Abstract
Fatigue is highly prevalent in the general population and is one of the most common side effects of cancer treatment. There is growing evidence that pro-inflammatory cytokines play a role in cancer-related fatigue, although the molecular mechanisms for chronic inflammation and fatigue have not been determined. The current study utilized genome-wide expression microarrays to identify differences in gene expression and associated alterations in transcriptional activity in leukocytes from breast cancer survivors with persistent fatigue (n = 11) and non-fatigued controls (n = 10). We focused on transcription of inflammation-related genes, particularly those responsive to the pro-inflammatory NF-κB transcription control pathway. Further, given the role of glucocorticoids as key regulators of inflammatory processes, we examined transcription of glucocorticoid-responsive genes indicative of potential glucocorticoid receptor (GR) desensitization. Plasma levels of cortisol were also assessed. Consistent with hypotheses, results showed increased expression of transcripts with response elements for NF-κB, and reduced expression of transcripts with response elements for glucocorticoids (p < .05) in fatigued breast cancer survivors. No differences in plasma levels of cortisol were observed. These data indicate that increased activity of pro-inflammatory transcription factors may contribute to persistent cancer-related fatigue and provide insight into potential mechanisms for tonic increases in NF-κB activity, specifically decreased expression of GR anti-inflammatory transcription factors.
Keywords: Gene expression, Inflammation, Fatigue, Cancer, Cortisol, NF-κB, Glucocorticoid receptor
1. Introduction
Fatigue is highly prevalent in the general population and a common side effect of cancer treatment (Bower et al., 2000; Cella et al., 2001). Growing evidence suggests an inflammatory basis for fatigue in healthy (Cho et al., 2009) and cancer populations (Alexander et al., 2009; Bower et al., 2002, 2009; Collado-Hidalgo et al., 2006; Orre et al., 2009; Schubert et al., 2007). The nuclear factor (NF)-κB transcription control pathway plays a key role in controlling expression of pro-inflammatory genes, yet no study has examined whether aberrant activation of NF-κB is associated with fatigue and what molecular processes might permit this activation. One potential mechanism involves reduced functional activity of the glucocorticoid receptor (GR), which inhibits NF-κB and other pro-inflammatory signaling pathways and is thought to contribute to persistent inflammation in chronic stress and other conditions (Cole et al., 2007; Miller et al., 2008b). In this study, we tested the hypothesis that persistent cancer-related fatigue is associated with: (1) increased transcription of inflammation-related genes, particularly those responsive to the pro-inflammatory NF-κB transcription control pathway; and (2) decreased transcription of glucocorticoid-responsive genes indicative of potential GR desensitization.
2. Methods
2.1. Subjects
Subjects were drawn from a larger project examining immune and behavioral correlates of fatigue in breast cancer survivors (Collado-Hidalgo et al., 2006). Participants in this study were recruited from the Los Angeles area through tumor registry listings, newspaper advertisements, flyers, and other media coverage. Eligibility criteria included: (1) originally diagnosed with early-stage breast cancer (Stage 0, I, or II) between 1 and 5 years previously; (2) completed all cancer treatment with the exception of tamoxifen/aromatase inhibitors; (3) no evidence of cancer recurrence; and (4) no chronic medical conditions involving the immune system or regular use of immunosuppressive medications. Among 314 women screened, we identified 50 eligible survivors (Collado-Hidalgo et al., 2006).
Fatigue was assessed with the vitality subscale of the SF-36, a reliable and valid 4-item scale that assesses how much of the time respondents felt “worn out”, “tired”, had “a lot of energy”, and felt “full of pep” over the past 4 weeks (Ware and Sherbourne, 1992). Scores on this scale range from 0 to 100, with scores below 50 indicating limitations or disability related to fatigue. For the current study, we utilized biological samples from 11 women who scored at or below 40 on the SF-36 vitality scale over two to three assessments, indicating severe and persistent fatigue. A comparison group of 10 non-fatigued women were identified who scored at or above 70 on the SF-36 vitality scale over two or more assessments. In previous research, we have shown that breast cancer survivors scoring below 50 on the SF-36 show alterations in psychosocial, immune, and neuroendocrine function, supporting the validity of this cut-point (Bower et al., 2000, 2002, 2003, 2005a,b).
Participants provided blood samples and completed questionnaires at morning appointments and were weighed and measured for determination of body mass index (BMI). The Structured Clinical Interview for DSM-IV (SCID) was administered to assess current and lifetime depression. The UCLA Institutional Review Board approved study procedures, and written consent was obtained from all participants.
2.2. Leukocyte gene expression
Total RNA was extracted from 107 peripheral blood mononuclear cells (Qiagen RNeasy; Qiagen Valencia CA), and subject to genome-wide transcriptional profiling using Affymetrix Human Gene 1.0 ST high-density oligonucleotide arrays (Affymetrix, Santa Clara CA) in the UCLA DNA Microarray Core as previously described (Cole et al., 2005). Transcript abundance was quantified by Robust Multiarray Averaging (Bolstad et al., 2003) and differentially expressed genes were identified based on ≥30% difference in average expression across groups (maintaining false discovery rate at 5%) (Benjamini and Hochberg, 1995). We tested the a priori hypothesis that inflammation-related genes would be up-regulated in fatigued survivors using GOstat bioinformatics software (Beissbarth and Speed, 2004).
RT-PCR analyses independently tested differential expression of six transcripts showing large differential expression across groups and consistency with the functional Gene Ontology annotations found to characterize the differentially expressed gene list as a whole (e.g., pro-inflammatory cytokines). Transcripts were assayed in triplicate on an iCycler instrument (Biorad, Hercules CA) using TaqMan Gene Expression Assays (Applied Biosystems, Foster City CA), and standard threshold cycle analyses with normalization to parallel-assayed ACTB mRNA.
2.3. Transcription control bioinformatics
TELiS promoter-based bioinformatics analyses (Cole et al., 2005) tested this study’s primary hypothesis that leukocytes from fatigued breast cancer survivors would show alterations in global gene expression profiles consistent with (1) increased activity of NF-κB (assessed by the TRANSFAC V$CREL_01 nucleotide motif in differentially expressing promoter), and (2) decreased activity of the GR (V$GRE_C). The ratio of response element frequencies in the promoters of up- vs. down-regulated genes was taken as a measure of differential activity of transcription control pathways, and (log) ratios were averaged over nine different parametric combinations of promoter length (−300, −600, and −1000 to +200 bp upstream of RefSeq-designated transcription start site) and motif detection stringency (TRANSFAC mat_sim values of .80, .90, and .95) to ensure robust results (Cole et al., 2005). Based on previous genomic analyses of glucocorticoid resistance in similarly sized samples (e.g., Miller et al., 2008b), we projected identifying >100 genes differentially expressed by more than 1 SD across groups, yielding .87 power to detect a 15% difference in GRE prevalence within promoters of up- vs. down-regulated genes.
2.4. Serum cortisol
Cortisol concentrations were assayed by enzyme-linked immunosorbent assay (Diagnostic Systems Laboratories Inc., Webster, TX), with a lower detection limit of 0.1 µg/dL and a 6.4% intra-assay coefficient of variation.
3. Results
The majority of participants were married, Caucasian, and postmenopausal, consistent with the parent study (Collado-Hidalgo et al., 2006). Fatigued women were younger (51.2 vs. 62.2 years old; p = .003), less likely to have received radiotherapy (64% vs. 100%; p = .043), and longer post-diagnosis than non-fatigued women (3.03 vs. 2.14 years post-diagnosis; p = .08). Fatigued women were also significantly more likely to have a current diagnosis of depression (50% vs. 0%; p = .01) and marginally more likely to have a past depression diagnosis (55% vs. 20%; p = .10). There were no other group differences in demographic, medical, or treatment-related characteristics.
3.1. Differential gene expression
A total of 437 transcripts showed >30% difference in expression in leukocytes from fatigued vs. non-fatigued survivors (119 relatively up-regulated in fatigued survivors, 318 up-regulated in non-fatigued survivors; see Fig. 1 and Supplementary Table 1). Prominent among genes showing up-regulation in fatigued survivors were genes encoding pro-inflammatory cytokines (IL1A, IL1B, IL6, OSM), genes involved in chemokine signaling (CXCL2, CXCR5, CCL20) and transcriptional activation (IER3, ZNF331, NR4A2, NR4A3), and the vascular growth factor VEGFA. Transcripts showing up-regulation in non-fatigued survivors included genes involved in cytotoxic T lymphocyte responses (TRGC2, TIGIT, CX3CR1, CMKLR1, GZMH), the apoptosis promoter NLRC4, the hemoglobin-regulator HP, the RNA splicing regulator CROP, and the glucoamylase MGAM. We found no differences in expression of consensus leukocyte subset marker genes (i.e., CD3Z, CD4, CD8A, CD19, CD14, CD16/FCGR3A, or CD56/NCAM1). Confirmatory RT-PCR analyses of four transcripts up-regulated in fatigued survivors and two up-regulated in non-fatigued survivors corroborated microarray findings (Supplementary Table 2).
Fig. 1.
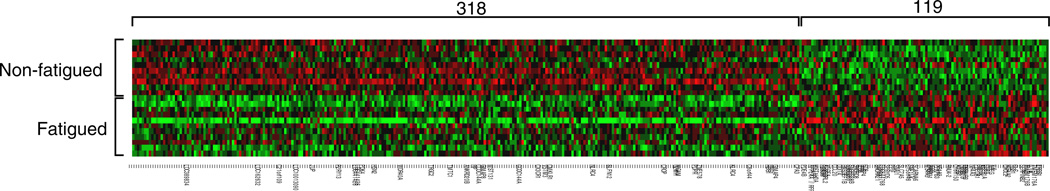
Differential gene expression in fatigued versus non-fatigued breast cancer survivors. Affymetrix Human Gene 1.0 ST high-density oligonucleotide arrays identified 119 genes relatively up-regulated in fatigued survivors, and 318 genes up-regulated in non-fatigued survivors (red indicates over-expression, green indicates under-expression) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.).
Consistent with the hypothesis that fatigue is associated with increased activity of immune- and inflammation-related genes, Gene Ontology analyses identified inflammation (GO annotation GO:0006954 and GO:0002526), immune response (GO:0006955; GO:0002376), and cytokine (GO:0005125) functional annotations as over-represented in genes showing selective up-regulation in fatigued survivors (all p < .001).
3.2. Role of NF-κB and GR signaling
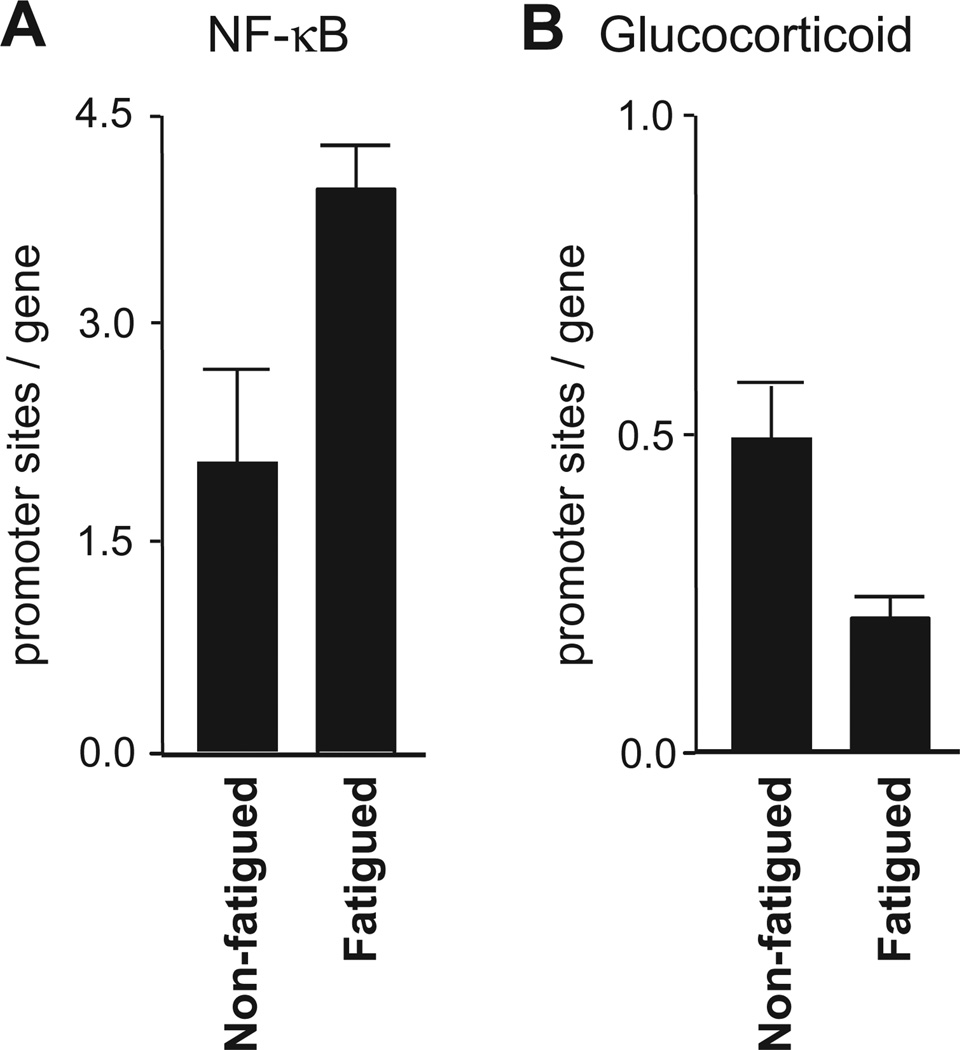
To test hypotheses about NK-κB and GR transcription factor activity, we conducted TELiS bioinformatic analyses of transcription factor response elements in the promoters of differentially expressed genes. Results showed a significantly greater density of NF-κB response elements in promoters of genes upregulated in fatigued vs. non-fatigued survivors (Fig. 2A; average difference 2.28-fold ±.09 across nine combinations of promoter length and scan stringency, p < .0001), and a significant under-representation of glucocorticoid response elements in the promoters of genes up-regulated in fatigued vs. non-fatigued survivors (Fig. 2B; average .45-fold ±.07 across nine combinations of promoter length and scan stringency, p = .007). This resulted in a net 5.10-fold skew in pro- vs. anti-inflammatory transcription factor-binding motifs in the promoters of genes up-regulated in fatigued survivors (difference from neutral 1.0-fold, p = .0004).
Fig. 2.
Transcriptional activity of NF-κB and GR signaling pathways. Results of TELiS bioinformatics analyses showed significantly greater density of NF-κB response elements (2A), and significantly lower density of glucocorticoid response elements (2B) in the promoters of genes up-regulated in fatigued versus non-fatigued breast cancer survivors.
Analyses of covariance were used to remove all variation in gene expression profiles attributable to age, time since diagnosis, and radiation treatment prior to promoter-based bioinformatic analyses. Results continued to show a 2.37-fold greater prevalence of NF-κB response elements and a 0.87-fold lower prevalence of glucocorticoid response elements in promoters of genes up-regulated in fatigued survivors (net pro- vs. anti-inflammatory skew 2.72-fold, p = .041). Bioinformatic indications of increased NF-κB also remained significant in analyses controlling for current depression and depression history (as measured by SCID; both differences >1.5-fold, p < .02). Indications of GR desensitization were attenuated by control for current depression (ns after adjustment) but continued to indicate reduced GR activity after controlling for history of depression (0.77-fold, p = .082).
To determine whether reduced GR transcriptional activity was mediated by decreased cortisol output, serum cortisol levels were assayed in blood samples drawn concurrently with transcriptionally profiled leukocytes. Results showed no significant difference between groups (mean = 29.9 µg/dl ± 4.3 for fatigued vs. 33.9 ± 3.6 in non-fatigued, p = .50). To determine whether reduced GR transcriptional activity might be secondary to reduced expression of the GR gene, we analyzed NR3C1 mRNA levels in microarray gene expression profiles. Average levels of GR transcripts differed by <1% across groups (p = .80).
4. Discussion
In the present study, leukocytes from breast cancer survivors experiencing persistent fatigue showed increased expression of genes encoding pro-inflammatory cytokines and other mediators of immunologic activation. Although these findings were generated in a small sample and should be cautiously interpreted, the present results support the hypothesis that peripheral inflammatory signaling may contribute to cancer-related fatigue by activating CNS-mediated “sickness behavior” (Miller et al., 2008a) and are consistent with previous data linking fatigue to elevated protein biomarkers of inflammation in cancer patients and survivors (Bower et al., 2002; Bower et al., 2009; Collado-Hidalgo et al., 2006).
The present study also extends basic links between fatigue and peripheral inflammatory signaling to identify alterations in the activity of upstream transcription control pathways that might potentially structure the observed differences in the expression of inflammation-related genes. Promoter-based bioinformatic analyses indicated increased activity of pro-inflammatory NF-κB/Rel transcription factors in leukocytes from fatigued breast cancer survivors. The basis for tonic upregulation of NF-κB activity is not clear, but it could potentially stem from decreased tonic inhibition of NF-κB by the anti-inflammatory GR transcription factor. Consistent with previous studies linking GR desensitization to increased NF-κB activity (Cole et al., 2007; Miller et al., 2008a, Miller et al., 2009), we found a marked diminution of GR-mediated transcriptional activity in leukocytes from fatigued breast cancer survivors. This effect occurred in the absence of differential HPA-axis cortisol output, suggesting that reduced ligand activation cannot account for the observed reduction in GR-mediated gene transcription, although more comprehensive evaluation of cortisol production (e.g., diurnal cortisol rhythm, total cortisol output) would be required to fully address this possibility. No decrease in GR (NR3C1) mRNA expression was observed, suggesting that reductions in GR number are also unlikely to account for the observed effects. This pattern of findings is consistent with several previous studies in finding reduced GR-mediated gene expression in the absence of decreased GR expression or cortisol levels. The most plausible mechanism for such effect involves inducible post-translational modification of the GR (e.g., phosphorylation), which can render the GR less sensitive to ligand-induced transcriptional activation and thereby induce a state of functional glucocorticoid resistance.
Results from analyses of covariance suggest that the general pro-inflammatory skew in observed gene expression profiles was independent of either current depression or depression history. These results are consistent with our previous work showing that fatigue-related elevations in plasma inflammatory markers cannot be accounted for by co-occurring symptoms of depression (Bower et al., 2002, 2009). However, the current findings also suggest that current depression may contribute more specifically to the observed GR desensitization. Future research in larger samples will be required to discriminate more clearly between cancer-related fatigue as a focal symptom and the broader array of symptom characteristics characterizing major depressive disorder.
The one previous study examining gene expression among individuals with cancer-related fatigue suggested that aberrant B lymphocyte activity might drive this symptom (Landmark-Hoyvik et al., 2009). The present study suggests a markedly different cellular mechanism in the up-regulation of key pro-inflammatory cytokines known to be expressed primarily by activated myeloid lineage cells. These differences might be explained by differences in the populations studied, the microarray assay platforms and related statistical methods, and the hypothesis-generating exploratory format of the previous study vs. the targeted hypothesis-testing approach of the present study. Nonetheless, the current findings require replication in a larger sample with a more comprehensive assessment of HPA activity.
Clinical trials have documented beneficial effects of cytokine antagonists on fatigue in patients with immune-related disorders (Tyring et al., 2006), and there is preliminary evidence that these therapies may also decrease fatigue in cancer patients (Monk et al., 2006). Discovery of altered NF-κB and GR transcription factor activity extends the range of therapeutic targets for ameliorating persistent cancer-related fatigue to include specific antagonists of NF-κB transcription factors and/or agents that inhibit or reverse acquired functional GR desensitization. In addition, behavioral therapies that have been associated with reduced inflammatory processes such as exercise (Fairey et al., 2005) and yoga (Kiecolt-Glaser et al., 2010; Pullen et al., 2008) may also have promise for treating inflammation-related fatigue in cancer patients.
Supplementary Material
Acknowledgments
This work was supported in part by the UCLA Cousins Center for Psychoneuroimmunology at the Semel Institute for Neuroscience and Human Behavior, Jonsson Comprehensive Cancer Center at UCLA, Jonsson Cancer Center Foundation, Breast Cancer Research Foundation, UCLA Older Americans Independence Center Inflammatory Biology Core, T32-MH19925, HL 079955, AG 026364, CA116778, RR00827, P30-AG028748, and General Clinical Research Centers Program.
Footnotes
Conflict of interest
The authors report no conflicts of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbi.2010.09.010.
References
- Alexander S, Minton O, Andrews P, Stone P. A comparison of the characteristics of disease-free breast cancer survivors with or without cancer-related fatigue syndrome. European Journal of Cancer. 2009;45(3):384–392. doi: 10.1016/j.ejca.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series. 1995;B57:289–300. [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Aziz N. Altered cortisol response to psychologic stress in breast cancer survivors with persistent fatigue. Psychosomatic Medicine. 2005a;67(2):277–280. doi: 10.1097/01.psy.0000155666.55034.c6. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Aziz N, Fahey JL. Fatigue and proinflammatory cytokine activity in breast cancer survivors. Psychosomatic Medicine. 2002;64(4):604–611. doi: 10.1097/00006842-200207000-00010. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Aziz N, Fahey JL, Cole SW. T-cell homeostasis in breast cancer survivors with persistent fatigue. Journal of the National Cancer Institute. 2003;95(15):1165–1168. doi: 10.1093/jnci/djg0019. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Desmond KA, Rowland JH, Meyerowitz BE, Belin TR. Fatigue in breast cancer survivors: occurrence, correlates, and impact on quality of life. Journal of Clinical Oncology. 2000;18(4):743–753. doi: 10.1200/JCO.2000.18.4.743. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Dickerson SS, Petersen L, Aziz N, Fahey JL. Diurnal cortisol rhythm and fatigue in breast cancer survivors. Psychoneuroendocrinology. 2005b;30(1):92–100. doi: 10.1016/j.psyneuen.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Tao ML, Hu W, Belin TR, Sepah S, Cole S, Aziz N. Inflammatory biomarkers and fatigue during radiation therapy for breast and prostate cancer. Clinical Cancer Research. 2009;15(17):5534–5540. doi: 10.1158/1078-0432.CCR-08-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella D, Davis K, Breitbart W, Curt G. Cancer-related fatigue: prevalence of proposed diagnostic criteria in a United States sample of cancer survivors. Journal of Clinical Oncology. 2001;19(14):3385–3391. doi: 10.1200/JCO.2001.19.14.3385. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Seeman TE, Bower JE, Kiefe CI, Irwin MR. Prospective association between c-reactive protein and fatigue in the coronary artery risk development in young adults study. Biological Psychiatry. 2009;66(9):871–878. doi: 10.1016/j.biopsych.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biology. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21(6):803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- Collado-Hidalgo A, Bower JE, Ganz PA, Cole SW, Irwin MR. Inflammatory biomarkers for persistent fatigue in breast cancer survivors. Clinical Cancer Research. 2006;12(9):2759–2766. doi: 10.1158/1078-0432.CCR-05-2398. [DOI] [PubMed] [Google Scholar]
- Fairey AS, Courneya KS, Field CJ, Bell GJ, Jones LW, Martin BS, Mackey JR. Effect of exercise training on C-reactive protein in postmenopausal breast cancer survivors: a randomized controlled trial. Brain, Behavior, and Immunity. 2005;19(5):381–388. doi: 10.1016/j.bbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Christian L, Preston H, Houts CR, Malarkey WB, Emery CF, Glaser R. Stress, inflammation, and yoga practice. Psychosomatic Medicine. 2010;72(2):113–121. doi: 10.1097/PSY.0b013e3181cb9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmark-Hoyvik H, Reinertsen KV, Loge JH, Fossa SD, Borresen-Dale AL, Dumeaux V. Alterations of gene expression in blood cells associated with chronic fatigue in breast cancer survivors. Pharmacogenomics Journal. 2009;9(5):333–340. doi: 10.1038/tpj.2009.27. [DOI] [PubMed] [Google Scholar]
- Miller AH, Ancoli-Israel S, Bower JE, Capuron L, Irwin MR. Neuroendocrine-immune mechanisms of behavioral comorbidities in patients with cancer. Journal of Clinical Oncology. 2008a;26(6):971–982. doi: 10.1200/JCO.2007.10.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, Ma R, Cole SW. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biological Psychiatry. 2008b;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Fok AK, Walker H, Lim A, Nicholls EF, Cole S, Kobor MS. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proceedings of the National Academy of Sciences. 2009;106(34):14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk JP, Phillips G, Waite R, Kuhn J, Schaaf LJ, Otterson GA, Guttridge D, Rhoades C, Shah M, Criswell T, Caligiuri MA, Villalona-Calero MA. Assessment of tumor necrosis factor alpha blockade as an intervention to improve tolerability of dose-intensive chemotherapy in cancer patients. Journal of Clinical Oncology. 2006;24(12):1852–1859. doi: 10.1200/JCO.2005.04.2838. [DOI] [PubMed] [Google Scholar]
- Orre IJ, Murison R, Dahl AA, Ueland T, Aukrust P, Fossa SD. Levels of circulating interleukin-1 receptor antagonist and C-reactive protein in long-term survivors of testicular cancer with chronic cancer-related fatigue. Brain, Behavior, and Immunity. 2009;23(6):868–874. doi: 10.1016/j.bbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Pullen PR, Nagamia SH, Mehta PK, Thompson WR, Benardot D, Hammoud R, Parrott JM, Sola S, Khan BV. Effects of yoga on inflammation and exercise capacity in patients with chronic heart failure. Journal of Cardiac Failure. 2008;14(5):407–413. doi: 10.1016/j.cardfail.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Schubert C, Hong S, Natarajan L, Mills PJ, Dimsdale JE. The association between fatigue and inflammatory marker levels in cancer patients: a quantitative review. Brain, Behavior, and Immunity. 2007;21(4):413–427. doi: 10.1016/j.bbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Tyring S, Gottlieb A, Papp K, Gordon K, Leonardi C, Wang A, Lalla D, Woolley M, Jahreis A, Zitnik R, Cella D, Krishnan R. Etanercept and clinical outcomes, fatigue, and depression in psoriasis: double-blind placebo-controlled randomised phase III trial. The Lancet. 2006;367(9504):29–35. doi: 10.1016/S0140-6736(05)67763-X. [DOI] [PubMed] [Google Scholar]
- Ware JE, Jr, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. conceptual framework and item selection. Medical Care. 1992;30(6):473–483. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.