiner software (developed by deCODE Genetics). CNVs were excluded according to low copy repeat content and frequency as for the Bulgarian sample. This resulted in 59 CNVs, an autosomal de novo rate of 2.2%. Given the difference in the platforms, we undertook a number of analyses to confirm that the Icelandic de novos are a suitable comparator group for the case de novos (see Results and Supplementary Material).
MGS/ISC/UK/Japan
MGS samples were analysed in the same way as the Bulgarian samples including MeZOD. Data for ISC2 UK3 and Japanese samples26 were taken from the original publications, and CNVs at loci of interest were manually verified in the available raw data (further information in Supplementary Material).
Gene set analyses
Sets
We collated experimentally defined proteomic data sets corresponding to the structures listed in Table 2. The details of how those gene sets were collated are provided in Supplementary Section 10. We also examined sets based upon the Gene Ontology system (GO sets) in the gene2go file available from the NCBI (National Center for Biotechnology Information) on 28 July 2010 (Supplementary Section 11).
Statistical approaches
A gene was considered ‘hit' if a CNV was overlapped according to the NCBI Build 36.3. Full details of mapping are given as Supplementary Section 10.
The impact of biases relating to gene-set analyses of CNVs have been discussed elsewhere.19 To overcome those biases, we fitted the following logistic regression models to the combined set of case and control (or control de novo) CNVs and compared the change in deviance between (1) and (2).
logit (pr(case))=CNV size + Total number of genes hit outside the gene set + number of genes hit in the gene set.
logit (pr(case))=CNV size + Total number of genes hit outside the gene set.
Significance was assessed by one-sided test of an excess of genes hit in the gene set by case CNVs. The inclusion of CNV size allows for case de novo CNVs being larger than typical CNVs (and thus likely to hit more genes). Inclusion of the total number of genes hit outside the gene set in the regression corrects for case CNVs hitting more genes overall (regardless of function) than control CNVs. Although explicitly adjusted for in the above analysis, to confirm that the results are not due to the fact that de novo CNVs are more likely to hit genes, we also performed an analysis restricted to CNVs that hit genes.
We used the same method to compare the number of genes in gene sets hit by case de novos with those hit by (1) 1367 CNVs from the 605 Bulgarian unaffected controls (2) 59 de novos found in Icelandic controls and (3) 14 control de novos from the unaffected sibs of autism probands.25 The analyses control for different sources of potential bias including array type (the Bulgarian controls) and the possibility that de novos have fundamentally different characteristics (other than size that is adjusted for) than control CNVs.
To investigate the impact of using ‘control' CNVs, we undertook a random placement analysis comparing the number of de novo CNVs hitting each gene set with that found when CNV locations were randomised, importantly ensuring that each random assignment hits at least one gene, and that the probability of a gene being hit was proportional to its length (Supplementary Section 12).
Partitioning the signals in gene sets
Gene sets are not fully independent, for example, some members of the synaptic vesicle set (Table 2) are also members of the postsynaptic density (PSD). To determine which among overlapping sets appeared to be responsible for a gene-set enrichment, we undertook conditional regression analyses as described in Supplementary Section 13.
Meta-analysis of case–controls
For meta-analysis combining cases and controls from multiple studies, we included in the above regression models a “study” term added as an N-level factor (where N=number of case/control sets being combined). This makes the analysis robust to differences between studies in chip, analytic method and other study-specific factors.
Results
We identified 34 confirmed de novo CNVs (Table 1), a rate in all cases of 5.1%. Detailed descriptions of individual de novo CNVs are given in Supplementary Section 6 and in the Discussion section. As in an earlier study,24 the de novo rate in those with a history of psychosis in a parent was lower (1.6%) than in those without such history (5.5%), although this was not statistically significant. Parents of probands with de novos were not older at the time of birth of their children than parents of probands without de novos: (27.8 vs 28.7 years, respectively, for fathers and 25.1 vs 25.1 years for mothers). Probands with de novo CNVs (Table 1) did not differ from the rest of the probands regarding age at onset (23.9 vs 23.8 years, P=0.9) and average school results (4.5 vs 4.7, P=0.5), and both sets of probands had similar numbers of children (0.52 vs 0.59, P=0.6). In those instances (21) where it was possible to determine the parental origin, more de novos occurred in the paternal (P=14) than the maternal (n=7) genome but this was not statistically significant (P=0.13). The nonsignificant excess of paternal de novos was largely attributable to CNVs that were not generated by nonallelic homologous recombination, eight such events being observed on chromosomes of paternal origin compared with two on those that were maternally derived, although this is not significantly different from chance (P=0.06).
Table 1. List of de novo CNVs found in the study.
| Cytoband | Location | Type | Size (bp) | Genes | Origin of mutation, F:M SNPs | Diagnosis | Schooling | Children | Age at onset/ age at sampling | Somatic |
|---|---|---|---|---|---|---|---|---|---|---|
| 1q21.1 | chr1:144101459–144503409 | Del | 401 950 | 16 genes, TAR region | 10F:0M | SA | V good, college | 2 | 40/49 | Psoriasis |
| 1q43 | chr1:235475280–235639644 | Del | 164 364 | RYR2 | 12F:0M | SZ, par | Good | — | 21/33 | Patent ductus arteriosusb |
| 2q21.2 | chr2:133504420–133879778 | Del | 375 358 | NAP5 | 7F:0M | SZ, par | V good | 1 | 18/25 | — |
| 3q13.12 | chr3:109330592–110198715 | Dupl | 868 123 | 9 genes | 11F:0M | SZ, cat | Pass | 2 | 33/48 | — |
| 3q29a | chr3:197185548–198825231 | Del | 1 639 683 | 21 genes, 3q29 syndrome | 56F:0M | SZ, par | Good, college | — | 19/45 | Congenital heart disease |
| 4q13.3 | chr4:70935504–70969553 | Del | 34 049 | HTN1, HTN3 | — | SZ, cat | Excellent, college | — | 24/33 | — |
| 4q21.21 | chr4:79944612–80081979 | Del | 137 367 | BMP2K, PAQR3 | — | SZ, par | Excellent | 1 | 32/33 | — |
| 6q12 | chr6:68675955–68761101 | Dupl | 85 146 | — | — | SZ, par | Excellent | — | 23/29 | Hypertension |
| 7p14.1 | chr7:38260614–38307187 | Del | 46 573 | TARP | — | SA | Excellent, university | — | 17/24 | Asthma |
| 7q11.23 | chr7:72390286–76445231 | Dupl | 4 054 945 | 38 genes, WBS region | 1F: 45M | SZ, dis | V good | — | 17/28 | — |
| 7q32.1 | chr7:127275795–127447967 | Del | 172 172 | C7orf54, SND1 | — | SA | V good | — | 25/35 | — |
| 8p23.2 | chr8:4121968–4299810 | Del | 177 842 | CSMD1 | 0F:23M | SZ, par | Excellent, university | — | 17/37 | — |
| 8p23.1 | chr8:10066862–10155414 | Del | 88 552 | MSRA | — | SZ, par | Pass | — | 15/41 | — |
| 9p22.3 | chr9:16310745–16327782 | Del | 17 037 | — | — | SA | Excellent, university | 1 | 34/37 | — |
| 9q31.3 | chr9:110859131–111433199 | Dupl | 574 068 | 4 genes | — | SZ, ns | Pass | — | 9/26 | — |
| 9q34.3 | chr9:139762152–139797423 | Dupl | 35 271 | EHMT1 | — | SZ, ns | V good | 2 | 20/37 | Overweight |
| 9q34.3 | chr9:139769564–139792102 | Del | 22 538 | EHMT1 | — | SZ, dis | Pass | 1 | 24/36 | Overweight |
| 11q14.1 | chr11:83472750–83842973 | Del | 370 223 | DLG2 | 0F:M12 | SZ, cat | V good, college | — | 18/22 | — |
| 11q14.1 | chr11:84006106–84226064 | Del | 219 958 | DLG2 | 10F:0M | SZ, par | Pass | 2 | 20/35 | — |
| 12q24.13 | chr12:111723795–111776045 | Del | 52 250 | RPH3A | — | SZ, par | Good | — | 21/33 | Patent ductus arteriosusb |
| 12q24.33 | chr12:130388037–130659530 | Dupl | 271 493 | — | — | SZ, par | Good | — | 27/33 | — |
| 13q14.11 | chr13:40319620–41182276 | Del | 862 656 | 8 genes | 15F:0M | SZ, par | Good | — | 22/33 | — |
| 14q13.2 | chr14:34464771–34627720 | Del | 162 949 | 3 genes | 1F:0M | SA, FH+ | Excellent | — | 16/24 | — |
| 15q11.2a | chr15:19548923–20852202 | Del | 1 303 279 | 8 genes | 0F:16M | SZ, par | Pass | — | 32/42 | — |
| 15q11.2a | chr15:20224751–20777909 | Dupl | 553 158 | 5 genes | 0F:10M | SA | Pass | 1 | 20/28 | — |
| 15q11.2a | chr15:20224751–20852202 | Dupl | 627 451 | 5 genes | 0F:8M | SA | Good | — | 29/31 | — |
| 15q11.2a | chr15:20302446–21038975 | Del | 736 529 | 6 genes | 19F:0M | SZ, par | V good | 1 | 17/28 | — |
| 15q13.1 | chr15:26785056–28289366 | Dupl | 1 504 310 | 4 genes | 0F:2M | SZ, dis | Pass | — | 23/43 | — |
| 15q13.3a | chr15:28707904–30326817 | Del | 1 591 596 | 7 genes | 93F:0M | SZ, par | Pass | — | 31/38 | — |
| 15q13.3a | chr15:28707904–30299500 | Del | 1 618 913 | 7 genes | 25F:0M | SZ, par | V good | — | 32/52 | — |
| 16p11.2a | chr16:29488112–30099396 | Dupl | 611 284 | 31 genes | 21F:0M | SZ, par | Excellent, college | 1 | 35/46 | Rheumatoid arthritis |
| 18p11.31 | chr18:3515935–4332609 | Del | 816 674 | DLGAP1, FLJ35776 | 61F:0M | SZ, par | Good | 1 | 18/25 | — |
| 20p12.1 | chr20:14694326–14863051 | Del | 168 725 | MACROD2 | 2F:0M | SZ, cat | V good | 1 | 32/37 | — |
| 21q21.1 | chr21:22698250–22778244 | Del | 79 994 | — | — | SA | Excellent, university | — | 29/30 | Single febrile convulsion |
Abbreviations: CNV, copy number variant; SA, schizoaffective disorder; SNP, single-nucleotide polymorphism; SZ, cat, catatonic schizophrenia; SZ, dis, disorganised schizophrenia; SZ, ns=schizophrenia, not otherwise specified; SZ, par, paranoid schizophrenia; TAR thrombocytopenia absent radius; WBS, Williams–Beuren syndrome.
Indicates a known schizophrenia locus.
Same patient who has two de novos.
Origin of mutation indicates whether the mutation had occurred on the paternal or maternal chromosome. The number of informative SNPs supporting parental origin are given in the order father(F):mother(M). Additional detail is given in Supplementary Section 6. Coordinates in the paper refer to the UCSC (University of California, Santa Cruz) human genome assembly hg18 (National Center for Biotechnology Information (NCBI) build 36). Final school results in Bulgaria are reported as 2=fail, 3=pass, 4=good, 5=very good, 6=excellent.
In order to estimate the de novo CNV rate in controls for comparison with cases, we compared the case de novo rate with that in controls from two sources, the Icelandic population controls and the unaffected sibs from a recent large study of autism25 (Supplementary Table S3). The de novo mutation rate in our cases was higher than in both sets of controls (2.2%, P=0.00015 and 1.6%, P=0.00008, respectively), both of which had a similar rate (P=0.28) despite differences in the density of markers in the control genotyping platforms.
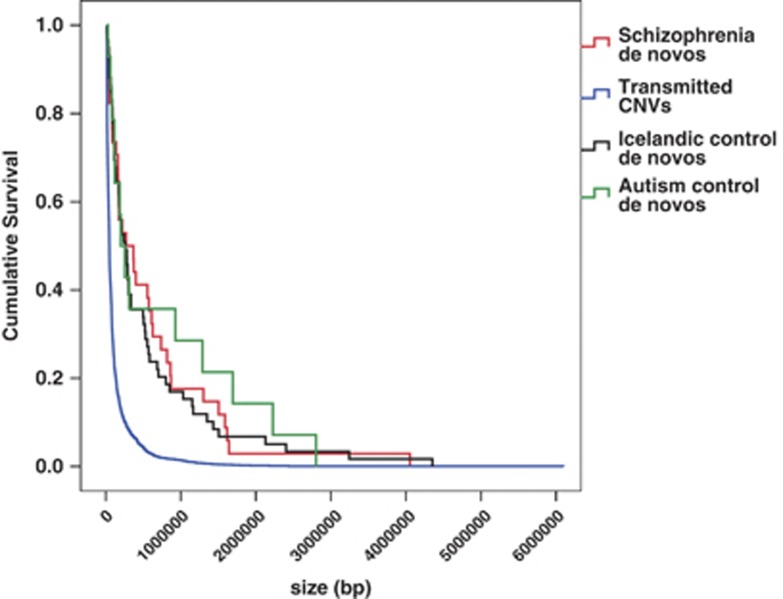
In order to exclude the possibility that the increased de novo rate seen in cases reflected the different platforms28 used in our cases and the control studies, we undertook sensitivity analyses. If the elevation in de novos in cases is an artefact of greater call sensitivity in the present study, the enrichment we observed should be biased towards smaller CNVs, larger CNVs being called reliably after exclusion of CNVs spanning complex repeat regions (as we have done).28 However, relative enrichment for de novos among cases was similar for large de novos >500 kb (2.1% vs 0.8%, P=0.0014) as it was for small de novos <200 kb (2.3% vs 0.9%, P=0.0035), and the overall size distribution of case de novos was not shifted towards smaller CNVs (Figure 2 and Supplementary Table S3) compared with the control de novos. In general, duplications are less easily detected by microarrays than deletions. To exclude the possibility that the excess of de novos in cases reflects a lower sensitivity of the control platforms to detect duplications, we also examined the duplication/deletion ratios in the data sets. These were not significantly different (P>0.35 for each sample), although contrary to the hypothesis of a selective loss of sensitivity to detect duplications, both sets of controls actually had a higher proportion of duplications (Icelandic=0.39, Autism controls=0.36) than the cases (0.29). Further details on the size distribution of the de novo CNV are given in Supplementary Section 8, and of the full sensitivity analyses in Supplementary Section 1. Finally, we note that the control rates were similar to those in our experimental group with an affected parent (who according to an earlier work24 do not have elevated rates of de novo mutation), suggesting that technical variation between our own and other studies does not make a major impact on our conclusions.
Figure 2.
Size of copy number variants (CNVs). Kaplan–Meier survival graph for the size of de novo CNVs in cases, Icelandic controls, Bulgarian controls and unaffected siblings of autism probands.25
Analysis of de novo loci in case–control studies
We examined the fully independent case–control data sets for rare CNVs at the novel loci affected by case de novos, including CNVs only of the same class that had been observed to have occurred as de novos (that is, deletions, duplications or both where relevant) (Supplementary Section 7) that intersected at least one exon of a gene (details in Supplementary Table S1). Even after an extremely conservative approach of excluding all CNVs (deletions and duplications) at known schizophrenia loci represented among our de novos (3q29, 15q11.2, 15q13.3 and 16p11.2), we found rates of 0.4% (32/7907) in cases and 0.21% (22/10 585) in controls, a twofold enrichment (Fisher one-tailed P=0.012). We did not obtain evidence for association to individual CNV loci at a level that would survive correction for multiple testing (N=19 excluding the known schizophrenia loci, giving a Bonferroni corrected threshold of P=0.0025). However, nominally significant associations (P uncorrected <0.05) were observed for deletions at DLG2 (P=0.02) and MSRA (P=0.03), whereas the EHMT1 locus just failed to reach this uncorrected threshold (P=0.055). Of interest, although not even nominally significant, we also observed an excess of CNVs in cases at two other loci known to be implicated in nonpsychiatric genomic disorders: deletions of the TAR (thrombocytopenia absent radius) region (P=0.11) and duplications of the WBS (Williams–Beuren syndrome) region (P=0.11)
Gene-set analyses
We initially undertook gene-set analyses based upon proteomics-based annotations (Table 2 and Supplementary Section 11). To avoid multiple testing involved in subgroup analysis, we present the findings for the full sample of de novos, although we note that exclusion of the single de novo in a proband with a family history of psychosis in a parent made essentially no difference to the results. Compared with Bulgarian control CNVs, we found a highly significant excess of PSD genes within case de novos (P=1.72 × 10−6; Table 2). As expected, the results where the analysis was restricted to CNVs hitting genes were similar to those of the primary analysis (data not shown).
Table 2. Enrichment of gene sets for de novo CNV hits in comparison with control CNVs.
| Gene set | N genes | N genes hit by CNVs P-value |
Genes hit by SCZ de novos | |||
|---|---|---|---|---|---|---|
| SCZ de novo (34) | Bulgarian control (1367) | Icelandic control de novo (59) | Autism control de novo (14) | |||
| PSD | 664 | 19 | 49 (1.72 × 10−6) | 13 (0.045) | 4 (0.11) | DLG1, DLG2, DLGAP1, RYR2, SND1, STX1A, MDH2, HSPB1, YWHAG, RPH3A, CYFIP1, TJP1, ALDOA, TAOK2, MAPK3 |
| ARC complex | 25 | 8 | 7 (3.78 × 10−8) | 1 (2.51 × 10−4) | 0 (0.0049) | DLG1, DLG2, DLGAP1, CYFIP1 |
| NMDAR complex | 59 | 8 | 6 (4.24 × 10−6) | 2 (0.0061) | 0 (0.01) | DLG1, DLG2, DLGAP1, STX1A, YWHAG, TJP1, MAPK3 |
| PSD-95 complex | 58 | 4 | 3 (1.17 × 10−5) | 1 (0.017) | 0 (0.033) | DLG1, DLG2, DLGAP1 |
| mGluR5 complex | 37 | 3 | 4 (0.026) | 2 (0.45) | 0 (0.15) | YWHAG, RPH3A, ALDOA |
| Presynapse | 426 | 8 | 25 (0.033) | 8 (0.32) | 2 (0.28) | STX1A, RPH3A, CYFIP1, ALDOA, MDH2 |
| Synaptic vesicle | 333 | 7 | 20 (0.014) | 8 (0.39) | 2 (0.31) | STX1A, RPH3A, CYFIP1, ALDOA |
| Active zone | 176 | 2 | 6 (0.29) | 3 (0.91) | 0 (0.26) | ALDOA, MDH2 |
| Nucleus | 160 | 5 | 10 (0.0024) | 2 (0.026) | 0 (0.018) | CYFIP1, TJP1 |
| Mitochondrion | 189 | 3 | 9 (0.41) | 1 (0.11) | 0 (0.093) | MDH2, BDH1, KIAA0564 |
| Cytoplasm | 263 | 4 | 11 (0.68) | 3 (0.55) | 0 (0.15) | EIF4H, YWHAG, MSRA, MVP |
| Endoplasmic reticulum | 94 | 1 | 3 (0.75) | 0 (0.18) | 0 (0.31) | POR |
| Endoplasmic reticulum/Golgi-derived vesicles | 94 | 0 | 0 | 0 | 0 | |
| Recycling endosomes | 65 | 0 | 2 (0.83) | 0 | 0 | |
| Early endosomes | 17 | 0 | 1 (0.82) | 0 | 0 | |
| Golgi | 31 | 0 | 1 (0.82) | 0 | 0 | |
| Plasma membrane | 50 | 0 | 2 (0.61) | 0 | 0 | |
Abbreviations: ARC, activity-regulated cytoskeleton-associated protein; CNV, copy number variant; NMDAR, N-methyl-D-aspartate receptor; PSD, postsynaptic density; SCZ, schizophrenia. Gene sets were tested for enrichment in 34 schizophrenia (‘SCZ') de novo CNVs compared with 1367 CNVs found in 605 Bulgarian controls (‘controls'), 59 de novo CNVs found in 2623 unaffected individuals from the Icelandic population (‘Icelandic control de novo') and 14 de novo CNVs found in unaffected siblings of autism proband from the study by Sanders et al.25 (‘Autism control de novo'). ‘N genes' refers to number of genes in the set. P-values are presented underneath the number of genes hit and correspond to one-tailed tests of an excess of gene hits in case CNVs. P-values in bold are significant. ‘N genes hit by CNVs' refers to the number of times any gene in the set is hit by a CNV. The unique genes hit in each set are given in the final column. Genes in bold are present in multiple subcellular components.
Significant enrichments were also observed in presynaptic vesicle and nuclear gene sets, but not after conditioning on the PSD (Pmin=0.66), whereas the PSD gene set remained significantly enriched for hits after conditioning on the other sets individually (Pmax=5.20 × 10−3) or combined (P=0.016) (Supplementary Section 13). The most parsimonious interpretation is that our findings specifically implicate the PSD, although we cannot exclude the possibility of effects across multiple functional sets.
To explore our findings in the context of a less restricted set of classifications, we performed enrichment analyses using the GO annotation. Of all categories, ‘the synapse' (GO: 45202) was by two orders of magnitude the most significantly enriched (P=9.6 × 10−9) (Supplementary Section 11 and Supplementary Table S4) but most of this signal was attributable to the PSD gene set (P=0.049 after removing PSD genes, Supplementary Table S5). Only one subcategory of ‘the synapse' GO: 45202 was enriched after PSD genes were removed: GO: 30672 ‘synaptic vesicle membrane' (P=0.036, Supplementary Table S5).
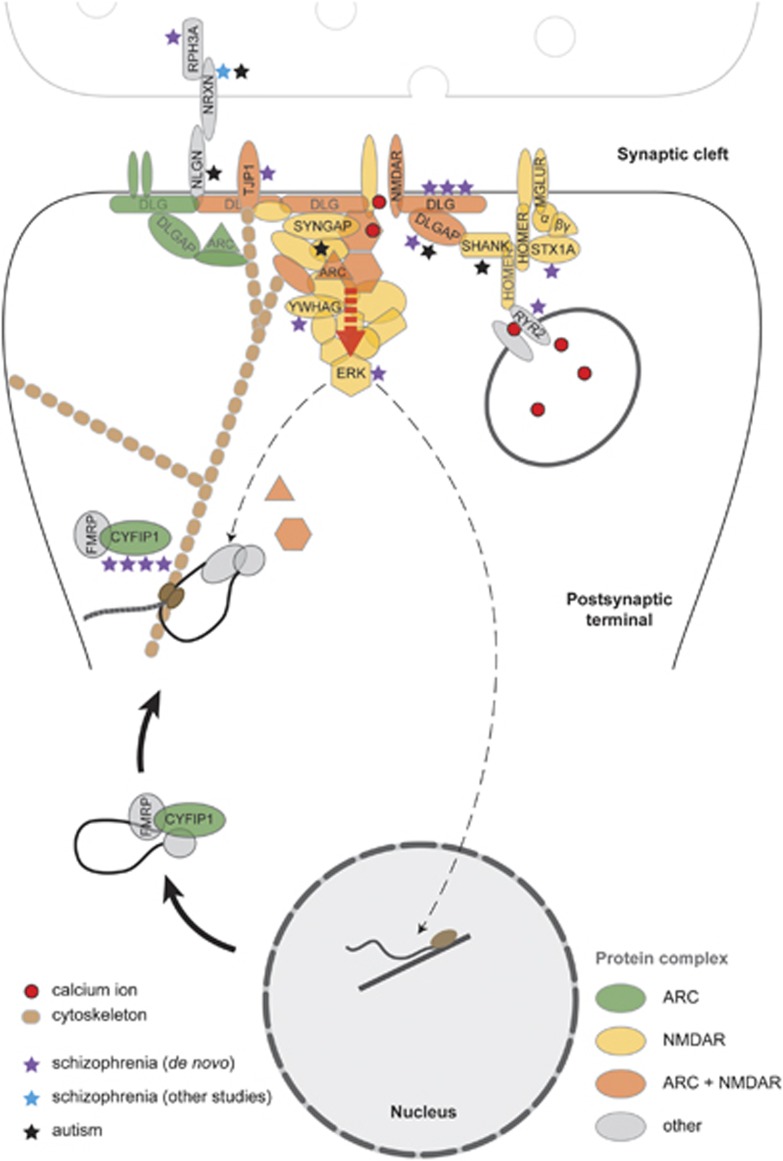
Aiming to localise more specifically the source of the PSD gene-set enrichment, we tested gene sets encoding PSD components (Table 2 and Figure 3). Activity-regulated cytoskeleton-associated protein (ARC; P=3.78 × 10−8), N-methyl-D-aspartate receptor (NMDAR; P=4.24 × 10−6) and PSD-95 complex (P=1.17 × 10−5) genes were highly significantly enriched among the de novos. However, conditional analyses revealed that the relatively small ARC and NMDAR sets explained both the PSD (conditional PPSD=0.231) and PSD-95 (conditional PPSD-95=0.603) enrichments but that the enrichments in ARC and NMDAR were partially independent of each other (conditional P=2.17 × 10−4 and P=0.019, respectively) (Supplementary Section 13). ARC and NMDAR sets also explained most of the enrichment for de novos in ‘the synapse', this GO category being only marginally enriched (P=0.017) after those genes were removed (Supplementary Section 11 and Supplementary Table S5). After removal of members of ARC and NMDAR sets, none of the subcategories comprising the synapse was significantly enriched except for ‘synaptic vesicle membrane' (P=4.22 × 10−4). These findings suggest enrichments in the PSD, and the great majority of that in the synapse GO gene set, is because of the enrichments in ARC and NMDAR, but that there is residual enrichment elsewhere in the synapse that is captured by ‘synaptic vesicle membrane' genes (GO: 30672).
Figure 3.
Disruption of postsynaptic signalling within activity-regulated cytoskeleton-associated protein (ARC) and N-methyl-D-aspartate receptor (NMDAR) complexes by copy number variants (CNVs). ARC and NMDAR bind to diverse structural and signalling molecules forming multiprotein complexes. Functional pathways encoded by these complexes are disrupted by de novo CNVs at multiple levels, as indicated by the purple asterisks (number of asterisks=number of de novos overlapping a gene or gene family). Calcium influx via the NMDAR, modulated by calcium release from internal stores (RYR2), drives downstream pathways whose association with the receptor is mediated by scaffold proteins (DLG1, DLG2, DLGAP1). Multiple pathways converge on ERK kinases (extracellular signal-regulated kinases), a focal point in the regulation of ARC transcription, dendritic localisation and local translation.47 ARC mRNA is transported to sites of synaptic activity in complexes containing CYFIP1, dissociation of which is required for ARC translation.49 CYFIP1 also regulates translation of CAMKII,49 a key component of NMDAR complexes. Although not identified in this study, deletions of synaptic adhesion protein NRXN1 (blue asterisk) have previously been found in schizophrenia.29 CNVs disrupting genes within these same functional pathways have also been identified in autism30(black asterisks).
To exclude unknown possible sources of confounding arising from the use of control CNVs, we also compared gene sets hit by de novo CNVs from cases with those hit in random assignments of gene-hitting CNVs of the same size, ensuring the probability of a gene being hit was proportional to its size. Again, we observed significant enrichment of PSD genes (P=0.0024), and a highly significant enrichment of the ARC (2.21 × 10−8) and NMDAR (2.95 × 10−4) complexes (Supplementary Section 12).
To exclude the possibility that our results reflect general properties of de novo CNVs, we compared case de novo CNVs with de novo CNVs identified in the control individuals from Iceland and from the Autism study by Sanders et al.25 Despite fewer control CNVs in these samples (N=59 and N=14), and therefore reduced power, the findings were consistent with our primary analysis in showing significant enrichment of ARC and NMDAR (Table 2) as were those of sensitivity analysis restricted to very large CNVs (>500 kb) (ARC P=1.27 × 10−4; NMDAR P=1.72 × 10−2).
Finally, we examined the ARC/NMDAR gene sets in the large case–control data sets. In this completely independent analysis, case CNVs were significantly enriched for members of the NMDAR (P=0.0015) but not ARC complexes (P=0.14). We note that in the de novo analysis, much of the additional signal for the ARC complex (over and above that of the NMDAR) comes from CNVs at 15q11.2 that span CYFIP1. Although there is strong published evidence for deletions at this locus being relevant to schizophrenia,3, 7 this locus was not significantly enriched in the MGS study,4 and was excluded by the filtering criteria adopted by the ISC,2 the two studies that combined comprise a large proportion of the case–control data set we use in this study.
Discussion
Aiming to identify novel candidate CNV loci for schizophrenia, and to illuminate aspects of the pathophysiology of the disorder through gene-set enrichment analyses, we have conducted the largest analysis of de novo CNVs in schizophrenia to date. Although not every observed case de novo CNV is likely to be pathogenic, the hypothesis that a substantial proportion of them are likely to be so is supported by several observations. First, eight of the de novos occurred at already known schizophrenia CNV loci (Table 1, marked with footnote ‘a'). Second, even after conservatively excluding those known loci, CNVs at the loci affected by case de novos occurred twice as frequently in cases in a meta-analysis of the largest available case–control CNV data sets. This elevation is much higher than the overall increase in CNV burden in cases in the large published studies.2, 4 Third, in the trios sample, the rate of de novo CNVs was more than twice that observed in other control samples (Supplementary Table S3), suggesting that at least 50% of the case de novos are relevant to the pathophysiology of schizophrenia.
Our estimate of the de novo rate is lower than initial reports in autism23 and schizophrenia24 but is comparable with more recent estimates in autism.25, 29, 30 Post hoc evaluation suggests it is unlikely that our filtering steps excluded large numbers of true de novo CNVs within our target size range. Of the 34 Agilent-validated de novos, 91% (N=31) had been rated (using MeZOD) as highly suggestive, whereas only 9% (N=3) had been called as ambiguous. Conversely, none of 33 putative de novos called by the Genotyping Console that were rejected by MeZOD were confirmed by Agilent.
It is notable that two previously documented schizophrenia loci, at 15q11.2 and 15q13.3, were each found more than once as de novos (Table 1). Two other loci were represented by two de novo CNVs each: EHMT1 (encoding Eu-HMTase1), a histone H3 Lys 9 (H3-K9) methyltransferase, and DLG2 (encoding discs, large homologue 2). Moreover, one de novo spanned each of the related genes DLG1 (whose orthologue in Drosophila is also dlg1) and DLGAP1 (encoding discs large associated protein 1).
At EHMT1 we observed a total of two de novos, three additional exonic CNVs in cases and one in a control. EHMT1 haploinsufficieny has been implicated as the cause of the 9q subtelomeric deletion syndrome (9qSTDS) characterised by moderate-to-severe mental retardation, childhood hypotonia and facial dysmorphisms, as well as a high prevalence of psychiatric symptoms in adulthood.31 A recent study has also reported strong evidence that deletions at EHMT1 are highly penetrant for phenotypes comprising developmental delay and a range of congenital anomalies.32 With this additional evidence for the involvement of this gene in neurodevelopmental phenotypes, our data point to EHMT1 as a schizophrenia susceptibility gene. Intriguingly, in Drosophila, ehmt coordinates epigenetic changes important in regulating cognition.33 Our findings at this locus thus suggest a role for epigenetic mechanisms in at least some cases of schizophrenia, and potentially point the way to novel therapeutic opportunities as the developmental effects of ehmt mutation on cognition are reversible.31
The DLG (discs large) family of membrane-associated guanylate kinases (MAGUKs), which were hit by multiple case de novo CNVs, are components of postsynaptic signalling complexes that are embedded within the larger group of over 1000 proteins that make up the PSD.34 They are associated with NMDA receptors and are highly concentrated in synapses. Remarkably, the orthologue of DLG2 in Drosophila (dlg1) is directly regulated by the orthologue of EHMT1 (emht also known as G9a).33 CNVs spanning DLG1 and DLG2 have been reported before in schizophrenia4, 6, 17 whereas other members of the family (DLG3 and DLGAP2) have been implicated in mental retardation35 and autism.30, 36 Together with our observation of multiple de novos spanning members of this family, and the nominally significant association of exonic CNVs at DLG2 in the case– control analysis, the findings strongly suggest that the CNVs we report in DLG-related genes are likely to be of pathogenic relevance to schizophrenia.
Although not strongly implicated by our study, a number of singleton de novo CNVs are also of note as they are at loci known to be associated with rare genomic disorders. The first is a deletion at 1q21.1 reported in TAR syndrome37 (which does not overlap the known 1q21.1 schizophrenia locus2, 7). We found the TAR region deleted in three more cases and only one control from the extended case–control samples. Again, deletions at this locus have very recently been strongly implicated in developmental delay.32 Another region is a duplication at the locus causing the 7q11.23 microduplication syndrome (which is deleted in Williams–Beuren Syndrome), the prominent features of which include autism and developmental delay.38 Duplications at the WBS region were found in three more cases and one control. Although this excess is not statistically significant (P=0.11, uncorrected), given duplications at this locus have also recently been identified increasing susceptibility for autism25 and developmental delay,32 it seems likely that the observations in the present study point to the involvement of this locus in schizophrenia as well. Further details about each of these loci are provided in Supplementary Section 6.
Given that our data suggest de novo CNVs are highly enriched for pathogenic loci, we sought evidence for convergence of de novo events onto specific biological pathways using a hypothesis-led, systems biology approach. Many of the CNVs robustly implicated in schizophrenia are also implicated in neurodevelopmental disorders in which cognitive impairment is common.39, 40, 41 Moreover, as discussed in the Introduction, it has been hypothesised that schizophrenia CNVs are enriched for genes encoding proteins associated with synaptic function. Our findings of apparent convergence of de novo CNVs onto genes encoding MAGUK proteins broadly support this synaptic hypothesis and suggested that more refined examination of synaptic genes is warranted.
Cognitive deficits are increasingly recognised as core features of schizophrenia, and it has long been known that antagonism of NMDA receptors at glutamatergic synapses can induce a schizophrenia-like psychosis that includes some of those deficits.42 This has led to a glutamate hypofunction hypothesis of schizophrenia. Glutamate receptors form multiprotein complexes with large sets of scaffold and signalling proteins including MAGUKs43 that are embedded in the PSD. It is clear that disruption of a number of synaptic proteins linked to glutamate receptor signalling alters cognitive function in rodents.44 The composition of the PSD has recently been identified in humans by some of the present authors,34 affording us an unprecedented opportunity to investigate the role of this complex in schizophrenia. Specifically, we tested the hypothesis that de novo CNVs in cases are enriched for genes encoding members of this complex.
We first compared the case de novos with a set of control CNVs drawn from the same population as the trios. Although those CNVs must have originally occurred as de novo mutations, predominantly transmitted CNVs clearly have different characteristics from the case de novos, most obviously size. Although our set-based analyses allow for size differences, to ensure our findings were robust to the control data set, we also compared gene sets hit by case de novo CNVs with those hit in random assignments of gene-hitting CNVs of the same size and obtained very similar gene-set enrichments. Finally, in order to exclude the possibility that our findings reflected general properties of de novo CNVs, we compared case de novo CNVs to two sets of control de novos, one drawn from the Icelandic population and the other from a much smaller sample of unaffected sibs of people with autism. Despite the wide disparities in the sources of the control CNVs, and the potential for different sources of bias, the results converge in pointing to the involvement of the synapse, the PSD, and more specifically, ARC and NMDAR complexes. Finally, and fully independent of those analyses, we show a significant enrichment for genes in the NMDAR complex in a meta-analysis of case–control data sets. We think it likely that the weaker finding for the NMDAR complex in the large case–control study compared with the relatively small de novo study, and the absence of association to ARC in the former, reflects the much lower power of the case–control design as a result of poorer enrichment for pathogenic CNVs.
This study adds to an accumulating body of evidence from human and animal genetic studies implicating disruption of synaptic processes in schizophrenia.45 By identifying an unprecedentedly large number of de novo CNVs in schizophrenia and demonstrating that these are likely to be highly enriched for pathogenic events, we have added substantially to the evidence implicating synaptic processes in schizophrenia. As well as implicating a set of functionally related synaptic proteins (EHMT1, DLG2, DLG1 and DLGAP1) we have identified a sufficient number of schizophrenia-enriched loci to identify potential points on convergence on specific synaptic complexes. Using gene sets that have been systematically annotated from individual, high-quality proteomic data sets and multiple analytic approaches carefully controlled for biases, we not only provide strong evidence for the importance of synaptic proteins, but also provide novel convergent support for the involvement of NMDAR, and to a lesser extent ARC protein, complexes in the aetiology and pathogenesis of the disorder, both of which are involved in NMDA signal transduction. NMDA receptor signalling regulates induction of multiple forms of synaptic plasticity,46 with local synthesis of ARC central to synaptic remodelling and the long-term maintenance of synaptic changes.47 Our finding that 12 out of the 34 case de novos impact on ARC and/or NMDAR complexes, supported by robust statistical analyses, suggest that disruption of NMDA signalling plays a key role in at least some cases of schizophrenia. As noted above, our findings do not exclude a role for mutations in other post- or pre-synaptic complexes, and given the close functional relationship between different synaptic components, we might expect pathology at a number of different points to play a role. Indeed, the robust association between NRXN1 deletions and schizophrenia48 points to presynaptic disruption in some cases, a hypothesis further supported by enrichment for case de novos in the GO category ‘synaptic vesicle membrane' after adjustment for ARC and NMDAR. Our findings delineate a circumscribed set of largely postsynaptic proteins and functions that warrant further functional analysis in model systems.
Acknowledgments
We acknowledge the families who participated in the study. Funding for recruitment was provided by the Janssen Research Foundation. Genotyping was funded by multiple grants to the Stanley Center for Psychiatric Research at the Broad Institute from the Stanley Medical Research Institute, The Merck Genome Research Foundation and the Herman Foundation. Work at Cardiff University was funded by Medical Research Council (MRC) Programme (Ref G0800509) and Centre Grants, the National Institutes of Mental Health (USA) (CONTE: 2 P50 MH066392-05A1) and a grant from the EU (EU-GEI). Genotyping and CNV analysis in the Icelandic trios by deCODE Genetics was funded in part by EU grants: HEALTH-2007-2.2.1-10-223423 (Project PsychCNV), PIAG-GA-2008-218251 (Project PsychGene) and IMI-JU-NewMeds. SGNG, ÀB, EF, LNVL, JC, NHK and MOC were supported by the Genes to Cognition Program funded by the Wellcome Trust, MRC Project (Ref G06706B), EU grants: (Project GENCODYS Nr 241995, Project EUROSPIN No. 242498 and Project SYNSYS No. 242167). ÀB is supported by EMBO and the European Commission. We acknowledge Edward Scolnick for helpful discussions.
The following psychiatrists in Bulgaria recruited families for the project:
District Hospital Blagoevgrad: Nikolay Ianakiev, Radoi Milev, Valentina Kovachka.
University Hospital Sofia: Vihra Milanova, Georgy Koichev, Stefan Krastev, Georgi Onchev, Elitza Djatova, Tsveteslava Galabova, Desislava Ivanova, Ivan Gerdjikov, Emil Kaludiev, Christian Kostov, Ivanka Krastnikova, Irina Lazarova, Nedialko Mihailov, Maria Nikolova, Iliana Pristavova, Kalina Shoilekova, Anton Slavchev, Vladimir Sotirov, Svetla Stambolova, Miroslava Stamenova, Vessela Stoianova, Milena Strashimirova, Snezhina Tacheva, Todor Todorov, Rositza Vladimirova.
Plovdiv University Hospital: Marin Roglev, Inko Kehaiov, Nadezhda Poryazova, Rayna Tsvetkova, Petia Blagoeva, Veselina Vlahova.
Radnevo Psychiatric Hospital: Todor Tolev, Kalinka Drebova, Penka Grozeva, Ivan Ivanov, Mariya Nikolova, Maya Pashova, Radka Patricheva, Minka Sokolovska, Antoniya Terzieva, Donka Terzieva, Vania Tesheva, Ilina Topcheva, Vladimir Trynov.
Psychiatric Hospital, Russe: Temenujka Mateva, Biliana Mechkunova, Plamen Panayotov, Rositsa Vacheva.
Psychiatric Dispensary, Sofia: Lubomir Jivkov, Svetlinka Koleva, Kalina Georgieva, Tatiana Kanova, Anna Maikova, Krasimira Marcheva, Vesselin Sekoulov, Diana Shkodrova, Vesselina Slavcheva, Tenko Tenev, Lora Tsoneva.
DNA handling and extraction was performed at the Department of Medical Genetics, University Hospital ‘Maichin Dom', Sofia (Head of Department Professor Draga Toncheva): Lyudmila Georgieva, Albena Dimitrova and Irina Zaharieva.
Author Contributions
GK, MJO, MCO'D, PS and SP designed the study. GK, MJO and MCO'D drafted the primary manuscript along with AJP, PH and SG. The primary genotyping analysis with Affymetrix arrays was organised and coordinated by SP, PS, JM and KC. MI, GK, DR, SP, MF, DI and RW analysed the raw intensity data. CNV analysis was performed by GK, DI, MF, ER and DR. MF and GK designed the Agilent custom arrays. DG, MI, LG and GK performed the Agilent validation experiments. HS, KS, PIO and YB were responsible for analysis of Icelandic trios. Synapse proteomic data sets were prepared by LNVL, ÀB, EF, MOC, JC and SGNG. AJP and SGNG identified the curated sets of genes for pathway analyses. PH and AJP analysed the involvement of gene sets in de novo CNVs. SP undertook an independent parallel analysis. GK and MJO planned and organised the recruitment of families, which was coordinated by GK, IN and DT. DI wrote the scripts used for de novo analysis, including the z-score method. All authors discussed the results, their interpretation and contributed to the final manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Owen MJ, Craddock N, O'Donovan MC. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch Gen Psychiatry. 2010;67:667–673. doi: 10.1001/archgenpsychiatry.2010.69. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium (ISC) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large CNVs in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OPH, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou H-H, Peoples A, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingason A, Kirov G, Giegling I, Hansen T, Isles AR, Jakobsen KD, et al. Maternally derived microduplications at 15q11-q13: implication of imprinted genes in psychotic illness. Am J Psychiatry. 2011;168:408–417. doi: 10.1176/appi.ajp.2010.09111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OPH, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophr Bull. 2009;35:851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Tam GWC, Redon R, Carter NP, Grant SGN. The role of DNA copy number variation in schizophrenia. Biol Psychiatry. 2009;66:1005–1012. doi: 10.1016/j.biopsych.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C, et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci USA. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Korn JM, McCarroll SA, Consortium TIS, Altshuler D, Sklar P, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function PLoS Genet 20106pii:e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy H, Stahl D, MacCabe JH. A systematic review and meta-analysis of the fertility of patients with schizophrenia and their unaffected relatives. Acta Psychiat Scand. 2010;123:98–106. doi: 10.1111/j.1600-0447.2010.01623.x. [DOI] [PubMed] [Google Scholar]
- Laursen TM, Munk-Olsen T. Reproductive patterns in psychotic patients. Schizophr Res. 2010;121:234–240. doi: 10.1016/j.schres.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Rees E, Moskvina V, Owen MJ, O'Donovan MC, Kirov G.De novo rates and selection of schizophrenia-associated copy number variants Biol Psychiatryadvance online publication, 18 August 2011[e-pub ahead of print]. [DOI] [PubMed]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Sanders Stephan J, Ercan-Sencicek AG, Hus V, Luo R, Murtha Michael T, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kirov G, Kinoshita Y, Yamanouchi Y, Kitajima T, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotech. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietiläinen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, Foulds N, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Kochinke K, Oortveld MAW, Marks H, Kramer D, de Jong EK, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayés A, van de Lagemaat LN, Collins MO, Croning MDR, Whittle IR, Choudhary JS, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:318–324. doi: 10.1086/422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca A-L, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Schulze H, Strau G, Ott C-E, Hall J, Trotier F, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, et al. Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet. 2009;52:94–100. doi: 10.1016/j.ejmg.2009.02.006. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Saab WM, D'Souza DC, Moghaddam B, Krystal JH. The NMDA antagonist model for schizophrenia: promise and pitfalls. Pharmacopsychiatry. 1998;31 (S 2:104–109. doi: 10.1055/s-2007-979354. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SGN. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Grant SG, Marshall MC, Page KL, Cumiskey MA, Armstrong JD. Synapse proteomics of multiprotein complexes: en route from genes to nervous system diseases. Hum Mol Genet. 2005;14:R225–R234. doi: 10.1093/hmg/ddi330. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ. The genetics of neurodevelopmental disease. Cur Opin Neurobiol. 2011;21:197–203. doi: 10.1016/j.conb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA. NMDA-receptor-dependent synaptic plasticity: multiple forms and mechanisms. Trends Neurosci. 1993;16:521–527. doi: 10.1016/0166-2236(93)90197-t. [DOI] [PubMed] [Google Scholar]
- Bramham C, Alme M, Bittins M, Kuipers S, Nair R, Pai B, et al. The Arc of synaptic memory. Exp Brain Res. 2010;200:125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G. The role of copy number variation in schizophrenia. Exp Rev Neurotherapeutics. 2010;10:25–32. doi: 10.1586/ern.09.133. [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.