Abstract
JBrowse is a web-based genome browser, allowing many sources of data to be visualized, interpreted and navigated in a coherent visual framework. JBrowse uses efficient data structures, pre-generation of image tiles and client-side rendering to provide a fast, interactive browsing experience. Many of JBrowse's design features make it well suited for visualizing high-volume data, such as aligned next-generation sequencing reads.
Keywords: genome browser, web, next-generation sequencing
INTRODUCTION
JBrowse is a web-based genome browser [1]: the user interface is displayed through a standard web browser (e.g. Microsoft Internet Explorer, Mozilla Firefox or Google Chrome), rather than a stand-alone program, allowing the user to view various forms of data related to a particular genome. Data input can be done via a wide variety of formats and databases: FASTA [2] for sequence files, BED [3], GFF [4] or BAM [5] for annotations of discrete features such as genes and experimentally detected transcripts, WIG [6] for quantitative per-base annotations, and Chado [7, 8] or Bio::DB [9] for relational database representations of these data types.
JBrowse is distinguished by running primarily on the web browser client: the user's machine does most of the work in arranging the browser's display, which both minimizes the amount of required client–server communication and enables many viewers to view the same centralized data without overloading the server. This, combined with its implementation in the JavaScript programming language, enables a more modern user interface while also making it an ideal tool for visualizing next-generation sequencing data. JBrowse is freely available under the GNU lGPL via GitHub: http://www.github.com/jbrowse/.
CONTEXT AND BACKGROUND
Most web pages, including some web-based genome browsers, are implemented using the Common Gateway Interface (CGI), which necessarily imposes a page-based model of user interaction. In many typical uses (e.g. searching the web, reading an online newspaper), this functions intuitively and does not interrupt user attention or interpretation of the page. Browsing a genome, however, is more like navigating a very large map: moves between locations and/or zoom levels must be done in such a way as to not disorient the user, but the space to be viewed is very large. In order to design a genome browser capable of offering an intuitive user interface over an internet connection, several hurdles need to be overcome.
In reloading a page after each navigation event (changing co-ordinates, changing zoom level, adding a track), interpreting the new display will often require the user to pick out landmark features common to the previous display inorder to re-orient themselves. JBrowse avoids this problem by adopting a Google maps-style browsing interface. Navigation event transitions occur without the current display disappearing. Instead, panning and scaling occur via smooth animated transitions.
Crucial to achieving this browsing style is the division of labor between client and server. Traditionally, in a CGI-based implementation, each page reload is handled by the server, which determines what to display, and then arranges it in a format that the client machine can read. With many users querying the same server, especially if the data to be queried is large, the central server can quickly become overloaded, slowing responses for all users. JBrowse avoids this problem by enabling the client machine (the computer running the user's web browser) to perform most of the computationally demanding tasks. When a user connects to a JBrowse server, files are sent from the server which are sufficient for the client to perform data querying and rendering operations. This frees up the server by distributing the workload onto the users, enabling more people to quickly connect to the same central server while simultaneously decoupling user interactions from client–server interactions, allowing for a more responsive browsing experience.
A genome browser may need to transmit data from server to client that is larger than what typical web browsing tasks would demand. Downloading a track describing the mapping of all next-generation reads to a particular chromosome, for example, would be extremely slow. JBrowse adopts a strategy whereby these data sets are divided by the server (ahead of time) into small regional chunks that are quick to download.
There are by now several alternative software packages that render genome annotations in the web browser client; notable examples are Anno-J [10] and Dalliance [11]. Compared with those tools, JBrowse is accessible to a much broader set of web users. Anno-J and Dalliance require recent web browser features—respectively, the HTML5 Canvas element and Scalable Vector Graphics support—that, while they allow richer graphical effects, do not exist in the widely used older versions of the Internet Explorer browser. JBrowse is also distinguished by its closer integration with the GMOD suite of tools (JBrowse began as an offshoot of the GBrowse genome browser, and tools to migrate existing GBrowse installations to JBrowse are in development).
FEATURES
Navigation
A reference ‘ruler’ at the top of the window indicates the chromosome, with the current viewing area encapsulated by a red box. A text box allows specification of the exact coordinates to be viewed, or the user can manually zoom and pan to the region of interest in one of several ways: arrow buttons move the view left or right, + and − buttons similarly zoom in and out. Small adjustments can be made by clicking and dragging the entire display left, right, up or down.
Manipulating tracks
A pane to the left of the data display stores the available tracks. Tracks can be moved in and out of the viewing area by a simple drag-and-drop maneuver. This allows the user to decide which of the tracks to use in populating the browser—unwanted tracks stay off the left without cluttering the display. Tracks can be dragged vertically within the viewing area to reorder them—especially useful when comparing multiple-related analyses.
Track types
Tracks are organized into two basic types: ‘feature’ tracks and ‘image’ tracks. Feature tracks describe discrete features with start and end points, such as coding regions or binding sites. Clicking on a feature can trigger a configurable option (e.g. clicking a protein-coding gene may open the corresponding PFAM page for that gene). ‘Sub-features’ such as introns and exons can be displayed via a special glyph style. Image tracks can display any image aligned to the genome; currently, they are primarily used to display fine-resolution histograms with a value defined at each position in the genome. These are well suited to displaying conservation, GC-content or other continuously varying data.
There is an API available for programmers who would like to implement their own image-track renderers, with example code that renders base pairing arcs indicative of RNA secondary structure.
Semantic zooming
In displaying genomic data at very different scales (e.g. whole chromosome versus tens of bases), summaries that may be informative at some levels may not be at others. For instance, a user may want to get an idea of the genome-scale distribution of a particular type of feature, and then zoom in to examine one particular such feature.
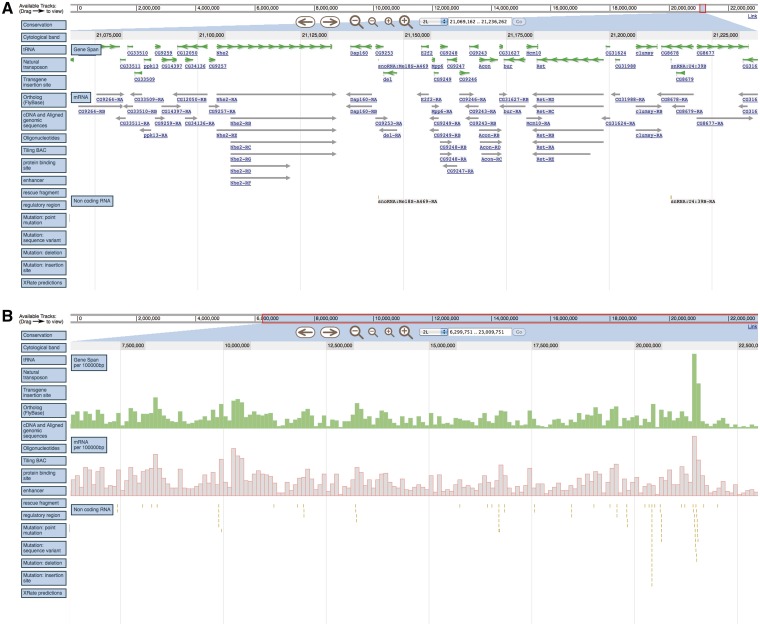
To handle this, JBrowse uses semantic zooming: different representations of the data are used at different zoom levels to maximize the ease of user interpretation [12]. For instance, at lower zoom levels, JBrowse collapses features into histograms showing the density distribution of a particular feature, rather than each individual feature. Similarly, DNA sequence is only showed at the highest zoom level, the only level where it can be readily interpreted. An example of this is shown in Figure 1, wherein the mRNA and GeneSpan tracks are both collapsed to histograms when moving from a ∼200 kb view to a 7 mb view.
Figure 1:
Semantic zooming uses different representations of the same data to maximize the ease of user interpretation across a broad range of zoom levels. When viewing a region of <200 kb in (A), individual genes and mRNA features are shown. When zooming out to a 7 mb region in (B), these two tracks are collapsed to histograms showing the frequency of each within 100 kb regions. Since non-coding RNA features are sufficiently sparse, they are still individually displayed at both zoom levels (as small vertical lines), but their individual labels are omitted for readability.
Feature name indexing
When working with large genomes with many features, there may be times when navigating using the aforementioned zoom/pan tools to find a particular feature may be cumbersome. JBrowse allows searching for features by entering part of the feature's name into the navigation text box.
Setup scripts
Currently, JBrowse is installed on a web server by running a series of scripts at the command line. First, the reference genome and its chromosomes are added, providing a base with which to index features. Annotations in the form of features and quantitative data, whose locations correspond to the reference genome, are added using one of several Perl scripts packaged with JBrowse. Finally, once all the features have been added, their names can optionally be indexed for efficient feature searching.
Once JBrowse is installed on a server, anyone with a web browser and access to the server can browse the data using the web interface; only the administrator is required to install JBrowse. Instructions on the installation of JBrowse (including dependencies) and usage of these setup scripts can be found at http://gmod.org/wiki/JBrowseDev/Current.
METHOD
Continuity of user attention is a priority in JBrowse. Central to accomplishing this goal are the issues of how data and tasks are split up between the client and server machines, and how these data are indexed on the server. Essentially, as much of the workload as possible is shifted to the client machine, and data on the server is indexed so as to allow rapid loading of the data relevant to a given region.
Indices of genomic features and data are generated on the server when the administrator sets up JBrowse. The underlying data structure is the Nested Containment List (NCList), an efficient hierarchical index for interval sets with sublinear access time [13]. When a user accesses a JBrowse page from a (possibly) remote location, static files are sent from the server to the client, which allow the client to perform the tasks inherent in browsing: determining which features overlap the current viewed region and then rendering the display based on these features.
Lazy loading
‘Lazy loading’ refers to the practice of postponing the retrieval (loading) of data from the server until the latest possible moment: when those data are needed to render a region that the user is currently (or will imminently be) viewing [14]. This strategy is key to the way JBrowse manages large next-generation data sets. To implement lazy loading, JBrowse uses the NCList index structure to break the data set into manageably sized ‘chunks’, each of which is only downloaded as and when it is necessary. This contrasts with indexing strategies used by many browsers such as GBrowse [15], UCSC [16] and Ensembl [17], as well as protocols such as DAS [18]. All these strategies similarly allow the data to be loaded in chunks, but further allow the chunk co-ordinates to be specified by the client (via range queries that the client sends to the server), meaning that the client must re-assemble the chunks and track overlaps. In JBrowse, the chunking is an inherent aspect of the data structure via which annotations are indexed, and the breakpoints are therefore predetermined by the server. This allows the server (or its administrator) to fine-tune the chunking so as to maximize the optimization of network transport, as well as minimizing computational work both on the client (which never needs to reassemble chunks, since the seams between chunks correspond to natural fragment boundaries in the annotation data structure) and on the server (which has to do fewer, or indeed no, dynamic queries).
BAM file processing
JBrowse includes a script to generate a feature track from a BAM-format index of read-to-reference genome alignments [5]. The script operates in an online fashion, avoiding the need to load enormous BAM files into memory at once.
UCSC human genome database
JBrowse also comes with a script that automatically downloads track data from the UCSC human genome database and uses it to initialize a JBrowse instance.
TECHNOLOGIES
The central challenge of a genome browser is to process and display a vast array of data in an intuitive, simple way that is easy for the user to understand and navigate. JBrowse utilizes various technologies with this goal in mind: nested containment lists for indexing features, Patricia/radix tries for indexing feature names and an online sort for processing BAM files.
Nested containment lists
Determining the features that overlap a given viewing region is a significant portion of the work the browser must do (whether it is done on the server or client). Since this step greatly affects the speed and smoothness of the browsing experience, it is important for it to be efficient. JBrowse uses NCList to represent features [13].
These data structures allow efficient [O(n + log(N)) for an interval set of cardinality N and n intervals returned by the query] determination of the intervals contained within a given query interval—in this case, the features contained within a viewing region.
Feature name indexing
Feature names are indexed in a Patricia trie or radix trie, allowing efficient searching by feature name.
FUTURE DIRECTIONS
The JSON data format used to store JBrowse track data is currently being redesigned to improve efficiency, flexibility and the facility to generate tracks dynamically. Following this redesign, the track format will be made public, enabling third-party applications (including web content management frameworks) to better interface to JBrowse.
A highly active area of JBrowse-related development is in the Web-Apollo project, which has adopted JBrowse as the web-based platform for the next version of the Apollo genome annotators’ curation tool. This software includes a number of extra features, such as the ability to see who else is working on (or viewing) a given genome at any one instant, and greater flexibility in drawing tracks using the HTML5 Canvas element.
Several enhancements have been added to the experimental forks of JBrowse, which are available from the JBrowse source code repository (GitHub). In particular, these include richer ‘widgets’ for organizing the list of undisplayed tracksthat can get very large for successful genome projects. One enhancement uses the Dojo tree widget with collapsible nodes to organize tracks hierarchically. Another uses the MIT Simile Exhibit widget to allow faceted browsing of track lists, whereby the track list can be dynamically queried using a number of orthogonal search parameters, to quickly locate the tracks of interest. The JBrowse team is working to integrate these enhancements, and the Web-Apollo enhancements mentioned above, back into the main JBrowse code base.
In the longer term, tools to assist migration from GBrowse to JBrowse are in development. Numerous small enhancements are also planned to the user interface, including (for example) display of paired-end reads, display of the sequence of reads (and highlighting of differences from the reference genome) and quantitative y-axis labels for histograms and WIG tracks.
Key points.
JBrowse is a web-based genome browser whose design and implementation make it well suited to the challenges brought on by the large sequencing data sets.
JBrowse is unique among web-based genome browsers in that it uses JavaScript to delegate a large amount of the workload to the client machine (breaking large data sets into manageable chunks). This enables browsing large data sets in a smooth, responsive environment.
FUNDING
National Institutes of Health/National Human Genome Research Institute (R01-HG004483).
References
- 1.Skinner ME, Uzilov AV, Stein LD, et al. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–8. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FASTA. format. http://en.wikipedia.org/wiki/FASTA (24 December 2011, date last accessed)
- 3.BED. format. http://genome.ucsc.edu/FAQ/FAQformat.html (24 December 2011, date last accessed)
- 4.GFF. format. http://www.sanger.ac.uk/resources/software/gff/ (24 December 2011, date last accessed)
- 5.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WIG. format. http://genome.ucsc.edu/goldenPath/help/wiggle.html (24 December 2011, date last accessed)
- 7.Zhou P, Emmert D, Zhang P. Using Chado to store genome annotation data. Curr Protoc Bioinformatics. 2006;Chapter 9:Unit 9.6. doi: 10.1002/0471250953.bi0906s12. [DOI] [PubMed] [Google Scholar]
- 8.Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–46. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 9.Stajich JE, Block D, Boulez K, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–8. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister R, O'Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Down T, Piipari M, Hubbard T. Dalliance: interactive genome viewing on the web. Bioinformatics. 2011;27:889. doi: 10.1093/bioinformatics/btr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlin K, Fox D. Proceedings of the 20th Annual Conference on Computer Graphics and Interactive Techniques. Anaheim, CA, USA: ACM; 1993. Pad: an alternative approach to the computer interface; pp. 57–64. [Google Scholar]
- 13.Alekseyenko A, Lee C. Nested containment list (NCList): a new algorithm for accelerating interval query of genome alignment and interval databases. Bioinformatics. 2007;23:1386–93. doi: 10.1093/bioinformatics/btl647. [DOI] [PubMed] [Google Scholar]
- 14.Kircher M. Lazy Acquisition. Munich, Germany: Siemens AG; 2001. pp. pp. 1–11. [Google Scholar]
- 15.Stein L, Mungall C, Shu S, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2003;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stalker J, Gibbins B, Meid P, et al. The Ensembl web site: Mechanics of a genome browser. Genome Res. 2004;14:951–5. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkinson A, Albrecht M, Birney E, et al. Integrating biological data–the Distributed Annotation System. BMC Bioinformatics. 2008;9(Suppl 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]