Abstract
So-called next-generation sequencing (NGS) has provided the ability to sequence on a massive scale at low cost, enabling biologists to perform powerful experiments and gain insight into biological processes. BamView has been developed to visualize and analyse sequence reads from NGS platforms, which have been aligned to a reference sequence. It is a desktop application for browsing the aligned or mapped reads [Ruffalo, M, LaFramboise, T, Koyutürk, M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics 2011;27:2790–6] at different levels of magnification, from nucleotide level, where the base qualities can be seen, to genome or chromosome level where overall coverage is shown. To enable in-depth investigation of NGS data, various views are provided that can be configured to highlight interesting aspects of the data. Multiple read alignment files can be overlaid to compare results from different experiments, and filters can be applied to facilitate the interpretation of the aligned reads. As well as being a standalone application it can be used as an integrated part of the Artemis genome browser, BamView allows the user to study NGS data in the context of the sequence and annotation of the reference genome. Single nucleotide polymorphism (SNP) density and candidate SNP sites can be highlighted and investigated, and read-pair information can be used to discover large structural insertions and deletions. The application will also calculate simple analyses of the read mapping, including reporting the read counts and reads per kilobase per million mapped reads (RPKM) for genes selected by the user.
Availability: BamView and Artemis are freely available software. These can be downloaded from their home pages:
http://bamview.sourceforge.net/; http://www.sanger.ac.uk/resources/software/artemis/.
Requirements: Java 1.6 or higher.
Keywords: genome browser, next-generation sequencing, visualization, Artemis, BamView
INTRODUCTION
The high demand for low-cost sequencing led to the introduction of new technologies that no longer rely on dideoxy terminator-based Sanger sequencing and are vastly parallelized and high-throughput. These so-called next-generation sequencing (NGS) techniques produce unprecedented amounts of read data and have enabled new opportunities for numerous types of experiments, such as genomic re-sequencing, genetic variation studies, RNA-Seq, exome sequencing and ChIP-Seq. NGS genome resequencing has been used, for example, to study transmission of methicillin-resistant Staphylococcus aureus (MRSA) on both the global and local level by sequencing the genomes of closely-related isolates at an unprecedented scale [1] and to show that homologous recombination has led to serotype switching and vaccine escape in a study of 240 isolates of Streptococcus pneumoniae PMEN1 [2]. Furthermore, it has enabled the re-sequencing of many human individuals in the 1000 Genomes Project to study population genetics and identify variations in different populations [3]. Using NGS to sequence cDNA produced from RNA (RNA-Seq), scientists are now able to analyse the transcriptome to a base pair resolution [4–8]. This enables correction and improvement of gene-coordinate annotation, investigation of splice variants or analysis of differential expression under different conditions. Visualizing and analysing these large data sets holds the key to furthering our understanding of many biological processes. For example manual examination of the NGS pileups to identify large structural variations and for SNP calling is an important part of verifying the output of variant calling pipelines. So, for these data sets to be interpretable, the visualization tools need to be able to examine the results in the context of the underlying biological data.
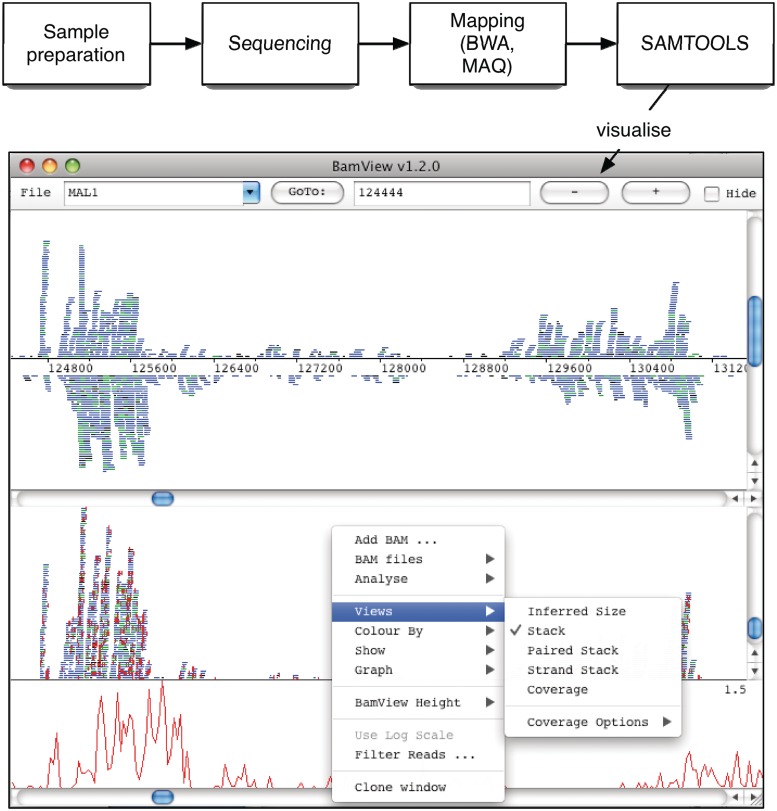
The BAM (Binary Alignment/Map) file format [9] is a generic and highly compressed format for storing alignments. This is the standard format for raw sequencing accepted and stored in the European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena/). Additionally this format has been widely adopted for storing all the read information from NGS data. This is a common output format of alignment programs [10] (e.g. BWA [11], MAQ, SSAHA2 [12]; Figure 1). Each record in the BAM file represents a read and contains a wealth of information about the read and the alignment. Collectively they can provide insight into structural annotation and variation.
Figure 1:
Schematic of the workflow from a NGS experiment to the visualization of the results produced. The BamView window shown here displays two panels produced using the clone option. The ‘Strand Stack’ view in the top panel shows the reads mapped to the forward and reverse strands above and below the sequence line. The bottom panel shows the ‘Stack’ view of the reads. The (red) vertical lines in this panel show the differences in the read sequence to the reference. The SNP density plot is shown as well as the pop-up menu with the available views and options.
There are a number of applications that have been developed for browsing, visualizing and interpreting NGS data. For example, EagleView [13], HawkEye [14] and Tablet [15] are designed to handle visualization of genome assemblies. LookSeq [16] and Genome browsers including Integrative Genome Viewer [17] (IGV), Integrated Genome Browser [18] (IGB) and GenoViewer (http://www.genoviewer.com), have been developed to visualize these read alignments in the context of genome annotation. MagicViewer [19] is also capable of displaying large-scale short read alignments in the context of annotations and provides a pipeline for genetic variation detection, annotation and visualization. Savant [20] is a genome and annotation visualization and analysis tool. BamView [21] have been developed specifically to visualize short-read alignment data. We developed BamView as an interactive Java application that can visualize large numbers of read alignments stored in BAM files, primarily so that it could be used as an integrated window in the widely used Artemis and Artemis Comparison Tool (ACT), although it can also be used as a portable, standalone, viewer. The strength of the BamView application lies in it being able to easily configure the five available views. It provides a wealth of options including the ability to filter reads on-the-fly, in and out of view based on various properties.
BACKGROUND
Artemis [22, 23] is an extensible DNA sequence browser that has been developed in the Pathogen Group at the Wellcome Trust Sanger Institute for over 10 years. It has been widely used as a genome viewer and annotation tool. The early development of Artemis coincided with an explosion in the availability of whole genome sequences including several closely related bacteria. Artemis filled the need for a tool to navigate and analyse sequence data and show the annotation in the context of the six possible translational reading frames. It as been used to annotate bacterial (e.g. Salmonella enterica [24], Burkholderia cenocepacia [25]) and small and medium-sized eukaryote genomes (e.g. Plasmodium knowlesi [26] and Schistosoma mansoni [27], respectively). To accommodate the need to compare such sequences, the Artemis code was leveraged to produce ACT [28], which enables the visualization of similarities and differences between sequences from individual bases to whole genomes [18, 19, 20, 29].
The development of Artemis and ACT has been influenced by the needs of the Pathogen Group at the Sanger Institute and in response to the suggestions and demands of the wider community via courses and an Artemis email group. As a result it has an abundance of functions to analyse and visualize sequences.
With the advent of NGS there has been a need to further develop Artemis to be able to visualize the large amounts of alignment data being produced. As a result of this requirement, BamView was implemented as a standalone application as well as a tool that can be integrated with Artemis. As ACT re-uses the components of Artemis, it shares much of the functionality and can also load in BAM files for visualization.
METHOD
Artemis, ACT and BamView are written in Java making them portable between MacOSX, UNIX and Windows platforms. These applications are designed to be straightforward and accessible for casual users, and highly configurable for the more advanced user. They are freely available for download.
BamView uses and is distributed with the Java library Picard (http://picard.sourceforge.net) to retrieve read alignment records from data files in BAM format. SAMtools [3] can be downloaded separately and is used to sort the reads in the BAM file by their left most coordinates and then index the BAM file. This creates an index file with a ‘bai’ suffix (e.g. in.bam.bai). The index is necessary to rapidly access reads mapped to the region of the reference genome being visualized. This makes it possible for BamView to rapidly query the file for data in a region rather than storing all the reads in memory, which would be prohibitive in many of cases because of the amount of data. Increasing the maximum memory that BamView uses may be necessary in the case of high coverage.
BamView can access the BAM and associated index files from a local file system or remotely over HTTP or FTP. Files can be loaded in either from the ‘File’ menu of the graphical user interface or from the command line (Table 1). Multiple BAM files can be loaded in again via the interface or from the command line. A file containing a list of BAM file names can also be used to load multiple files.
Table 1:
Summary of the BamView standalone command line options
| Option | Parameter type | Description |
|---|---|---|
| -h | Prints the command line options. | |
| -a | File | BAM/SAM file to display. |
| -r | File | Path to the reference sequence file (FASTA, EMBL, GenBank). |
| -n | Integer | Number of bases to display in the view. |
| -c | String | Chromosome name. |
| -v | IS, S, PS, ST or C | View used on opening: IS (inferred size), S (stack, default), PS (paired stack), ST (strand), C (coverage). |
| -b | Integer | Base position to open at. |
| -o | Show the read orientation. | |
| -pc | Plot the average coverage. | |
| -ps | Plot the SNP density (only with −r). |
If no options are specified then a prompt for the BAM file is shown.
BamView can be useful simply for visualizing the distribution of mapped reads (Figure 1), so loading the reference sequence is optional. However, if the reference sequence is provided then single nucleotide polymorphisms (SNP's) in relation to the reference sequence can be examined.
Alternatively, Artemis and BamView can be launched from a web page link using Java Web Start. This can be set up to supply arguments, such as input files, to the application to automatically open up remote reference sequence and BAM files, enabling the data to be easily shared (see the examples at. http://www.sanger.ac.uk/resources/software/artemis/ngs/).
FEATURES
The BamView panel provides a wealth of features and functionality (Table 2). BAM file(s) can be visualized in various views, which can be selected via a pop-up menu, which is activated by right-clicking on the BamView window. From this menu it is also possible to hide the reads from an individual BAM or load in additional BAM files for display. In the standalone BamView the ‘+’ and ‘−’ buttons (or down and up arrow) are used to zoom in and out to different levels of resolution along the nucleotide axis. Reads can be selected which can be used to highlight read pairs or facilitate tracking reads when zooming.
Table 2:
Summary of the features and functionality in BamView
| Read views |
|
|
| Read filtering |
|
|
| Zoom | From large sequence regions to the nucleotide level. | |
| Clone panels | The read panel can be cloned and each clone configured independently. | |
| Highlight |
|
|
| Display | SNPs are shown as vertical red lines on the reads. | |
| Read summary |
|
|
| Base colour | At nucleotide resolution bases can be coloured by their mapping quality. | |
| Graphs |
|
|
| Analyses |
|
|
| File format | BAM sorted and indexed. It can be on a local and remote (HTTP/FTP) file system. | |
| When reference sequences are concatenated together (e.g. in a multiple FASTA) BamView will offset the read positions correctly by matching the names (e.g. locus_tag or label) of the features to the sequence name in the BAM. | ||
| Use cases |
|
|
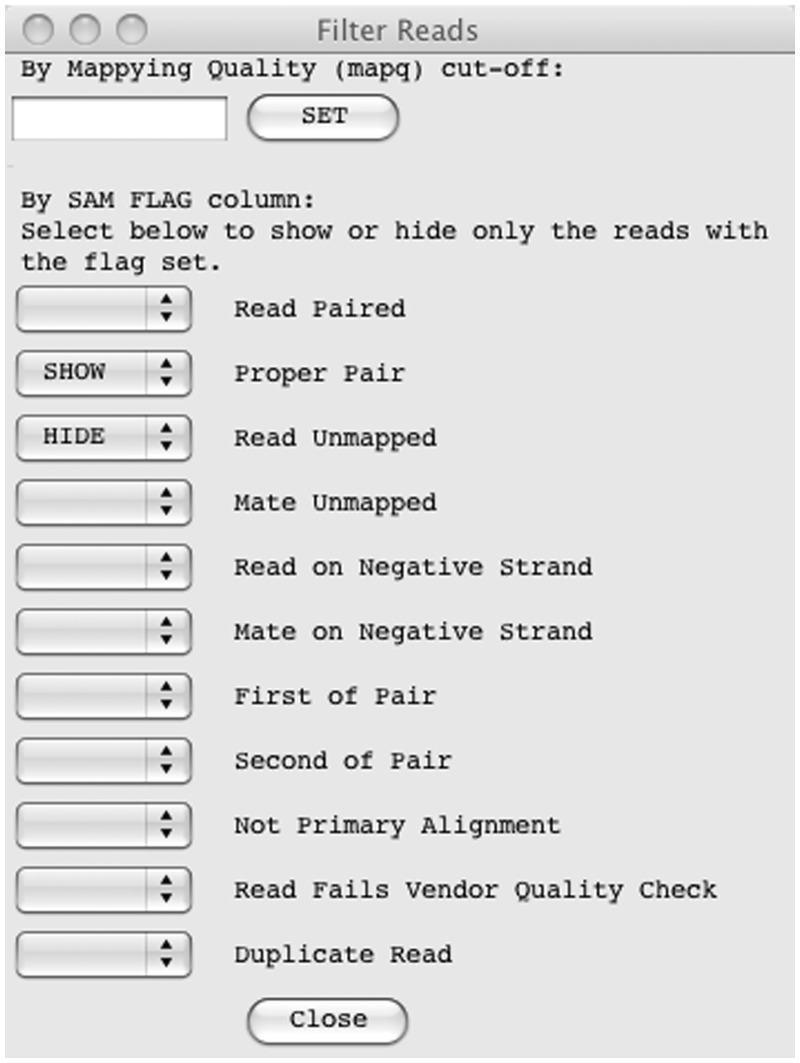
An important requirement for the interface is the ability to filter reads based on their mapping quality (MAPQ column in the BAM file), see Figure 2. This means it will show only the reads above a given mapping quality, which can remove misleading read alignments. A MAPQ cut-off can be set from the ‘Filter Reads …’ option in the pop-up menu. Reads can also be filtered using the reads flag present in the BAM file. As well as being able to filter out reads by selecting ‘HIDE’ from the check-box, there is also an option to ‘SHOW’ only the reads with that flag set, so BamView can, for example, show only reads with the ‘proper pair’ flag set. The default filter is to hide unmapped reads.
Figure 2:
The filter options for sequence reads. A mapping quality cut-off (MAPQ) can be applied and reads can be explicitly filtered in or out based on their flag. The filter shown will hide all unmapped reads and only show the reads that are defined as a proper pair.
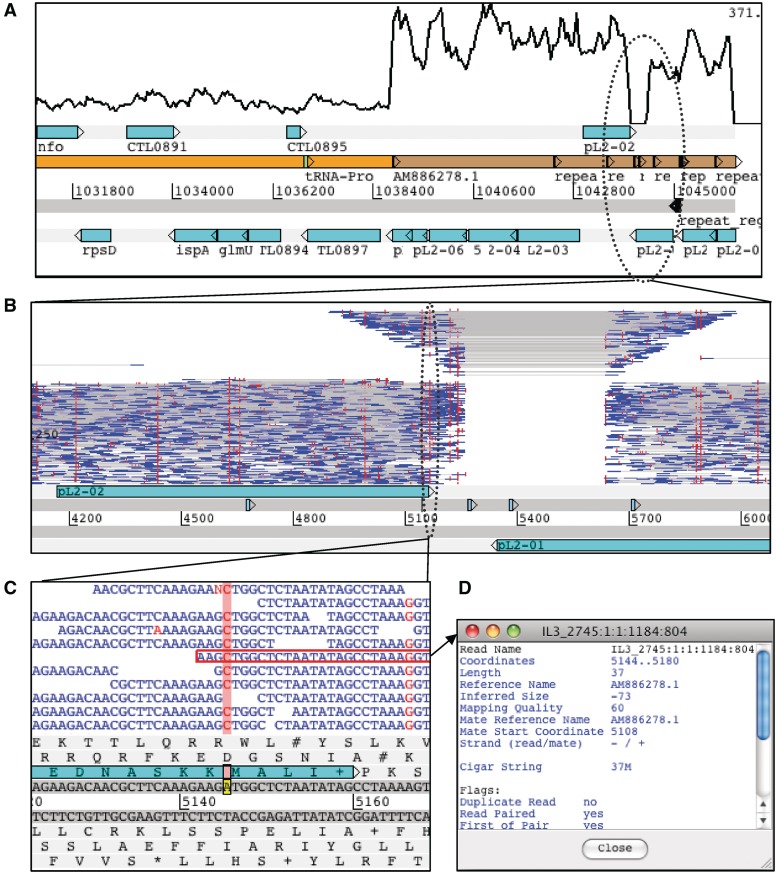
Figure 3 shows an example of the Chlamydia trachomatis genome and plasmid. By default the BamView panel shows a coverage plot for each BAM file (Figure 3A) when zoomed out, although it is still possible to select one of the other views from the menu. The coverage plots are calculated from the number of mapped read bases at a position, which are averaged over a window size that can be configured via the menus. As a result of the plasmid having a higher copy number an increase in read coverage can clearly be seen at the boundary of the genome and plasmid sequence. It can also be seen that a region of the plasmid shows zero read coverage. There are two possibilities as to why there are no reads mapped here (i) the region may have significantly diverged from the reference or (ii) it may be a deletion. To investigate this region further it is possible to zoom in and switch view to show the read pairs plotted by their inferred insert size (Figure 3B). There is also an option to plot the read pairs as the log of their inferred insert size as in this example. It can be seen that the inferred insert size of the reads increases either side of the unmapped region, characteristic of the pattern seen at a deletion. BamView illustrates when part of a read is mapped to each side of the deletion by joining the read blocks with a grey line. The significance of this indel is that this new variant C. trachomatis with the 377-bp deletion escaped routine diagnostic tests, which were based on the presence of this sequence [30, 31]. This led to a significant increase in new variant cases in Sweden [32] where it is found.
Figure 3:
Chlamydia trachomatis genome and plasmid sequences in Artemis at different levels of resolution. (A) Coverage is plotted in the BamView panel showing an increase at the boundary between the genome (orange) and plasmid (brown) sequence. The plasmid has a higher copy number and so has a higher read coverage. (B) Shows a smaller region (∼18 kb) of the plasmid with the reads plotted in the BamView panel along the y-axis by the log of the read-pair's inferred size. Vertical (red) lines on the reads indicate differences to the reference sequence. This view reveals the presence of an indel by the increase in the inferred size and drop in coverage. (C) Showing a region at the nucleotide level and highlighting a position where there appears to be a SNP. (D) One of the reads has been highlighted and the detailed SAM information is opened in a separate window.
BamView can also be used to analyse SNPs. The red vertical lines indicate where the read sequence differs from the reference sequence (Figure 3B) that may be a result of true divergence or an error in the reference sequence. Random SNP marks are likely to be caused by sequencing errors. However at positions where there is a consensus between reads then this may be a candidate SNP. Zooming in to the nucleotide level shows the bases for the reads (Figure 3C) and the SNP bases are marked red. The bases of the reads can be coloured by their quality scores (blue <10; green <20; orange <30; black ≥30). Additionally insertions in the reads are marked as purple vertical lines. Deletions are marked with horizontal lines joining the sections of the read they belong to. Clicking on a read highlights the read pair with a red border. On placing the mouse over a read and right clicking there is a menu option to show the information for the read and its mate (Figure 3D).
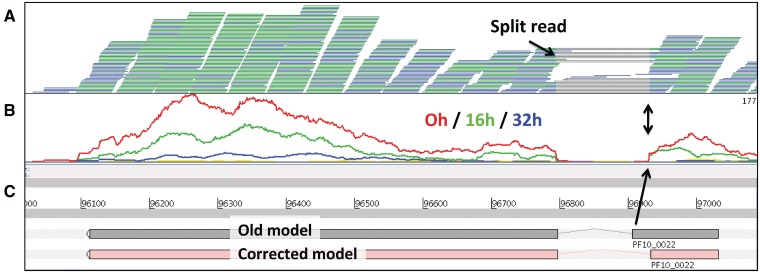
Another application for Artemis BamView is to correct reference sequence annotation using RNA-Seq data. Figure 4 shows RNA-Seq expression data of seven time points of the blood stage of the malaria parasite Plasmodium falciparum [4]. Reads that are mapped in the correct orientation (paired end reads orientated inwards, →←) and at the correct distance—often termed proper pairs—are coloured blue. Each read record in the BAM file has a flag indicating whether it is part of a proper pair or not. To maximize the screen space, reads with matching start and end positions are collapsed into one line and are coloured green. Figure 4A shows reads that are mapped using TopHat [35]. It can be seen in this example that individual reads are split over splice sites, as they show gray lines representing the joins between the mapped blocks in the read sequence. Artemis can be used to correct gene models if the splice site is wrong, as is the case for this gene (Figure 4C). Other applications of RNA-Seq splice site data in BamView would be to show alternative splicing, detect UTRs or new genes or new non-coding RNA.
Figure 4:
Correction of annotation with RNA-Seq data in Artemis. (A) The gene has two exons and the splice site can be identified by split reads in the BamView top panel. The gray lines represent split reads and show splice sites. The split reads provide evidence for a different splice site (black arrows). Therefore this gene was corrected [4]. (B) The middle panel shows the coverage plots for three different time points. This gene is expressed mostly at time point 0 h and the coverage plots also pinpoint the position of the splice site. Two different representations of gene PF10_0022 of Plasmodium falciparum are shown (C), an incorrect old version and a corrected version.
Figure 4B shows the coverage view with a separate plot for each time point. It can be seen that the expression levels vary over time for this gene. Genomic features (e.g. CDS's) can be selected and the number of mapped reads in those selected regions can be calculated (from the ‘Analyse’ menu). The number of reads is then presented for each feature with a value for each of the BAM files loaded in, i.e. for each time point in this example. Additionally, the reads per kilobase per million mapped reads (RPKM) can be calculated on the fly and written out in tab-delimited format. This output can then be used for further processing, such as differential expression analysis with DESeq [34]. The calculations for the read count and RPKM take into account the filter options that have been applied, so that different filter parameters will result in different values; the filters are set by default to commonly used values.
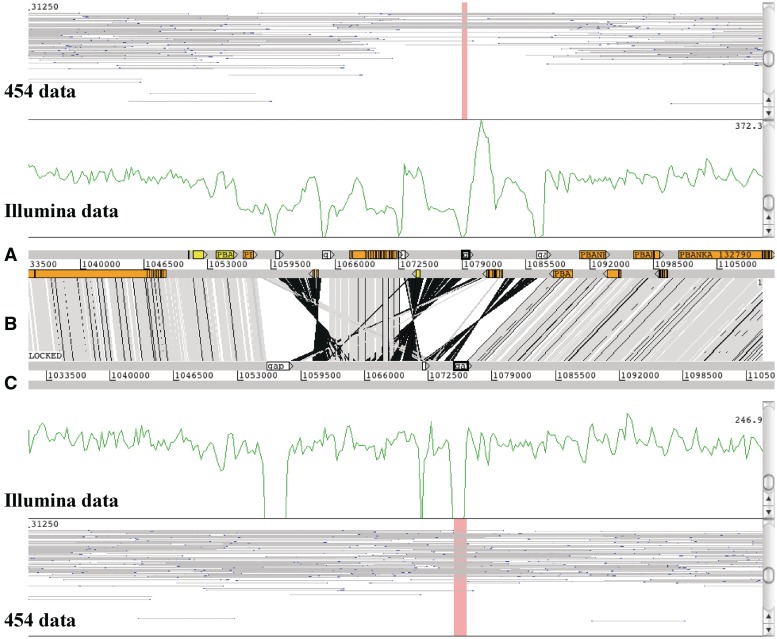
In ACT, the BamView panel also has various uses, such as identifying miss-assemblies (Figure 5) as breakpoints in mapped reads and collapsed repeats from regions where coverage doubles. In the example shown (Figure 5), the top genome is the assembly of Plasmodium berghei from GeneDB [35] and below that is a de novo assembly using NGS data. The BamView panels show (i) 454 reads with a 20 k insert size library and (ii) 500 bp Illumina library. A miss-assembly of the 454 data can clearly be seen as fewer proper read-pairs mapped. The Illumina data indicates the miss-assembly with an uneven coverage. Comparing this with the de novo assembly, the mapped Illumina reads show an even coverage (disregarding the gaps) and the 454 data have more proper read-pairs mapped over the region. Furthermore the gaps are spanned by reads pairs, indicating that there are no scaffolding errors and so we can be confident that this region has no further errors.
Figure 5:
ACT example of two Plasmodium berghei genome assemblies. A BamView panel is loaded for each assembly to identify a miss-assembly in the top sequence (A). The bottom sequence (C) is the de novo assembly. The vertical gray bars (B) are BLAST matches (with a 99% identity cut-off) that indicate synteny and the black bars inversions. The white boxes on the forward strand indicate sequencing gaps, the other boxes in the top genome are gene models. In each BamView panel the view is cloned to show a coverage plot of mapped Illumina reads and the insert size view of mapped 454 reads. In the corrected genome (C) the coverage is more even and does not drop except in the gap regions. The outer BamView show mapped 454 reads with an insert size of 20 k. The read filter was used to just show proper pairs. Far fewer read pairs confirm the top assembly. As the new assembly is more covered with 454 mate pairs, the assembly seems to be correct.
FUTURE DIRECTIONS
BamView within Artemis and ACT provides a powerful tool for visualizing, interpreting and analysing next generation sequence data sets. These applications are in continuous development. However, as more and more sequencing data becomes available, scientific focus will move to the variation identified by these sequence data. Therefore, additional ways of viewing variation information are likely to be required, such as the new Variant Call Format (VCF, http://vcftools.sourceforge.net/specs.html) view in Artemis that is in development. This can be viewed in conjunction with the BamView read alignments so that layers of information can be harnessed to elucidate sequence detail.
Key Points.
BamView is a powerful application that can be used at both the novice and expert levels.
Multiple local or remote BAM files can be loaded in and investigated which is useful for sharing data across collaborations.
BamView is highly configurable and provides five different views of mapped sequence reads. Depending on the type of sequencing performed the different views can be used to investigate assemblies, small variations, large structural variations and exon boundaries.
Reads can be viewed at the base level and colour coded to show the base qualities. This can be useful in confirming the evidence for calling SNP sites.
Reads can be filtered by their mapping quality, or various other flags.
Integrated into Artemis and ACT, the BamView panel is used to view next-generation data in the context of the annotation and greatly aids interpretation of biological processes. Within Artemis and ACT, further analyses are provided in the form of read counts and RPKM values for selected features, which is useful in investigating expression levels for genes.
FUNDING
This work was supported by the Wellcome Trust through their funding of the Pathogen Genomics group at the Wellcome Trust Sanger Institute (grant number WT 076964).
Biographies
Tim Carver was a postdoc at the University of Cambridge before joining the Medical Research Council in 2000. From 2004 he has been the lead developer for Artemis and ACT at the Wellcome Trust Sanger Institute.
Simon Harris joined the Wellcome Trust Sanger Institute in 2008 as a bacterial phylogeneticist and develops new techniques for using next generation sequence data in bacterial pathogenomics.
Thomas Otto has worked since 2008 as senior computer biologist at the Wellcome Trust Sanger Institute. His expertise lies in algorithm development for biological problems and next generation sequencing that are mostly applied to Plasmodium.
Matthew Berriman has worked on the sequencing and analysis of parasite genomes at the Wellcome Trust Sanger Institute since 2000.
Julian Parkhill has worked on the sequencing and analysis of microbial genomes at the Wellcome Trust Sanger Institute since 1997.
Jacqueline McQuillan has a PhD in Software Engineering and joined the Pathogen Genomics group at the Wellcome Trust Sanger Institute as a postdoctoral fellow in 2008. From 2010, she has managed the informatics team that develops pipelines and analysis support for the Pathogen Genomics group.
References
- 1.Harris SR, Feil EJ, Holden MTG, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croucher NJ, Harris SR, Fraser C, et al. Rapid Pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altshuler D, Durbin RM, Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto TD, Wilinski D, Assefa S, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Biol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daines B, Wang H, Wang L, et al. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–24. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 7.Bruno VM, Wang Z, Marjani SL, et al. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–8. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu T, Lu G, Fan D, et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–49. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruffalo M, LaFramboise T, Koyutürk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 2011;27:2790–6. doi: 10.1093/bioinformatics/btr477. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–9. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Marth GT. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–43. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schatz MC, Phillippy AM, Shneiderman B, Salzberg SL. Hawkeye: an interactive visual analytics tool for genome assemblies. Genome Biol. 2007;8:R34. doi: 10.1186/gb-2007-8-3-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milne I, Bayer M, Cardle L, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–32. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotech. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicol JW, Helt GA, Blanchard SG, et al. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–1. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou H, Zhao F, Zhou L, et al. MagicViewer: integrated solution for next-generation sequencing data visualization and genetic variation detection and annotation. Nucleic Acids Res. 2010;38:W732–6. doi: 10.1093/nar/gkq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiume M, Williams V, Brook A, et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–44. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carver T, Böhme U, Otto TD, et al. BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformatics. 2010;26:676–7. doi: 10.1093/bioinformatics/btq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford K, Parkhill J, Crook J, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 23.Carver T, Berriman M, Tivey A, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhill J, Dougan G, James KD, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–52. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 25.Holden MTG, Seth-Smith HMB, Crossman L, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009;191:261–77. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pain A, Böhme U, Berry AE, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–8. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carver TJ, Rutherford KM, Berriman M, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 29.Peacock CS, Seeger K, Harris D, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–47. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seth-Smith HMB, Harris SR, Persson K, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripa T, Nilsson P. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill. 2006;11:E061109.2. doi: 10.2807/esw.11.45.03076-en. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann B, Törner A, Low N, et al. Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerging Infect. Dis. 2008;14:1462–5. doi: 10.3201/eid1409.080153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertz-Fowler C, Peacock CS, Wood V, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–43. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]