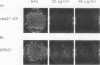
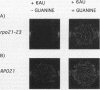
Abstract
Little is known about the regions of RNA polymerase II (RNAPII) that are involved in the process of transcript elongation and interaction with elongation factors. One elongation factor, TFIIS, stimulates transcript elongation by binding to RNAPII and facilitating its passage through intrinsic pausing sites in vitro. In Saccharomyces cerevisiae, TFIIS is encoded by the PPR2 gene. Deletion of PPR2 from the yeast genome is not lethal but renders cells sensitive to the uracil analog 6-azauracil (6AU). Here, we show that mutations conferring 6AU sensitivity can also be isolated in the gene encoding the largest subunit of S. cerevisiae RNAPII (RPO21). A screen for mutations in RPO21 that confer 6AU sensitivity identified seven mutations that had been generated by either linker-insertion or random chemical mutagenesis. All seven mutational alterations are clustered within one region of the largest subunit that is conserved among eukaryotic RNAPII. The finding that six of the seven rpo21 mutants failed to grow at elevated temperature underscores the importance of this region for the functional and/or structural integrity of RNAPII. We found that the 6AU sensitivity of the rpo21 mutants can be suppressed by increasing the dosage of the wild-type PPR2 gene, presumably as a result of overexpression of TFIIS. These results are consistent with the proposal that in the rpo21 mutants, the formation of the RNAPII-TFIIS complex is rate limiting for the passage of the mutant enzyme through pausing sites. In addition to implicating a region of the largest subunit of RNAPII in the process of transcript elongation, our observations provide in vivo evidence that TFIIS is involved in transcription by RNAPII.
Full text
PDF




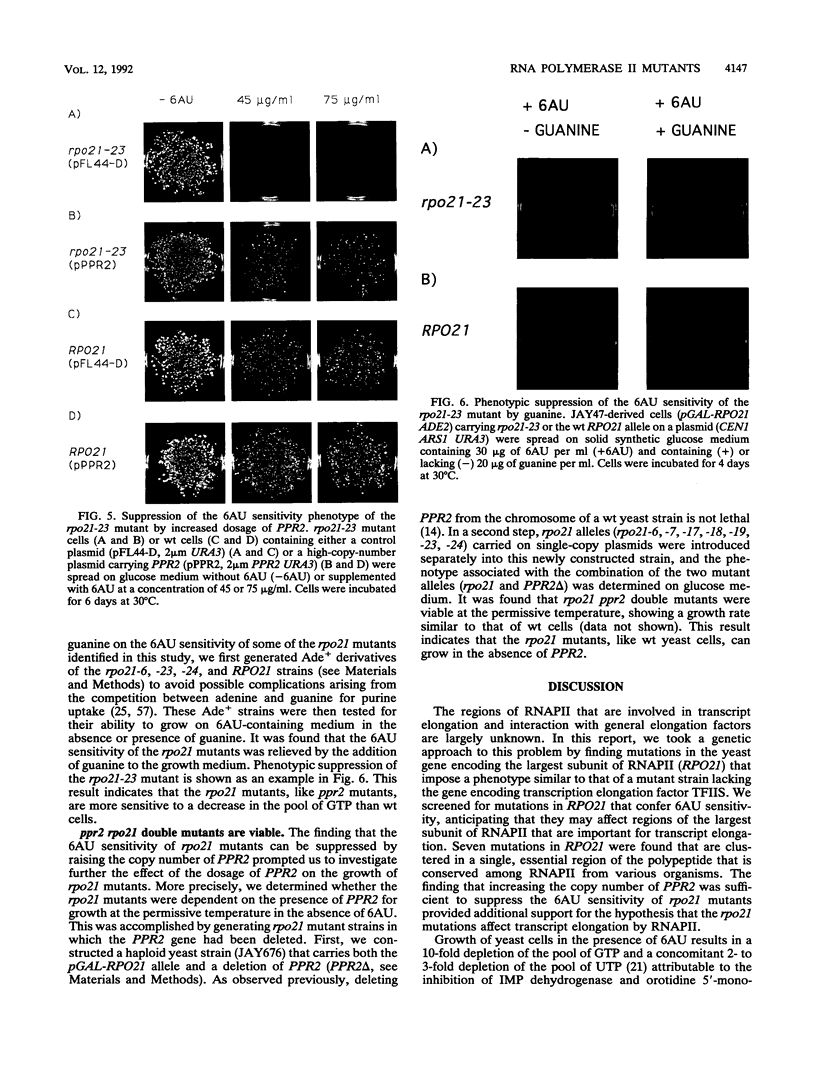

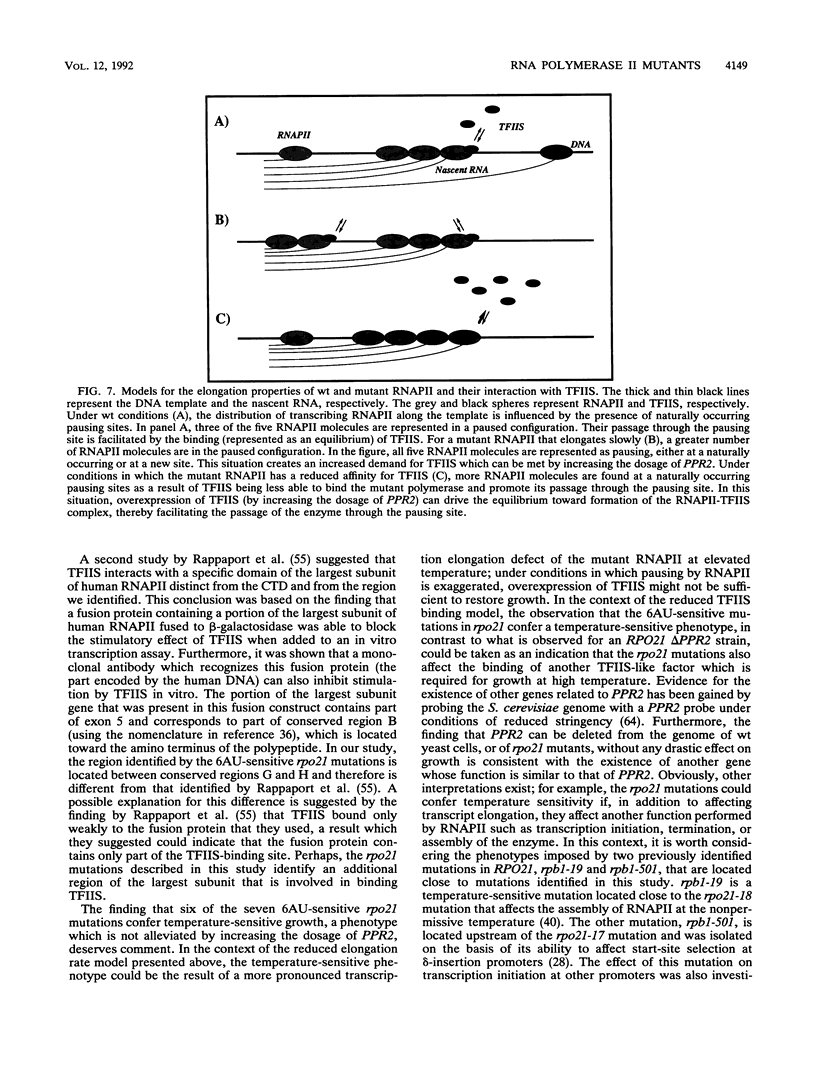



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agarwal K., Baek K. H., Jeon C. J., Miyamoto K., Ueno A., Yoon H. S. Stimulation of transcript elongation requires both the zinc finger and RNA polymerase II binding domains of human TFIIS. Biochemistry. 1991 Aug 6;30(31):7842–7851. doi: 10.1021/bi00245a026. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y., Hay N. Attenuation may regulate gene expression in animal viruses and cells. CRC Crit Rev Biochem. 1985;18(4):327–383. doi: 10.3109/10409238509086785. [DOI] [PubMed] [Google Scholar]
- Archambault J., Drebot M. A., Stone J. C., Friesen J. D. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Apr;232(3):408–414. doi: 10.1007/BF00266244. [DOI] [PubMed] [Google Scholar]
- Bengal E., Flores O., Krauskopf A., Reinberg D., Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991 Mar;11(3):1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Killeen M., Sopta M., Ortolan L. G., Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol. 1988 Apr;8(4):1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton Z. F., Ortolan L. G., Greenblatt J. Proteins that bind to RNA polymerase II are required for accurate initiation of transcription at the adenovirus 2 major late promoter. EMBO J. 1986 Nov;5(11):2923–2930. doi: 10.1002/j.1460-2075.1986.tb04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. B., Dykstra C. C., Sugino A. Isolation, DNA sequence, and regulation of a Saccharomyces cerevisiae gene that encodes DNA strand transfer protein alpha. Mol Cell Biol. 1991 May;11(5):2576–2582. doi: 10.1128/mcb.11.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M. A., Tourkine N., Belin D., Vassalli P., Jeanteur P., Blanchard J. M. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol Cell Biol. 1991 May;11(5):2826–2831. doi: 10.1128/mcb.11.5.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway J. W., Conaway R. C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989 Feb 5;264(4):2357–2362. [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985 Oct 25;260(24):13190–13198. [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. Properties of mutationally altered RNA polymerases II of Drosophila. J Biol Chem. 1982 Feb 25;257(4):1945–1952. [PubMed] [Google Scholar]
- Davies C. J., Trgovcich J., Hutchison C. A., 3rd Homologue of TFIIS in yeast. Nature. 1990 May 24;345(6273):298–298. doi: 10.1038/345298a0. [DOI] [PubMed] [Google Scholar]
- Exinger F., Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992 Jul;22(1):9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Flores O., Ha I., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990 Apr 5;265(10):5629–5634. [PubMed] [Google Scholar]
- Flores O., Maldonado E., Burton Z., Greenblatt J., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. RNA polymerase II-associating protein 30 is an essential component of transcription factor IIF. J Biol Chem. 1988 Aug 5;263(22):10812–10816. [PubMed] [Google Scholar]
- Flores O., Maldonado E., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989 May 25;264(15):8913–8921. [PubMed] [Google Scholar]
- Gardner W. J., Woods R. A. Isolation and characterisation of guanine auxotrophs in Saccharomyces cerevisiae. Can J Microbiol. 1979 Mar;25(3):380–389. doi: 10.1139/m79-059. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986 Nov;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANDSCHUMACHER R. E. Orotidylic acid decarboxylase: inhibition studies with azauridine 5'-phosphate. J Biol Chem. 1960 Oct;235:2917–2919. [PubMed] [Google Scholar]
- Hekmatpanah D. S., Young R. A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991 Nov;11(11):5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima S., Hirai H., Nakanishi Y., Natori S. Molecular cloning and characterization of cDNA for eukaryotic transcription factor S-II. J Biol Chem. 1988 Mar 15;263(8):3858–3863. [PubMed] [Google Scholar]
- Horikoshi M., Sekimizu K., Hirashima S., Mitsuhashi Y., Natori S. Structural relationships of the three stimulatory factors of RNA polymerase II from Ehrlich ascites tumor cells. J Biol Chem. 1985 May 10;260(9):5739–5744. [PubMed] [Google Scholar]
- Horikoshi M., Sekimizu K., Natori S. Analysis of the stimulatory factor of RNA polymerase II in the initiation and elongation complex. J Biol Chem. 1984 Jan 10;259(1):608–611. [PubMed] [Google Scholar]
- Hubert J. C., Guyonvarch A., Kammerer B., Exinger F., Liljelund P., Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO J. 1983;2(11):2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J Biol Chem. 1991 Aug 5;266(22):14478–14485. [PubMed] [Google Scholar]
- Jokerst R. S., Weeks J. R., Zehring W. A., Greenleaf A. L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989 Jan;215(2):266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Nierman W. C., Chamberlin M. J. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981 Mar 25;256(6):2787–2797. [PubMed] [Google Scholar]
- Kipling D., Kearsey S. E. TFIIS and strand-transfer proteins. Nature. 1991 Oct 10;353(6344):509–509. doi: 10.1038/353509a0. [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991 Sep;11(9):4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Cloning of a eukaryotic regulatory gene. Mol Gen Genet. 1981;184(3):394–399. doi: 10.1007/BF00352511. [DOI] [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall T. K., Guo H., Price D. H. Drosophila RNA polymerase II elongation factor DmS-II has homology to mouse S-II and sequence similarity to yeast PPR2. Nucleic Acids Res. 1990 Nov 11;18(21):6293–6298. doi: 10.1093/nar/18.21.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Greenblatt J. Related RNA polymerase-binding regions in human RAP30/74 and Escherichia coli sigma 70. Science. 1991 Aug 23;253(5022):900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- McGeady M. L., Wood T. G., Maizel J. V., Vande Woude G. F. Sequences upstream from the mouse c-mos oncogene may function as a transcription termination signal. DNA. 1986 Aug;5(4):289–298. doi: 10.1089/dna.1986.5.289. [DOI] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Jeanteur P., Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991 May;11(5):2832–2841. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., Bennetzen J., Sentenac A., Fromageot P. Single-stranded DNA transcription by yeast RNA polymerase B. Biochim Biophys Acta. 1981 Dec 28;656(2):220–227. doi: 10.1016/0005-2787(81)90090-3. [DOI] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2443–2451. doi: 10.1128/mcb.6.7.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. H., Sluder A. E., Greenleaf A. L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989 Apr;9(4):1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. H., Sluder A. E., Greenleaf A. L. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J Biol Chem. 1987 Mar 5;262(7):3244–3255. [PubMed] [Google Scholar]
- Rappaport J., Cho K., Saltzman A., Prenger J., Golomb M., Weinmann R. Transcription elongation factor SII interacts with a domain of the large subunit of human RNA polymerase II. Mol Cell Biol. 1988 Aug;8(8):3136–3142. doi: 10.1128/mcb.8.8.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport J., Reinberg D., Zandomeni R., Weinmann R. Purification and functional characterization of transcription factor SII from calf thymus. Role in RNA polymerase II elongation. J Biol Chem. 1987 Apr 15;262(11):5227–5232. [PubMed] [Google Scholar]
- Reichert U., Winter M. Gene dosage effects in polyploid strains of Saccharomyces cerevisiae containing gua-1 wild-type and mutant alleles. J Bacteriol. 1975 Dec;124(3):1041–1045. doi: 10.1128/jb.124.3.1041-1045.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987 Mar 5;262(7):3331–3337. [PubMed] [Google Scholar]
- Reines D., Chamberlin M. J., Kane C. M. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989 Jun 25;264(18):10799–10809. [PubMed] [Google Scholar]
- Rosen C. A. Regulation of HIV gene expression by RNA-protein interactions. Trends Genet. 1991 Jan;7(1):9–14. doi: 10.1016/0168-9525(91)90015-i. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Ruet A., Sentenac A., Fromageot P., Winsor B., Lacroute F. A mutation of the B220 subunit gene affects the structural and functional properties of yeast RNA polymerase B in vitro. J Biol Chem. 1980 Jul 10;255(13):6450–6455. [PubMed] [Google Scholar]
- Saltzman A. G., Weinmann R. Promoter specificity and modulation of RNA polymerase II transcription. FASEB J. 1989 Apr;3(6):1723–1733. doi: 10.1096/fasebj.3.6.2649403. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A., Fromageot P. Interaction of a new polypeptide with yeast RNA polymerase B. J Biol Chem. 1980 Jan 10;255(1):12–15. [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K., Kobayashi N., Mizuno D., Natori S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976 Nov 16;15(23):5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Nakanishi Y., Mizuno D., Natori S. Purification and preparation of antibody to RNA polymerase II stimulatory factors from Ehrlich ascites tumor cells. Biochemistry. 1979 Apr 17;18(8):1582–1588. doi: 10.1021/bi00575a031. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- SivaRaman L., Reines D., Kane C. M. Purified elongation factor SII is sufficient to promote read-through by purified RNA polymerase II at specific termination sites in the human histone H3.3 gene. J Biol Chem. 1990 Aug 25;265(24):14554–14560. [PubMed] [Google Scholar]
- Sluder A. E., Greenleaf A. L., Price D. H. Properties of a Drosophila RNA polymerase II elongation factor. J Biol Chem. 1989 May 25;264(15):8963–8969. [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989 Oct 5;341(6241):410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Sopta M., Carthew R. W., Greenblatt J. Isolation of three proteins that bind to mammalian RNA polymerase II. J Biol Chem. 1985 Aug 25;260(18):10353–10360. [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Stotz A., Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990 Oct 30;95(1):91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- Winsor B., Lacroute F., Ruet A., Sentenac A. Isolation and characterisation of a strain of Saccharomyces cerevisiae deficient in in vitro RNA polymerase B(II) activity. Mol Gen Genet. 1979 Jun 7;173(2):145–151. doi: 10.1007/BF00330304. [DOI] [PubMed] [Google Scholar]
- Yoo O. J., Yoon H. S., Baek K. H., Jeon C. J., Miyamoto K., Ueno A., Agarwal K. Cloning, expression and characterization of the human transcription elongation factor, TFIIS. Nucleic Acids Res. 1991 Mar 11;19(5):1073–1079. doi: 10.1093/nar/19.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Young R. A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- de Mercoyrol L., Job C., Job D. Studies on the inhibition by alpha-amanitin of single-step addition reactions and productive RNA synthesis catalysed by wheat-germ RNA polymerase II. Biochem J. 1989 Feb 15;258(1):165–169. doi: 10.1042/bj2580165. [DOI] [PMC free article] [PubMed] [Google Scholar]