Abstract
Objectives
To assess if genetic variation in the PACE4 gene, PCSK6, influences the risk for symptomatic knee OA.
Methods
Ten PCSK6 single nucleotide polymorphisms (SNP) were tested for association in a discovery cohort of radiographic knee OA (n= 156 asymptomatic and 600 symptomatic cases). Meta-analysis of the minor allele at rs900414 was performed in three additional independent cohorts (total n=674 asymptomatic and 2068 symptomatic). Pcsk6 knockout (KO) mice and wildtype C57BL/6 mice were compared in a battery of algesiometric assays, including hypersensitivity in response to intraplantar substance P; pain behaviours in response to intrathecal substance P; and pain behaviour in the abdominal constriction test.
Results
In the discovery cohort of radiographic knee OA, an intronic SNP at rs900414 was significantly associated with symptomatic OA. Replication in three additional cohorts confirmed that the minor allele at rs900414 was consistently increased among asymptomatic compared to symptomatic radiographic knee OA cases in all four cohorts. A fixed-effects meta-analysis yielded an odds ratio =1.35 (95% CI 1.17, 1.56; p-value 4.3×10−5 and no significant between-study heterogeneity). Studies in mice revealed that Pcsk6 knockout (KO) mice were significantly protected against pain in a battery of algesiometric assays.
Conclusions
These results suggest that a variant in PCSK6 is strongly associated with protection against pain in knee OA, offering some insight as to why in the presence of the same structural damage, some individuals develop chronic pain and others are protected. Studies in Pcsk6 null mutant mice further implicate PACE4 in pain.
Keywords: Knee osteoarthritis, pain, PACE4, genetic association, SNP
INTRODUCTION
Osteoarthritis (OA) is the most common joint disorder, affecting primarily the knees, hips, hands, and spine [1]. Joint damage in OA includes cartilage erosion, subchondral bone sclerosis, bone remodelling with osteophyte formation, and episodes of synovitis. Diagnosis is based on radiographical changes - joint space narrowing, subchondral bone sclerosis and osteophytes - accompanied by clinical symptoms, most prominently pain. A recent American College of Rheumatology task force concluded that pain is the most common symptom of patients with rheumatic disorders, including OA [2]. Pain is the major reason for seeking medical care for OA, and knee OA continues to be the leading cause of impaired mobility in the elderly in the United States [3].
As in most chronic pain conditions, the correlation between the extent of tissue damage and symptoms appears weak [4, 5]. Particularly for knee OA, multiple studies have examined the association between radiographic severity and pain, concluding that the correlation is low [6]. It is well documented that people with severe radiographic OA may have no pain at all, while conversely, people suffering from OA pain may have no detectable structural joint changes [7–9]. This statement is flawed by current limitations in detecting and quantifying structural changes in different joint tissues, as well as in assessing spontaneous pain and pain-dependent measures in subjects with OA. Recent studies are finding a relationship between specific radiographic changes and pain experienced by patients [10, 11]. Nevertheless, the correlation remains unimpressive, especially when the influence of psychological and social factors and co-morbidity are taken into account [12, 13], suggesting that the molecular pathways that mediate joint structural changes and those that lead to chronic pain associated with those changes diverge as the pathology progresses.
Heritability studies in OA have focused almost exclusively on the genetic contribution to the risk for radiographic OA of the knee, hip or hands [14–16]. Recent years have seen an explosion of candidate gene and genome-wide association studies that have identified susceptibility genes for radiographic OA [17]. In contrast, there has been a paucity of studies on heritability and genetic determinants of symptomatic versus asymptomatic OA, although it has been established that robust inter-individual variation in the susceptibility to pain exists, and this is partially genetically determined [18, 19]. A handful of pain-relevant genetic variants have been provisionally identified [18, 19], and some candidate “pain genes” are currently being analyzed in symptomatic versus asymptomatic OA cohorts (see discussion).
We recently reported a role for the serine protease, PACE4 (paired amino acid converting enzyme 4), in the pathogenesis of OA [20, 21]. PACE4 belongs to the family of proprotein convertases that process latent precursor proteins into biologically active products through cleavage at paired basic amino acid processing sites, which is essential for proteolytic maturation of a wide variety of proteins, including growth factors, hormones, neuropeptides, and zymogens [22]. PACE4 removes the prodomain of the proaggrecanases, pro-ADAMTS-4 and pro-ADAMTS-5, in OA cartilage, thus activating these proteases. Active ADAMTS-4 and ADAMTS-5 are responsible for proteolytic degradation of the major cartilage macromolecule, aggrecan, which is a key pathological event in OA [23]. This suggests that PACE4 represents a novel target for the development of OA therapeutics [20]. To assess whether PACE4 may also play a direct role in OA pain, we undertook analysis of genetic variation in the gene that encodes PACE4, PCSK6. The aim of the current study was two-fold: first, to assess if genetic variants in the PCSK6 gene are implicated in the risk for symptomatic knee OA; and second, based on results of the human genetic assessment, to explore a role for PACE4 in pain sensitivity, using Pcsk6 null mutant mice.
METHODS
Discovery Cohort
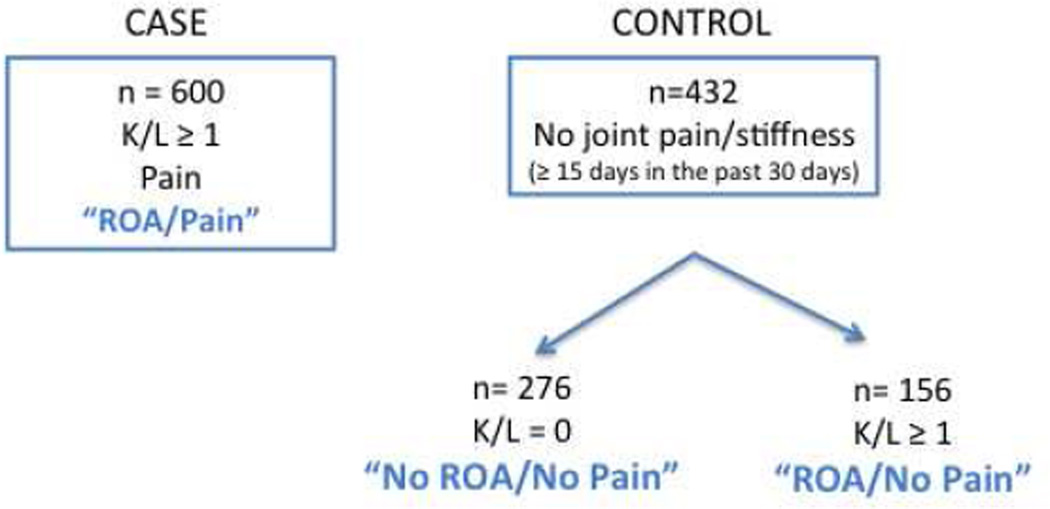
The Discovery Cohort consisted of 600 OA cases and 432 controls, all Caucasian subjects. OA cases were derived from three Pfizer-sponsored phase II/III clinical trials in the US and the UK, with radiologically confirmed painful radiographic knee OA (designated “ROA/Pain”). The entire control cohort (n= 432) was collected from a single site in Canada, enrolling asymptomatic controls. Of those 432 asymptomatic subjects, 276 were radiographically negative in both knees, K/L = 0 (designated “No ROA/No Pain”) and 156 showed radiographic evidence of knee OA (K/L ≥ 1) in the absence of pain (designated “ROA/No Pain”) (Fig. 1). Details on X-ray findings and pain assessment are listed in the online supplementary section. Descriptive characteristics of the Discovery Cohort are in Table 1A.
Figure 1.
Schematic representation of the Discovery Study layout.
Table 1A.
Descriptive characteristics of study subjects included in the discovery study
| Symptomatic knee OA* | n=600 | “ROA/Pain” |
| Age, years mean (SD) | 60.5 (8.3) | |
| Gender, % female | 61.7% | |
| BMI kg/m2 mean (SD) | 32.4 (7.2) | |
| WOMAC mean (SD) | 9.2 (4.1) | |
| Asymptomatic knee OA* | n= 156 | “ROA/No Pain” |
| Age, years mean (SD) | 61.3 (5.5) | |
| Gender, % female | 57.7% | |
| BMI kg/m2 mean (SD) | 27.3 (4.4) | |
| WOMAC mean (SD) | NA | |
| No radiographic knee OA | n= 276 | “No ROA/No Pain” |
| No pain | ||
| Age, years mean (SD) | 59.2 (4.2) | |
| Gender, % female | 77.9% | |
| BMI kg/m2 mean (SD) | 25.1 (3.7) | |
| WOMAC mean (SD) | NA |
| Table 1B. Descriptive characteristics of study subjects included in the meta-analysis | ||||
|---|---|---|---|---|
| Discovery | GOAL | COS | Chingford | |
| Symptomatic knee OA** | n=352 | n=1569 | n=85 | n=62 |
| Age, years mean (SD) | 62.8 (9.0) | 68.4 (7.2) | 65.6 (8.3) | 65.7(5.8) |
| Gender, % female | 61.4% | 49.1% | 66.7% | 100% |
| BMI kg/m2 mean (SD) | 32.7(6.8) | 30.8 (5.4) | 29.1 (5.1) | 26.7 (3.4) |
| WOMAC mean (SD) | 10(3.1) | 5.3 (4.7) | N/A | N/A |
| CC | 6.3% | 7.5% | 7.6% | 9.7% |
| CT | 40.9% | 42.0% | 42.4% | 37.1% |
| TT | 52.8% | 50.5% | 50.0% | 53.2% |
| Asymptomatic knee OA** | n= 127 | n=298 | n=92 | n=157 |
| Age, years mean (SD) | 61.4(5.7) | 67.7 (7.3) | 64.0 (9.8) | 65.7(5.9) |
| Gender, % female | 59.8% | 43.0% | 54.6% | 100% |
| BMI kg/m2 mean (SD) | 27.5 (4.6) | 28.5 (4.9) | 26.1 (4.7) | 26.2 (4.4) |
| WOMAC mean (SD) | N/A | 0.6(1.8) | N/A | N/A |
| CC | 21.3% | 9.6% | 12.0% | 12.3% |
| CT | 35.4% | 47.4% | 48.2% | 35.2% |
| TT | 43.3% | 43.0% | 39.8% | 52.5% |
| No radiographic knee OA no pain | n=296 | n=743 | N/A | n=527 |
| Age, years mean (SD) | 59.3 (4.2) | 67.7 (7.3) | N/A | 62.9 (5.7) |
| Gender, % female | 76.0% | 43.0% | 100% | |
| BMI kg/m2 mean (SD) | 25.2 (3.7) | 28.5 (4.9) | N/A | 24.7 (3.6) |
| WOMAC mean (SD) | N/A | 0.6(1.8) | N/A | N/A |
| CC | 14.6% | 7.3% | N/A | 9.3% |
| CT | 42.3% | 39.8% | N/A | 41.0% |
| TT | 43.1% | 52.9% | N/A | 49.7% |
radiographic knee OA defined as K/L ≥ 1
radiographic knee OA defined as K/L ≥ 2
Replication Cohorts
Four independent study cohorts were included in the meta-analysis: (1) The Discovery Cohort; (2) The Genetics of Osteoarthritis and Lifestyle (GOAL) Study; (3) The Clearwater Osteoarthritis Study (COS); (4) The Chingford Study. For consistency between cohorts, the radiographic definition of knee OA was K/L grade ≥2. Details on each of the study cohorts, X-ray findings, and pain assessment are listed in the online supplementary section. Descriptive characteristics of all 4 cohorts are summarized in Table 1B.
Laboratory Methods
Genomic DNA was extracted from peripheral blood (Discovery Cohort, GOAL and Chingford) or from buccal swabs (COS) using standard protocols. For the Discovery Cohort, SNPs were genotyped using the Taqman assay with supplied or custom probes and primers (Table S1) (Applied Biosystems, CA USA). Reactions were analyzed using a 7900HT Sequence Detection System. DNA samples from all subjects in the Discovery Cohort were genotyped on the same day and randomly placed across plates. A subset was tested by sequencing to ascertain that the genotype data were accurate. Genotyping of the rs900414 SNP in the replication cohorts was carried out by Kbioscience Ltd, Hertfordshire UK (Table 1B), by using the KASPar chemistry, a competitive allele-specific PCR SNP genotyping system. The polymorphism was in Hardy-Weinberg equilibrium in controls in all three additional cohorts (p>0.05).
Statistical Analysis
Association between SNPs and knee OA was initially assessed in the Discovery Cohort by logistic regression models using two different kinds of binary phenotypic variables"Pain” and “Radiographic OA”. The “Pain” variable was defined as “symptomatic OA (K/L≥1 with pain, designated “ROA/Pain”, n=600)” vs. “asymptomatic OA (K/L≥1 without pain, designated “ROA/No Pain” n=156)”. The “Radiographic OA” variable was defined as symptomatic OA (“ROA/Pain”, n=600)” vs. “K/L=0 and no pain, designated “No ROA/No Pain”, (n=276)” (Fig.1). In the logistic regression models, genotype at rs900414 was treated as a 3-level categorical variable. Two covariates, sex and BMI, were included. For the meta-analysis, radiographic OA was defined as K/L ≥ in the Discovery Cohort, to be consistent with other cohorts. Also for consistency, an additive mode of inheritance was assumed for rs900414 and this variable was treated as a numeric variable with values 0, 1 and 2. In addition, two more covariates, age and K/L grade, were included in the logistic regression models. The natural logarithm of the odds ratio (OR) and its standard error were estimated independently within each study adjusted for age, gender, BMI and, in the case of asymptomatic versus symptomatic cases, for K/L grade (the maximum for radiographic OA cases, the pre-surgical grade for the index knee in TKR cases). Meta-analyses of data from these different cohorts were performed using R version 2.10.1 (The R Foundation for Statistical Computing; http://www.rproject.org). Heterogeneity was evaluated using the variance for heterogeneity τ2 and Cochran’s Q test. Random effects models were not used as no significant heterogeneity was seen for any of the analyses (τ2<0.02, p>0.20).
Mice
Subjects were naïve, adult (2–6 month old) C57BL/6J mice of both sexes [24], either wild type (WT) or Pcsk6 null mutant (knockout; KO) mice, maintained on a fully congenic C57BL/6 background. Pcsk6 KO mice were obtained from Dr. Daniel Constam (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) on a hybrid 129S6×C57BL/6J background [25] and backcrossed onto C57BL/6 for at least 10 generations. Mice were maintained in a controlled environment as described [26]. They were assessed for their sensitivity on a battery of acute/tonic nociceptive assays similar to that described in [26], including:
Mechanical Allodynia in response to intraplantar administration of substance P
Mice were placed in Plexiglas cubicles (5 cm wide × 8.5 cm long × 6 cm high) on a metal grid floor above an automated von Frey apparatus (Ugo Basile, Italy). Activation of the apparatus initiated the extension of a metal probe to contact the plantar surface of the hind paw. The computer was programmed to apply a 10-s ramp to a maximum force of 20 g. Withdrawal of the hind paw from the probe caused a retraction of the probe and displayed the threshold force. Three baseline withdrawal thresholds per hind paw were measured. Mice were then given an intraplantar injection (20 µl volume) of substance P (2.5, 5 or 10 µg) (Sigma, St. Louis MO) into the right hind paw. Post-injection withdrawal thresholds were measured at 5-min intervals for 20 min. For the purposes of constructing dose-response curves, percentages of maximum possible allodynia were calculated by comparing total allodynia displayed by each mouse over the 20-min testing period (using the method of trapezoids) to that of a subject with the same average baseline and maximum postinjection responses (i.e., withdrawal thresholds of 0 g) at every time point. Using this dependent measure, half-maximal effective doses (ED50 values) and 95% confidence intervals (CIs) were calculated using the method of Tallarida and Murray [27], implemented by FlashCalc 40.1® software (M. Ossipov, University of Arizona). Statistical significance at p<0.05 was established via non-overlapping 95% CIs.
Thermal Hyperalgesia in response to intraplantar administration of substance P
Thermal hyperalgesia in response to intraplantar substance P was assessed as described [28, 29].
Spontaneous pain behavior in response to intrathecal administration of substance P
Mice were placed in a clear Plexiglas cylinder (15 cm diameter×30 cm high) for observation. After habituation, mice were lightly restrained in cloth and given intrathecal injections of substance P (5, 15 or 45 ng/mouse in 5 µl saline) with a 25 µl Hamilton microsyringe and a 30-gauge needle. This triggers a caudally directed scratching, biting and licking response [30]. Immediately following intrathecal injection, mice were returned to their observation cylinders. Directed activity toward either hindpaw (scratching, biting or licking) was counted for the next 60 s. Data were analyzed by two-way ANOVA (genotype×dose).
Abdominal Constriction Test
Writhing in response to intraperitoneal acetic acid was measured as previously described [31].
RESULTS
Discovery Study
The Discovery Cohort was used to test 10 SNPs that were roughly equally spaced across the entire PCSK6 gene for association with OA and pain using logistic regression adjusting for sex and BMI. The genetic analysis was performed using radiographic OA (K/L ≥1) without pain and with pain as separate phenotypes. The strongest association was detected at SNP rs900414 with the dichotomized phenotype of “ROA/Pain” (n= 600) vs. “ROA/No Pain” (n=156), with an unadjusted p-value of 0.0002 (Table 2) and a p-value adjusted for multiple testing of 0.008. The adjusted p-value was obtained through permutation testing. An unadjusted p-value of 0.025 was obtained at this same SNP for the association with the phenotype of “ROA/Pain” (n=600) vs. “No ROA/No pain” (n = 276), but this association was not significant after adjusting for multiple testing. Due to the limited LD across this region, the % of variation captured with these SNPs is very low (<10%). In order to get a better definition of LD around 900414, an additional 6 SNPs surrounding rs900414 were selected to further refine the association, and rs900414 remained the most significant (Table S2). Genotype numbers for rs900414 by cases and controls in the Discovery Cohort are shown Table S3. For the risk of experiencing pain in the presence of radiographic knee OA, the OR between genotype T/T and C/C at rs900414 was 3.12 with a 95% confidence interval of (1.75, 5.59), and the OR between genotype T/C and C/C was 3.48 with a 95% confidence interval of (1.90, 6.36) (Table S3). This marker was out of Hardy-Weinberg Equilibrium (HWE) in the “ROA/No pain” group only (p=0.001) while maintaining HWE in both the “ROA/Pain” and “No ROA/No Pain” populations, which may further corroborate that SNP rs900414 is associated with OA pain, because this “ROA/No Pain” group was selected for subjects with radiographic knee OA who had no pain. Moreover, we found that this SNP was not in strong LD (r2<0.4) with any other SNP nearby (Fig.S1).
Table 2.
PACE4 SNPs in the Discovery Cohort. Observed Minor Allele Frequencies (MAF)
| PACE4 LocusLocus | Minor (Major) Allele | "ROA/Pain" | "ROA/No Pair" | p-value (unadj) | "No ROA/No Pain" | p-value (unadj) |
|---|---|---|---|---|---|---|
| MAF | MAF | ROA/Pain vs ROA/No Pain | MAF | ROA/Pain vs No ROA/No Pain | ||
| Rs7182915 | C(G) | 21.36% | 22.73% | 0.1883 | 21.98% | 0.2218 |
| Rs4965878 | G(A) | 14.40% | 14.74% | 0.8751 | 15.07% | 0.2049 |
| Rs12902244 | G(A) | 10.12% | 10.65% | 0.7523 | 13.10% | 0.5120 |
| Rs1532364 | T(C) | 24.29% | 22.08% | 0.1492 | 19.49% | 0.1578 |
| Rs4246334 | G(A) | 28.47% | 34.00% | 0.3706 | 27.44% | 0.5298 |
| Rs20543 | T(C) | 37.50% | 35.62% | 0.7636 | 37.78% | 0.6455 |
| Rs4965831 | A(G) | 45.03% | 43.55% | 0.9613 | 45.57% | 0.3937 |
| Rs11630012 | A(C) | 18.29% | 17.74% | 0.9500 | 18.98% | 0.7383 |
| Rs900414 | C(T) | 29.12% | 38.14% | 0.0002 | 35.77% | 0.0249 |
| Rs2073595 | A(G) | 17.58% | 20.97% | 0.4126 | 21.59% | 0.4759 |
Meta-analyses
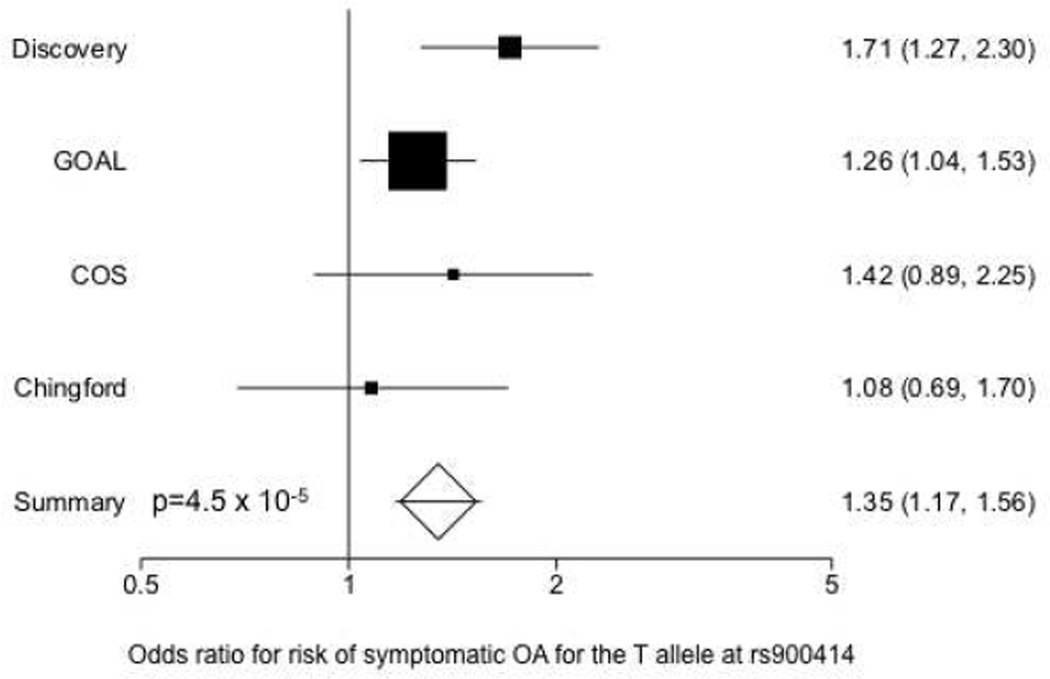
We assessed whether the association of rs900414 with asymptomatic versus symptomatic knee OA could be replicated in three more independent cohorts. In a meta-analysis of all four cohorts, radiographic knee OA was defined as K/L ≥ 2 with or without symptoms. In all four cohorts, the minor allele at rs900414 was significantly associated with symptomatic OA and consistently increased among asymptomatic ROA cases, compared to symptomatic knee OA (Figure 2). An unadjusted fixed-effects meta-analysis yielded a per allele OR=1.35 with a 95% CI (1.17, 1.56) and a p-value of 4.5 × 10−5. The heterogeneity variance was τ2=0.007 (p=0.28). When adjusted for age, sex, BMI and K/L grade, the OR remained 1.33, with a 95% CI (1.12, 1.57), a p-value= 0.00093, and a heterogeneity variance τ2=0.016 (p= 0.222). When analysed separately, the association was significant both in K/L=2 and K/L ≥ 3 radiographic knee OA (not shown). In order to ensure that the high OR in the discovery cohort was not biasing the effect size in the meta-analysis, a separate analysis was performed on the three replication cohorts combined. Meta-analysis of the three replication cohorts alone was still statistically significant, although smaller, with an OR=1.26 (95% CI 1.06,1.48; p=0.0065) for the unadjusted analysis and OR=1.21 (95% CI 1.00,1.46; p=0.0488) for the analysis adjusted for age, sex and BMI.
Figure 2.
Forest plot of fixed-effect OR estimates and 95% CI for the association between the T allele at rs900414 of the PCSK6 gene and the risk of symptomatic versus asymptomatic knee osteoarthritis.
Algesiometric assays in WT and Pcsk6 KO mice
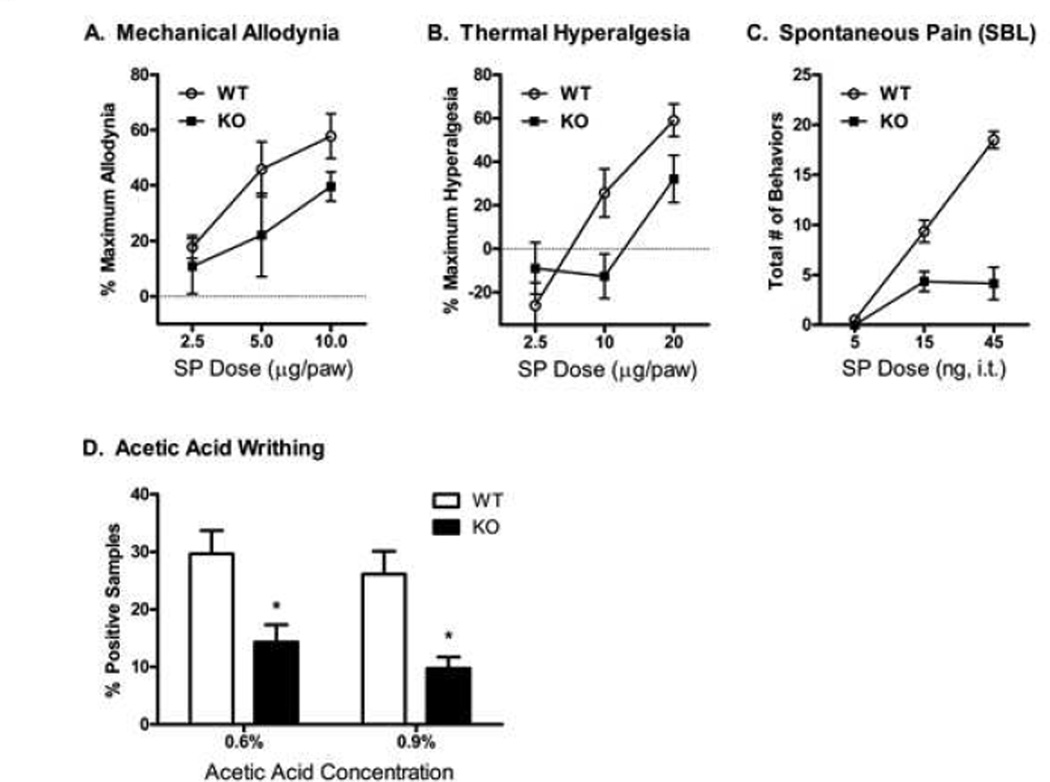
Based on the observed genetic association of the PCSK6 gene with the absence or presence of pain in knee OA, we tested the hypothesis that PACE4 may influence pain sensitivity, comparing WT and Pcsk6 KO mice on a battery of acute/tonic nociceptive assays [26]. We present those assays displaying a significant KO phenotype in Fig. 3; all other data are in Table S4. Intraplantar injection of substance P resulted in a dose-dependent induction of mechanical allodynia in WT mice (Fig. 3A) (ED50: 7.0 µg; 95% CI: 4.7 – 10.3 µg), while Pcsk6 KO mice were significantly protected (ED50: 17.4 µg; 95% CI: 11.2 – 82 µg). Similarly, substance P induced a dosedependent thermal hyperalgesia (Fig. 3B) in WT mice (ED50: 16.8 µg; 95% CI: 11.7 – 24.1 µg), whereas Pcsk6 KO mice were significantly protected (ED50: 195 µg; 95% CI: 35.0 – >1000 µg). For both mechanical and thermal allodynia, no changes from baseline were noted in the contralateral hind paw (not shown). Intrathecal injection of substance P resulted in a dose-dependent induction of spontaneous pain behaviors (scratching, biting and licking; Fig. 3C) in WT but not in KO mice (genotype×dose: F2,56 = 16.9 p<0.001). Finally, KO mice showed significantly less pain behavior than WT mice in the abdominal constriction test (Fig. 3D), both at 0.6% and at 0.9% acetic acid (both p<0.05).
Figure 3.
Algesiometric assays in wild type and Pcsk6 null mutant C57BL/6 mice. (A) Mechanical allodynia after intraplantar injection of substance P in WT mice (ED50: 7.0 µg; 95% CI: 4.7 – 10.3 µg) and Pcsk6 null mice (ED50: 17.4 µg; 95% CI: 11.2 – 82 µg); (B) Thermal hyperalgesia in response to intraplantar substance P in WT mice (ED50: 16.8 µg; 95% CI: 11.7 – 24.1 µg) and Pcsk6 KO mice (ED50: 195 µg; 95% CI: 35.0 – >1000 µg). ; (C) Spontaneous pain behaviours, including scratching, biting, and licking, in response to intrathecal substance P; (D) Writhing in response to intraperitoneal injection of acetic acid. Data are represented as mean ± SEM.
DISCUSSION
Twin study-derived heritability estimates ranging from 35% to 65% have been reported for knee pain [32], low back pain and neck pain [33]. The presence or absence of chronic pain in patients with joint damage is likely to be partly determined by a combination of genetic variations. Currently, however, only three reports in the literature describe SNPs that are differentially associated with symptomatic versus asymptomatic OA. In one study, van Meurs et al. [34] describe a functional variant in the catechol-O-methyltransferase (COMT) gene associated with hip pain in women with radiographic hip OA. This SNP (V158M) results in lower activity of COMT, and had been previously associated with increased pain sensitivity in other chronic musculoskeletal pain states, including fibromyalgia [35]. The association of this COMT variant with painful hip OA was only present in women, and is awaiting replication. In another data set, Reimann et al. [36] identified a SNP in the SCN9A gene, which encodes the α subunit of the Nav1.7 voltage-gated sodium channel, a protein that has been strongly implicated in pain [37]. The SNP confers an R1150W amino acid substitution that was associated with altered pain thresholds in several diseases, including higher pain scores in OA [36]. This association failed to replicate when tested in three independent cohorts with knee OA, although the high pain variant was significantly associated with increased risk of chronic widespread pain [38]. Finally, it was recently reported that the Ile585Val variant in the gene encoding transient receptor potential cation channel, subfamily V, member 1 (TRPV1), a genotype that is generally associated with lower pain sensitivity, is significantly associated with a decreased risk of painful knee OA [39].
Given the number of false positive genetic association reports, replication of genetic studies is extremely important [17]. Meta-analysis of results from different cohorts provides a quantitative approach for combining results of various studies to estimate the true genetic risk conferred by any variant in human disease [40]. The current study suggests that the rs900414 SNP in PCSK6 is strongly associated with protection against pain in knee OA, K/L ≥ 2, with an OR= 1.33, a 95% CI (1.16, 1.52) and a p-value of 3.52 × 10−5. The overall sample size was relatively small, particularly for asymptomatic cases, but the association is stronger than any of the hitherto reported associations of known pain genes with OA symptoms, and was replicated in four independent knee OA cohorts. An important limitation to this kind of study is that “asymptomatic” is defined as absence of symptomatic OA, which in 2 of the 4 cohorts is defined as “joint pain on more than 50% of the month”. While this is a generally accepted inclusion criterion for symptomatic OA, it means that there may pain in these asymptomatic subjects, and going forward, studying genetic variation in large cohorts should aim at carefully describing different levels of pain.
PCSK6 is upstream from CHSY1, the gene that encodes chondroitin sulphate, which is commonly used in OA for its anti-inflammatory properties. Therefore the extent of LD between the SNPs tested in our study, and in particular rs900414, was assessed by HapMap, and was found not to extend to CHSY1.
PACE4 is broadly expressed, notably in the liver and central nervous system – especially the pituitary, cerebellum, and spinal cord [41–43] (and unpublished observations in humans). The human PCSK6 gene, located on chromosome 15q26.3 (chromosome 7 in mice), spans at least 250 kb and is distributed over 25 exons, with strikingly large introns, making it the largest gene in this family [44]. Different splice variants of PACE4 have been identified, with different distribution [45]. For example, the full length PACE4A is encoded by a cDNA that lacks exon 23, resulting in a secreted form of the enzyme, while the cDNA containing exon 23 encodes the intracellular PACE4E isoform. Exon 23 encodes a hydrophobic cluster that causes retardation of PACE4 secretion [46]. The rs900414 SNP is located in the intron between exons 22 and 23, and might thus be involved in controlling splice variation. Despite the lack of an obvious functional correlate, we decided to investigate pain in Pcsk6 KO mice. No genotype differences were observed in any acute thermal or mechanical assays (Table S4), except for a marginal difference on the manual von Frey test. The use of tonic, chemical/inflammatory assays revealed genotype differences in the abdominal constriction and substance P assays, but not in the formalin and capsaicin tests. The transduction mechanisms in the latter tests involve TRPA1 and TRPV1 channels, respectively [47–49], and the lack of a null mutant phenotype in these tests would seem to preclude a direct interaction between PACE4 and these receptors. On the other hand, findings may suggest a direct interaction between PACE4 and the substance P/neurokinin-1 (NK1) signalling pathway. Substance P and its NK1 receptor may play a role in mediating pain in the abdominal constriction test. Writhing sensitivity in Tac1 (preprotachykinin-1; the substance P precursor) KO mice was decreased significantly in one study [50] and was also decreased, albeit non-significantly, in another [51]; to our knowledge the Tacr1 (neurokinin-1 receptor) KO mouse has never been tested on the abdominal constriction test.
In conclusion, the current study identifies a variant in the PCSK6 gene that confers protection against pain in subjects with radiographic knee OA. This is important for two reasons. First, identifying genetic variation in symptomatic versus asymptomatic subpopulations in subjects with radiographic knee OA will enable a much-needed stratification of patient cohorts for the design of clinical trials in OA. Second, in combination with the murine findings, this study suggests a functional role for PACE4 in pain generation. Further studies will define this role, and the potential interaction between PACE4 and substance P/NK1. Taken together, the current findings offer some insight as to why, in the presence of the same structural damage, some individuals develop chronic pain and others are protected. Such insight may well lead to the development of new analgesic strategies. The finding that PACE4 has now been reported to play a role in symptoms associated with OA as well as in cartilage degradation may indicate a common pathway for these pathological processes.
Supplementary Material
Figure S1. LD heat plot of SNPs tested in the Discovery Cohort, based on 921 individuals. LD is represented as r2. These LD estimates are not the same as those from HapMap for the SNP under study. In HapMap B36 (release 28) there are 174 Caucasian samples and after excluding missing genotypes for rs3784515 there are 165 genotypes, while for rs900414, genotypes are available for only 90 Caucasian samples. LD measurements between rs3784515 and rs3784519 are r2 =0.88, between rs3784515 and rs900414 r2 =0.042, between rs3784519 and rs900414 r2 =0.09. The marker, T-Indel, represents an insertion/deletion polymorphism of a T at position 99,587,992.
ACKNOWLEDGEMENTS
The authors would like to thank Elizabeth Arner for critically reading the manuscript. AM Malfait is funded by an Arthritis Foundation Innovative Research Grant and by R01AR060364-01 (NIAMS).
Footnotes
The authors report no Competing Interests. Please note that AM. Malfait and M. Tortorella were Pfizer employees until 2009; A. Seymour and F. Gao were Pfizer employees until the spring of 2011. MP. Hellio and L. Wood are Pfizer employees. R. Maciewicz is an Astra-Zeneca employee.
REFERENCES
- 1.Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Report of the American College of Rheumatology Pain Management Task Force. Arthritis Care Res (Hoboken) 2010;62:590–599. doi: 10.1002/acr.20005. [DOI] [PubMed] [Google Scholar]
- 3.Guccione AA, Felson DT, Anderson JJ, et al. The effects of specific medical conditions on the functional limitations of elders in the Framingham Study. Am J Public Health. 1994;84:351–358. doi: 10.2105/ajph.84.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet. 2005;365:965–973. doi: 10.1016/S0140-6736(05)71086-2. [DOI] [PubMed] [Google Scholar]
- 5.Hunter DJ, McDougall JJ, Keefe FJ. The symptoms of osteoarthritis and the genesis of pain. Rheum Dis Clin North Am. 2008;34:623–643. doi: 10.1016/j.rdc.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedson J, Croft PR. The discordance between clinical and radiographic knee osteoarthritis: a systematic search and summary of the literature. BMC Musculoskelet Disord. 2008;9:116. doi: 10.1186/1471-2474-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis MA, Ettinger WH, Neuhaus JM, et al. Correlates of knee pain among US adults with and without radiographic knee osteoarthritis. J Rheumatol. 1992;19:1943–1949. [PubMed] [Google Scholar]
- 8.Hannan MT, Felson DT, Pincus T. Analysis of the discordance between radiographic changes and knee pain in osteoarthritis of the knee. J Rheumatol. 2000;27:1513–1517. [PubMed] [Google Scholar]
- 9.Lawrence JS, Bremner JM, Bier F. Osteo-arthrosis. Prevalence in the population and relationship between symptoms and x-ray changes. Ann Rheum Dis. 1966;25:1–24. [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan R, Peat G, Thomas E, et al. Symptoms and radiographic osteoarthritis: not as discordant as they are made out to be? Ann Rheum Dis. 2007;66:86–91. doi: 10.1136/ard.2006.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neogi T, Felson D, Niu J, et al. Association between radiographic features of knee osteoarthritis and pain: results from two cohort studies. BMJ. 2009;339:b2844. doi: 10.1136/bmj.b2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayis S, Dieppe P. The natural history of disability and its determinants in adults with lower limb musculoskeletal pain. J Rheumatol. 2009;36:583–591. doi: 10.3899/jrheum.080455. [DOI] [PubMed] [Google Scholar]
- 13.Dieppe P. Developments in osteoarthritis. Rheumatology (Oxford) 2011;50:245–247. doi: 10.1093/rheumatology/keq373. [DOI] [PubMed] [Google Scholar]
- 14.Lanyon P, Muir K, Doherty S, Doherty M. Assessment of a genetic contribution to osteoarthritis of the hip: sibling study. BMJ. 2000;321:1179–1183. doi: 10.1136/bmj.321.7270.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacGregor AJ, Antoniades L, Matson M, et al. The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum. 2000;43:2410–2416. doi: 10.1002/1529-0131(200011)43:11<2410::AID-ANR6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 16.MacGregor AJ, Li Q, Spector TD, Williams FM. The genetic influence on radiographic osteoarthritis is site specific at the hand, hip and knee. Rheumatology (Oxford) 2009;48:277–280. doi: 10.1093/rheumatology/ken475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdes AM, Spector TD. Genetic epidemiology of hip and knee osteoarthritis. Nat Rev Rheumatol. 2011;7:23–32. doi: 10.1038/nrrheum.2010.191. [DOI] [PubMed] [Google Scholar]
- 18.Diatchenko L, Nackley AG, Tchivileva IE, et al. Genetic architecture of human pain perception. Trends Genet. 2007;23:605–613. doi: 10.1016/j.tig.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix-Fralish ML, Mogil JS. Progress in genetic studies of pain and analgesia. Annu Rev Pharmacol Toxicol. 2009;49:97–121. doi: 10.1146/annurev-pharmtox-061008-103222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malfait AM, Arner EC, Song RH, et al. Proprotein convertase activation of aggrecanases in cartilage in situ. Arch Biochem Biophys. 2008;478:43–51. doi: 10.1016/j.abb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Tortorella MD, Arner EC, Hills R, et al. ADAMTS-4 (aggrecanase-1): N-terminal activation mechanisms. Arch Biochem Biophys. 2005;444:34–44. doi: 10.1016/j.abb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Seidah NG, Mayer G, Zaid A, et al. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol. 2008;40:1111–1125. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Little CB, Fosang AJ. Is cartilage matrix breakdown an appropriate therapeutic target in osteoarthritis--insights from studies of aggrecan and collagen proteolysis? Curr Drug Targets. 2010;11:561–575. doi: 10.2174/138945010791011956. [DOI] [PubMed] [Google Scholar]
- 24.Mogil JS, Chanda ML. The case for the inclusion of female subjects in basic science studies of pain. Pain. 2005;117:1–5. doi: 10.1016/j.pain.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Constam DB, Robertson EJ. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 26.Mogil JS, Ritchie J, Sotocinal SG, et al. Screening for pain phenotypes: analysis of three congenic mouse strains on a battery of nine nociceptive assays. Pain. 2006;126:24–34. doi: 10.1016/j.pain.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Murray RB, Gmerek DE, Cowan A, Tallarida RJ. Use of programmable protocol timer and data logger in the monitoring of animal behavior. Pharmacol Biochem Behav. 1981;15:135–140. doi: 10.1016/0091-3057(81)90352-x. [DOI] [PubMed] [Google Scholar]
- 28.Hargreaves K, Dubner R, Brown F, et al. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32:77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- 29.Mogil JS, Wilson SG, Bon K, et al. Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. Pain. 1999;80:67–82. doi: 10.1016/s0304-3959(98)00197-3. [DOI] [PubMed] [Google Scholar]
- 30.Hylden JL, Wilcox GL. Intrathecal substance P elicits a caudally-directed biting and scratching behavior in mice. Brain Res. 1981;217:212–215. doi: 10.1016/0006-8993(81)90203-1. [DOI] [PubMed] [Google Scholar]
- 31.Langford DJ, Crager SE, Shehzad Z, et al. Social modulation of pain as evidence for empathy in mice. Science. 2006;312:1967–1970. doi: 10.1126/science.1128322. [DOI] [PubMed] [Google Scholar]
- 32.Zhai G, Stankovich J, Ding C, et al. The genetic contribution to muscle strength, knee pain, cartilage volume, bone size, and radiographic osteoarthritis: a sibpair study. Arthritis Rheum. 2004;50:805–810. doi: 10.1002/art.20108. [DOI] [PubMed] [Google Scholar]
- 33.MacGregor AJ, Andrew T, Sambrook PN, Spector TD. Structural, psychological, and genetic influences on low back and neck pain: a study of adult female twins. Arthritis Rheum. 2004;51:160–167. doi: 10.1002/art.20236. [DOI] [PubMed] [Google Scholar]
- 34.van Meurs JB, Uitterlinden AG, Stolk L, et al. A functional polymorphism in the catechol-Omethyltransferase gene is associated with osteoarthritis-related pain. Arthritis Rheum. 2009;60:628–629. doi: 10.1002/art.24175. [DOI] [PubMed] [Google Scholar]
- 35.Gursoy S, Erdal E, Herken H, et al. Significance of catechol-O-methyltransferase gene polymorphism in fibromyalgia syndrome. Rheumatol Int. 2003;23:104–107. doi: 10.1007/s00296-002-0260-5. [DOI] [PubMed] [Google Scholar]
- 36.Reimann F, Cox JJ, Belfer I, et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci U S A. 2010;107:5148–5153. doi: 10.1073/pnas.0913181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer TZ, Waxman SG. Familial pain syndromes from mutations of the NaV1.7 sodium channel. Ann N Y Acad Sci. 2010;1184:196–207. doi: 10.1111/j.1749-6632.2009.05110.x. [DOI] [PubMed] [Google Scholar]
- 38.Valdes AM, Arden NK, Vaughn FL, et al. Role of the Na(V)1.7 R1150W amino acid change in susceptibility to symptomatic knee osteoarthritis and multiple regional pain. Arthritis Care Res (Hoboken) 2010 doi: 10.1002/acr.20375. (EPub) [DOI] [PubMed] [Google Scholar]
- 39.Valdes AM, De Wilde G, Doherty SA, et al. The Ile585Val TRPV1 variant is involved in risk of painful knee osteoarthritis. Ann Rheum Dis. 2011;70:1556–1561. doi: 10.1136/ard.2010.148122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70:349–355. doi: 10.1136/ard.2010.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong W, Marcinkiewicz M, Vieau D, et al. Distinct mRNA expression of the highly homologous convertases PC5 and PACE4 in the rat brain and pituitary. J Neurosci. 1995;15:1778–1796. doi: 10.1523/JNEUROSCI.15-03-01778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiefer MC, Tucker JE, Joh R, et al. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991;10:757–769. doi: 10.1089/dna.1991.10.757. [DOI] [PubMed] [Google Scholar]
- 43.Seidah NG, Chretien M, Day R. The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 44.Tsuji A, Hine C, Tamai Y, et al. Genomic organization and alternative splicing of human PACE4 (SPC4), kexin-like processing endoprotease. J Biochem. 1997;122:438–452. doi: 10.1093/oxfordjournals.jbchem.a021772. [DOI] [PubMed] [Google Scholar]
- 45.Tsuji A, Higashine K, Hine C, et al. Identification of novel cDNAs encoding human kexin-like protease, PACE4 isoforms. Biochem Biophys Res Commun. 1994;200:943–950. doi: 10.1006/bbrc.1994.1541. [DOI] [PubMed] [Google Scholar]
- 46.Mori K, Kii S, Tsuji A, et al. A novel human PACE4 isoform, PACE4E is an active processing protease containing a hydrophobic cluster at the carboxy terminus. J Biochem. 1997;121:941–948. doi: 10.1093/oxfordjournals.jbchem.a021677. [DOI] [PubMed] [Google Scholar]
- 47.Caterina MJ, Rosen TA, Tominaga M, et al. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature. 1999;398:436–441. doi: 10.1038/18906. [DOI] [PubMed] [Google Scholar]
- 48.Macpherson LJ, Xiao B, Kwan KY, et al. An ion channel essential for sensing chemical damage. J Neurosci. 2007;27:11412–11415. doi: 10.1523/JNEUROSCI.3600-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNamara CR, Mandel-Brehm J, Bautista DM, et al. TRPA1 mediates formalin-induced pain. Proc Natl Acad Sci U S A. 2007;104:13525–13530. doi: 10.1073/pnas.0705924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao YQ, Mantyh PW, Carlson EJ, et al. Primary afferent tachykinins are required to experience moderate to intense pain. Nature. 1998;392:390–394. doi: 10.1038/32897. [DOI] [PubMed] [Google Scholar]
- 51.Zimmer A, Zimmer AM, Baffi J, et al. Hypoalgesia in mice with a targeted deletion of the tachykinin 1 gene. Proc Natl Acad Sci U S A. 1998;95:2630–2635. doi: 10.1073/pnas.95.5.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. LD heat plot of SNPs tested in the Discovery Cohort, based on 921 individuals. LD is represented as r2. These LD estimates are not the same as those from HapMap for the SNP under study. In HapMap B36 (release 28) there are 174 Caucasian samples and after excluding missing genotypes for rs3784515 there are 165 genotypes, while for rs900414, genotypes are available for only 90 Caucasian samples. LD measurements between rs3784515 and rs3784519 are r2 =0.88, between rs3784515 and rs900414 r2 =0.042, between rs3784519 and rs900414 r2 =0.09. The marker, T-Indel, represents an insertion/deletion polymorphism of a T at position 99,587,992.