Abstract
Data visualization is an essential component of genomic data analysis. However, the size and diversity of the data sets produced by today’s sequencing and array-based profiling methods present major challenges to visualization tools. The Integrative Genomics Viewer (IGV) is a high-performance viewer that efficiently handles large heterogeneous data sets, while providing a smooth and intuitive user experience at all levels of genome resolution. A key characteristic of IGV is its focus on the integrative nature of genomic studies, with support for both array-based and next-generation sequencing data, and the integration of clinical and phenotypic data. Although IGV is often used to view genomic data from public sources, its primary emphasis is to support researchers who wish to visualize and explore their own data sets or those from colleagues. To that end, IGV supports flexible loading of local and remote data sets, and is optimized to provide high-performance data visualization and exploration on standard desktop systems. IGV is freely available for download from http://www.broadinstitute.org/igv, under a GNU LGPL open-source license.
Keywords: visualization, next-generation sequencing, NGS, genome viewer, IGV
INTRODUCTION
Next-generation sequencing (NGS) and array-based profiling methods now generate large quantities of diverse types of genomic data and are enabling researchers to study the genome at unprecedented resolution. Analysis of these large, diverse, data sets has become the rate-limiting step in many studies. Although much of the analysis can be automated, human interpretation and judgment, supported by rapid and intuitive visualization, is essential for gaining insight and elucidating complex biological relationships. We describe the Integrative Genomics Viewer (IGV) [1], a high-performance desktop tool for interactive visual exploration of diverse genomic data. Even for very large data sets, IGV supports real-time interaction at all scales of genome resolution, from whole genome to base pairs. IGV is designed to be accessible to a wide range of users, including bench biologists and bioinformaticians. While new users appreciate the user-friendly and intuitive interface, more experienced users can also take advantage of the many advanced features and preferences.
There are a number of other desktop applications available for visualization of genomic data, particularly NGS data, including Tablet [2], BamView [3], Savant [4] and Artemis [5]. In comparison to these tools, a notable characteristic of IGV is its breadth. IGV was developed to support a diverse range of data types, including NGS and array-based platforms, such as expression and copy-number arrays. These different data types can be flexibly integrated, and combined with clinical and other sample metadata to dynamically group, sort and filter data sets. Another distinguishing feature of IGV is the ability to view data in multiple genomic regions simultaneously in adjacent panels, for example to view correlated events in distal regions.
HISTORICAL BACKGROUND
IGVs direction and focus are driven by our collaborations with investigators from a wide variety of large and small biomedical research projects. IGV development started in 2007 in response to a need by The Cancer Genome Atlas (TCGA) [6] project to visualize integrated copy number data, expression, mutation and clinical data. The size of these data sets posed a challenge to desktop visualization tools available at the time. To handle these large data sets in IGV, we introduced a data-loading scheme that includes indexing of data files as well as on-demand loading. This strategy enabled viewing and interactive exploration of up to several hundred samples from the largest copy-number array available at the time, comprising approximately half a million probes.
The next major application of IGV, visualization of ChIP-seq data from whole-genome sequences to find de novo long intergenic noncoding RNAs (lincRNAs) [7], provided motivation to extend the data size limit even further. We developed a binary multiresolution tiled data format to support data sets of up to hundreds of gigabytes in size. At this time, we also added support for viewing genome annotations. In August 2008, we deployed the first public release of the IGV software and web site (www.broadinstitute.org/igv).
Support for visualization of short-read sequence alignments followed in May 2009. In this effort, we collaborated with members of the 1000 Genomes Project [8] involved in development of the SAM/BAM alignment format [9]. Another collaboration with the 1000 Genomes Project, led to IGV support for visualizing genome variation data in the VCF format [10] in the IGV 2.0 release of May 2011.
In IGV 2.0, we also introduced a flexible, ‘multilocus’ mode for viewing multiple genomic regions, side by side and thereby eliminated the restriction of viewing only a single contiguous genomic region at a time. This new view mode is particularly useful when investigators seek to test hypotheses and draw inferences based on related events that are widely separated in genomic coordinates. IGVs multilocus views are described further in the ‘Features’ section below.
METHODS AND TECHNOLOGIES
IGV is a desktop application written in the Java programming language and runs on all major platforms (Windows, Mac and Linux). Below, we describe in more detail some components of the IGV implementation, including our data-tiling approach for supporting large data sets and IGVs support for different categories of file formats. We also provide a high-level overview of IGVs software architecture.
Data tiling
A primary design goal of IGV is to support interactive exploration of large-scale genomic data sets on standard desktop computers. This poses a difficult challenge as NGS and recent array-based technologies can generate data sets from gigabytes to terabytes in size. Simply loading these entire data sets into memory is not a viable option. In addition, researchers search for meaningful events at many different genomic resolution scales, from whole genome to individual base pairs. The problem is analogous to that faced by interactive geographical mapping tools, which provide views of very large geographical databases over many resolution scales. Tools such as Google Maps solve this problem by precomputing images representing sections of maps at multiple resolution scales, and providing fast access to the images as needed to construct a view. We considered such an approach for IGV, based on precomputed images of genomic data. However, millions of images would be required to support all resolution scales for a large genome, thus making image management difficult without introducing the requirement of a database. Furthermore, the representation of the data would be fixed when the images are computed, making it difficult to provide interactive graphing options. Consequently, we adopted a different approach that is based on precomputing summarizations of data at multiple resolution scales, with rendering of the data deferred to runtime. We refer to this as ‘data tiling’, to distinguish it from ‘image tiling’.
IGVs data tiling implementation is built on a pyramidal data structure that can be described as follows. For each resolution scale (‘zoom level’), the genome is divided into tiles that correspond to the region viewable on the screen of a typical user display. The first zoom level consists of a single tile that covers the entire genome. The next zoom level contains a single tile for each chromosome. The number of tiles then increases by a factor of 2 for each level, so the next zoom level consists of two tiles per chromosome, then four, etc. Each tile is subdivided into ‘bins’, with the width of a bin chosen to correspond to the approximate genomic width represented by a screen pixel at that resolution scale. The value of each bin is calculated from the underlying genomic data with a summary statistic, such as ‘mean’, ‘median’ or ‘maximum’. By organizing data in this way, tile sizes for each zoom level are constant and small, containing only the data needed to render the view at the resolution supported by the screen display. Hence, a single tile at the lowest resolution, which spans the entire genome, has the same memory footprint as a tile at a high-resolution zoom level, which might span only a few kilobases. As the user moves across the genome and through zoom levels, IGV only retrieves the tiles required to support the current view and discards tiles no longer in view to free memory. This method supports browsing very large data sets at all resolution scales with minimal memory requirements.
For large genomes, precomputing tiles for all zoom levels would be inordinately expensive with respect to disk space. For example, the human genome requires approximately 23 zoom levels, or on the order of 223 tiles, to cover the whole genome to base pair resolution.
In practice, IGV uses a hybrid approach; combining precomputed lower-level zoom levels with high-resolution tiles computed on the fly. This is possible as the high-resolution tiles cover relatively small portions of the genome. The number of precomputed zoom levels required to achieve good performance varies by data density and genome size. In our experience, seven levels give acceptable performance for even the highest density human genome data.
File formats
To support the multiresolution data model described earlier, we developed a corresponding file format. The ‘tiled data format’, or TDF, stores the pyramidal data tile structure and provides fast access to individual tiles. TDF files can be created using the auxiliary package ‘igvtools’. We note however that IGV does not require conversion to TDF before data can be loaded. In fact, IGV supports a variety of genomic file formats, which can be divided into three categories: (i) nonindexed, (ii) indexed and (iii) multiresolution formats:
Nonindexed formats include flat file formats such as GFF [11], BED [12] and WIG [13]. Files in these formats must be read in their entirety and are only suitable for relatively small data sets.
Indexed formats include BAM and Goby [14] for sequence alignments. Additionally, many tab-delimited feature formats can be converted to an indexed file using Tabix [15] or ‘igvtools’. Indexed formats provide rapid and efficient access to subsets of the data for display, but only when zoomed in to a sufficiently small genomic region. Zooming out requires ever-larger portions of the file to be loaded. Thus, indexed formats can efficiently support views only for a limited range of resolution scales. This range depends on the genomic density of the underlying data and can span tens of kilobases for NGS alignments, hundreds of megabases for typical variant (SNP) files, or whole chromosomes for sparse feature files. IGV uses heuristics to determine a suitable upper limit on the genomic range that can be loaded quickly with a reasonable memory footprint. If zoomed out beyond this limit, the data are not loaded.
Multiresolution formats, such as our TDF described earlier and the bigWig and bigBed formats [16], include both an index for the raw data, and precomputed indexed summary data for lower resolution (zoomed out) scales. Multiresolution formats can efficiently support views at any resolution scale.
Software architecture
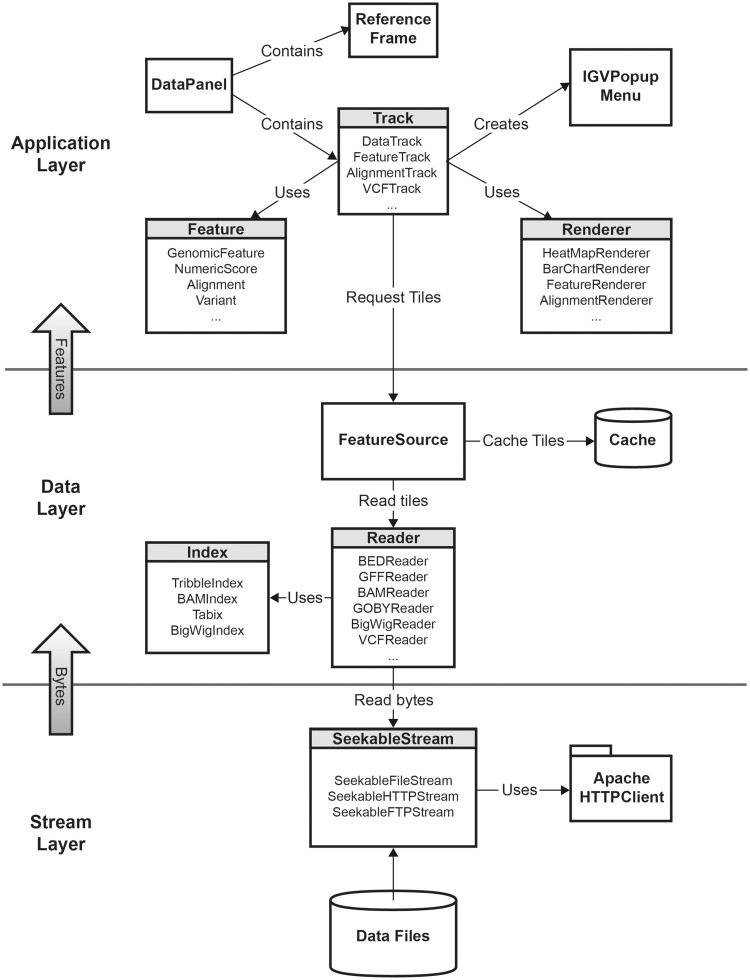
The IGV software structure is designed around a core set of interfaces and extendable classes. These components can be separated into three conceptual layers as illustrated in Figure 1: (i) a top-level application layer, (ii) a data layer and (iii) a stream layer. These are described in more detail below:
The application layer includes the main IGV window and user interface elements, along with controllers for user interaction events. It also contains representations of genomic features and data. IGV displays these in horizontal rows known as ‘tracks‘. Tracks are displayed in a data panel, which is implemented as a class derived from Java Swing components. The data panel is responsible for the coordination of track layout and rendering, and managing mouse events. It handles certain globally shared mouse actions, such as zooming and panning, and delegates other events to the objects representing tracks. Tracks are responsible for handling these events, as well as requesting features as needed from the data layer and drawing these features on the panel. Most track implementations delegate the drawing task to a renderer object. Renderers are designed to be pluggable, and can be swapped at runtime, for example to switch graph types in response to a menu action.
The data layer reads and parses the different genomic file formats and supplies the application layer with data tiles on demand. It also implements caching of tiles, for improved efficiency if a previously visited genomic region is requested again.
The stream layer is responsible for supporting random access to sections of files accessed by any of the protocols supported by IGV, i.e. local file, HTTP, HTTPS or FTP. Random file access is necessary to take advantage of the indexed and multiresolution file formats. For local files, this is straightforward using Java’s RandomAccessFile class, or alternatively positionable file channels. Remote files presented a challenge, as there are no Java built-in functions or libraries that support this access pattern. Initially, we solved this problem using a web service. However, this approach was not ideal, as users who wished to host IGV files were also required to install and run the web service on their systems. Consequently, we designed and implemented a set of classes to provide a uniform interface for random file access for all the protocols. IGVs implementation for the HTTP protocol uses byte range requests from the standard HTTP specification. For the FTP protocol, IGV uses the mechanism for restarting downloads that is supported by most FTP servers via the ‘REST’ command.
Figure 1:
IGV class diagram, illustrating the IGV software structure.
FEATURES
IGV is a desktop application for the visualization and interactive exploration of genomic data in the context of a reference genome. A key characteristic of IGV is its focus on the integrative nature of genomic studies. It allows investigators to flexibly visualize many different types of data together—and importantly also integrate these data with the display of sample attribute information such as clinical and phenotypic information. To support interactive exploration of data, IGV provides direct manipulation navigation in the style of Google Maps. For instance, you click and drag to pan the view across the genome and double-click on a region to zoom in for a more detailed view. It supports real-time interaction at all scales of genome resolution, from whole genome to base pairs, even for very large data sets. The Broad IGV data server hosts many genome annotation files and data sets from a variety of public sources (including from TCGA, 1000 Genomes Project, ENCODE Project [17] and others). However, the primary emphasis is on supporting biomedical researchers who wish to load, visualize and explore their own data sets aligned to the selected reference genome. Researchers can also make their data sets available to others for view in IGV, sharing them with colleagues or the community at large.
Launching IGV
IGV is available on all platforms that support Java. Installation and launching are accomplished with a click of a button on the IGV web site at www.broadinstitute.org/igv. Alternatively, users can download a ZIP archive to install the application locally. IGV can also be launched from links embedded in web pages or other documents.
The IGV window
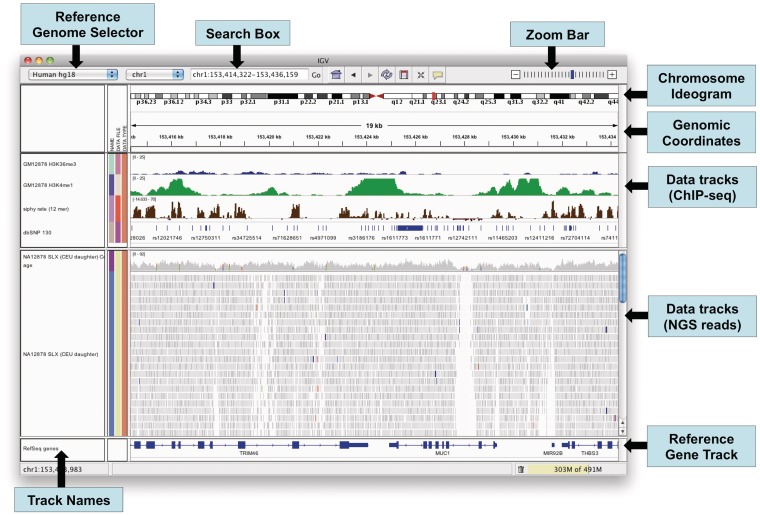
The IGV window is divided into a number of controls and panels as illustrated in Figure 2. At the top is a command bar with controls for selecting a reference genome, navigating and defining regions of interest. Just below the command bar is a header panel with an ideogram representation of the currently viewed chromosome, along with a genome coordinate ruler that indicates the size of the region in view. The ideogram also displays a red rectangle that outlines the region in view. The remainder of the window is divided into one or more data panels and an attribute panel. Data are mapped to the genomic coordinates of the reference genome and are displayed in the data panels as horizontal rows called ‘tracks’. Each track typically represents one sample, experiment or genomic annotation. If any sample or track attributes have been loaded, they are displayed as a color-coded matrix in the attribute panel as illustrated in Figure 3. Each column in the matrix corresponds to an attribute, and a track’s attribute values are displayed as a row of colored cells adjacent to the track.
Figure 2:
The IGV application window.
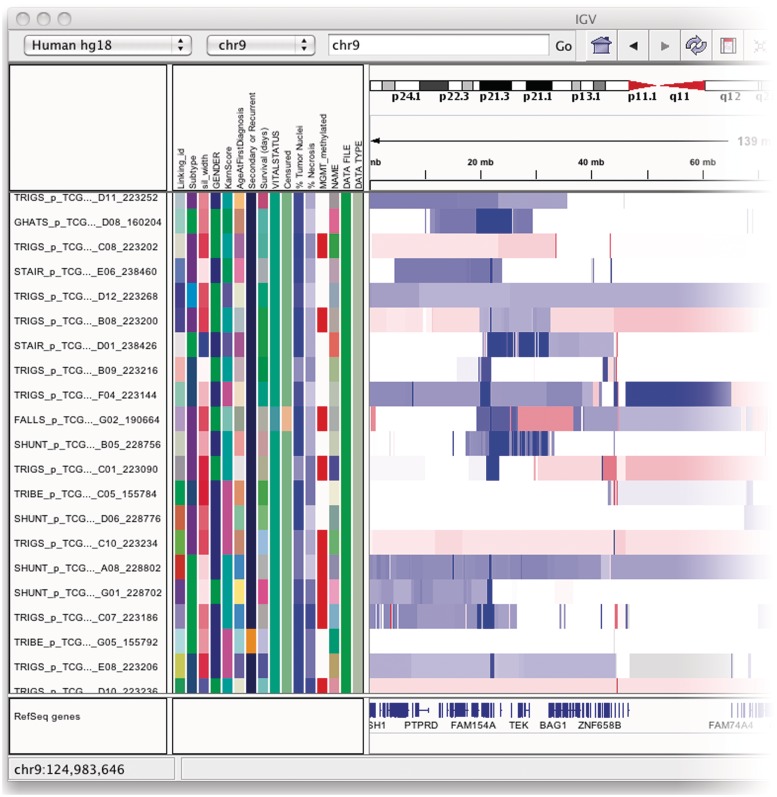
Figure 3:
The attribute panel displays a color-coded matrix of phenotypic and clinical data. Clicking on a column header will sort the tracks by the corresponding attribute.
Reference genome
A reference genome must be selected before loading data. IGV provides dozens of hosted reference genomes to choose from, but also provides the option of importing others from the sequence data. The minimal requirement for importing a genome is a FASTA file containing chromosome or contig sequences. Other genome information is optional, including: (i) cytoband information for the chromosome ideogram in the IGV window, (ii) annotations defining the features displayed in the gene track for the genome and (iii) chromosome alias information that defines synonyms for the sequence names defined in the FASTA file, e.g. ‘1’ for ‘chr1’. We note that this option is intended primarily for finished assemblies. IGV is not designed for visualization of unassembled genomes.
When the view is sufficiently zoomed in, IGV displays the reference genome sequence as a separate track in the data panel. Depending on the zoom level, the nucleotides are represented as colored bars or letters. By default, the forward strand is displayed. Clicking on a strand indicator for the track toggles the strand direction. Another option enables the display of three-frame translation of codons for the current strand.
Loading data
IGV was designed to accommodate any data that can be mapped to genomic coordinates. It currently supports more than 30 different file formats, including many of the common formats for genome annotations, sequence alignments, variant calls and microarray data. Importantly, users can also load metadata, such as clinical, phenotypic or other attribute information, to annotate the genomic data. Data files are loaded into IGV by: (i) using the built-in file browser to select a file on the local file system, (ii) entering the URL of a file accessible over a network via HTTP or FTP, (iii) entering the URL of a Distributed Annotation System (DAS) feature source [18] or (iv) selecting entries from the ‘File > Load from Server’ menu. By default, the menu provides access to data and annotation files that are hosted at the Broad Institute for viewing in IGV. This can easily be changed to point to any set of web-accessible files. For example, the menu could provide access to shared files on a research project’s central server.
Viewing data
IGV supports simultaneous viewing of multiple data sets, with the same or different types of data. A track’s default appearance and available view options will vary depending on the data type. The following sections describe some of the commonly viewed types.
NGS data
IGV includes a large number of specialized features for exploring NGS read alignments, including features tailored for variant visualization and validation, splicing of RNA transcripts and methylation from bisulfite sequencing. IGV supports several read alignment file formats, including SAM, BAM and Goby. When more than one file is loaded into IGV, it can display the reads from each file in a separate panel or merge them together as if they came from the same file.
Due to the magnitude of the data stored in NGS alignment files, IGV displays varying level of data detail depending on the zoom level. This is necessary for both application performance, as described in the ‘Methods and Technologies’ section above, and to help investigators make sense of the massive amount of data. When viewing a whole chromosome it is not useful to display all the reads, as individual reads are not distinguishable in the view at this level of zoom. Therefore, when zoomed all the way out, only a bar chart of the read coverage is displayed. IGV provides tools to precompute this coverage. When zoomed in past a user-settable visibility threshold (by default, 30 kb), the individual read alignments come into view and are displayed as horizontal bars. At this zoom level, IGV dynamically computes the read coverage in the viewed region. Zooming further reveals the individual read bases.
IGV uses color and transparency to highlight interesting events in the alignment data and to visually deemphasize others—in the coverage chart, at the read level and for individual bases. Various properties can be used to change the read color scheme on the fly, and to interactively group, sort and filter the reads. These features can be used alone or in combination, using one or more properties. The properties include sample identifier, strand, read group, mapping quality, base call at a particular position, pair distance and orientation, custom tags and more.
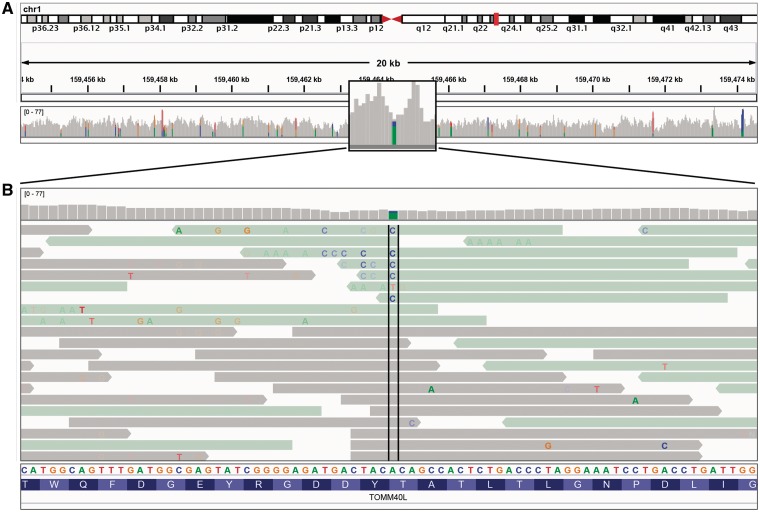
Zooming in past the alignment visibility threshold will also add color bars to the gray coverage track at locations where a large number of read bases mismatch the reference—helpful in identifying putative SNPs. The relative size and color of these bars indicates the allele frequency of each base at that location. An example of this can be seen in Figure 4A. In Figure 4B, the view has been zoomed in further to show the reads and individual bases in the region of one of the putative SNPs identified in the coverage track. Individual read bases that match the reference genome are displayed in the same color as the read, while mismatches are color-coded by the called base and are assigned a transparency value proportional to the base call quality (phred) score. This is the default coloring scheme for read bases, and has the effect of emphasizing high-quality mismatches. In this example, the read alignments have been colored and sorted by read strand. Visual inspection quickly reveals a number of factors that indicate this is not a true SNP. First, the reads harboring the putative SNP clearly have a large number of additional mismatched bases. Also, it is suspicious that all mismatches occur on the negative read strand, and that the mismatches tend to occur towards the end of the read.
Figure 4:
Read alignment views at 20 kb and base pair resolution. IGV displays varying level of data detail depending on the zoom level, and uses color and transparency to highlight interesting events in the data. (A) Reads are summarized as a coverage plot. Positions with a significant number of mismatches with respect to the reference are highlighted with color bars indicative of both the presence of mismatches and the allele frequency. (B) Individual base mismatches are displayed with alpha transparency proportional to quality. In this example, the reads have been sorted and colored by strand.
A number of options are available for paired-end alignments to help elucidate structural variants, such as deletions, insertions and rearrangements. To highlight potential inter-chromosomal rearrangements, alignments whose mate falls on a different chromosome are assigned a color indicating the mate chromosome. This makes it easy to distinguish potential rearrangements from noise caused by misalignments, as rearrangements will appear as a pileup of reads that are consistent in color and orientation. Intra-chromosomal events, such as insertions, deletions, inversions and duplications, can affect both the genomic distance between the mate alignments, as well as their orientation. To highlight these events, IGV samples the alignment file to dynamically determine the expected distance and orientations, and then uses color to flag aberrant pairs.
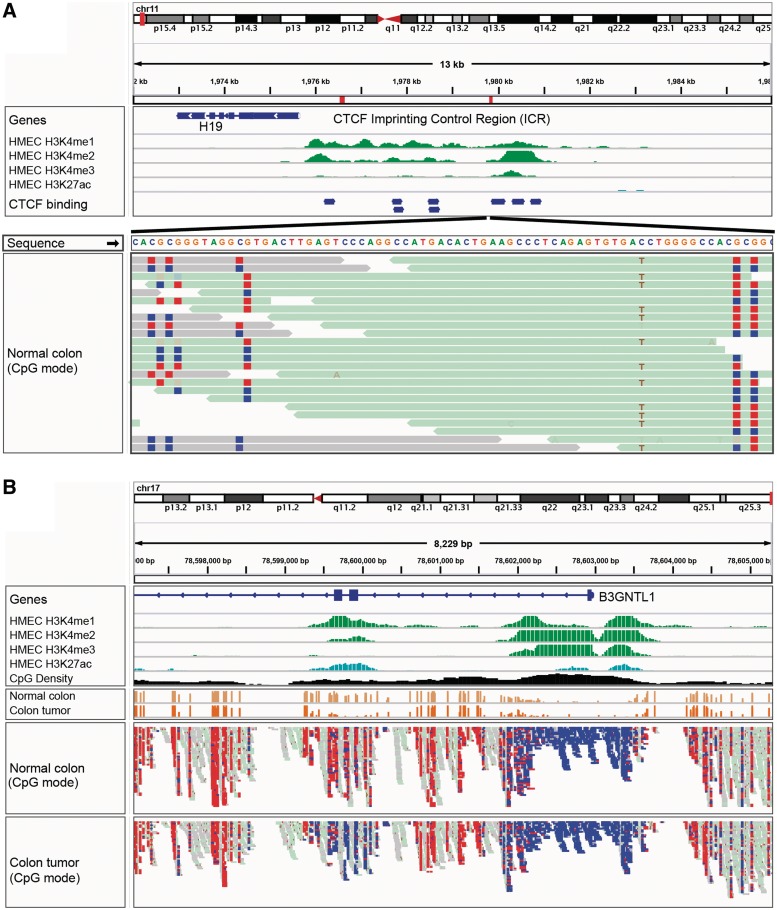
Another coloring scheme is used to view bisulfite sequencing data. In this mode, the rules for what constitutes a mismatch to the reference genome are adjusted to account for the expected cytosine to uracil conversions. Figure 5 illustrates a view of Whole-Genome Bisulfite Sequencing (WGBS) data from a colorectal tumor and a matched normal sample [19]. Red indicates hypermethylated sites, and blue indicates hypomethylation.
Figure 5:
IGV bisulfite sequencing view. (A) Two views of the IGF2/H19 Imprinting Control Region (ICR), illustrating allele-specific methylation of CTCF binding sites. The top view shows a 13-kb region of ChIP-seq histone marks from the ENCODE normal epithelial tissue (HMEC) cell line. The second view shows WGBS read alignments from normal colonic mucosa [19], zoomed in to 75 bp. CpG dinucleotides are shown as blue (unmethylated) and red (methylated) squares. A heterozygous C/T SNP is also apparent, and the T allele is overwhelmingly associated with reads that have methylated CpGs (from the paternal chromosome). (B) The enhancer region surrounding exons 2 and 3 of the B3GNTL1 gene is apparent from the ENCODE tracks showing characteristic enhancer histone marks in a normal epithelial (HMEC) cell line. The bisulfite sequencing view of the read alignments shows that this enhancer is methylated (red – lighter) in normal colon mucosa, but almost completely unmethylated (blue – darker) in the matched colon tumor sample [19]. The cancer-specific de-methylation of this enhancer is consistent with the upregulation of the B3GNTL1 transcript in the tumor.
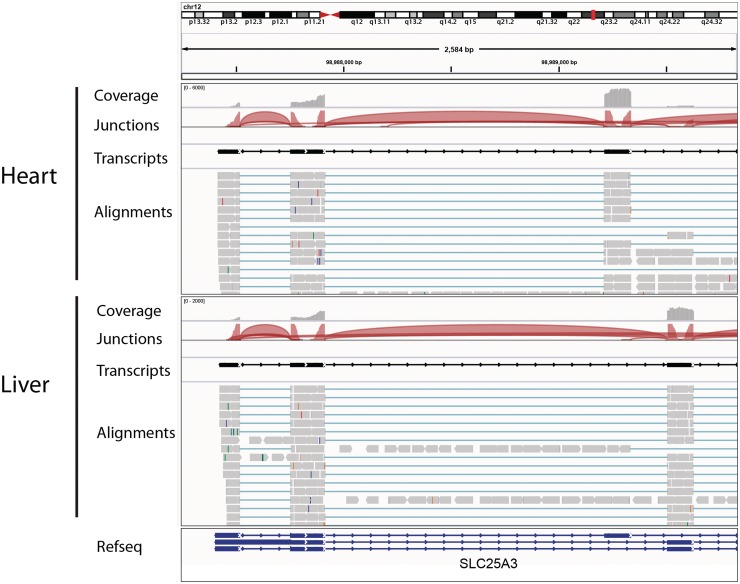
Figure 6 shows how IGV displays RNA-seq read alignments, connecting segments of reads that are split across exons with thin horizontal lines. This example demonstrates several RNA-seq data tracks for normal tissue samples from two different organs, including tracks for coverage, junctions, transcripts and the read alignments. The junction tracks highlight alternative splicing, also visible in the alignments themselves.
Figure 6:
Visualization of RNA-seq data from heart and liver tissue samples. Each panel includes tracks for total coverage, junction coverage, predicted transcripts and read alignments. Reads that span junctions are connected with thin blue lines. In the junction track, the height of each arc is proportional to the total number of reads spanning the junction. There is clear evidence of alternative splicing between the two tissues.
Variant calls
IGV provides extended support for viewing variants stored in the VCF format. This format allows for the encoding of variant calls (SNPs, indels and genomic rearrangements) as well as the supporting genotype information for individual samples. Samples can also be annotated with attribute information, including pedigree and family information. IGV uses these annotations to group, sort and filter samples, for example, to group samples by pedigree or population group.
Copy number and expression data
Copy number and expression data can be loaded from a variety of file formats. These data types are displayed as heatmaps by default. Heatmaps are very space efficient in comparison to bar charts and other graph types, as the height of each track can be reduced to a single pixel. This is important for these data types as experiments are often performed on high-throughput arrays and it is not uncommon to view hundreds or thousands of samples simultaneously.
Expression data require special treatment as the expression values are usually not specified in genomic coordinates, but rather are associated with gene names or chip probe identifiers. These data must be mapped to genomic locations prior to display and IGV provides several options for this step: (i) Automatically map data values associated with gene names, based on information in the gene track of the reference genome. (ii) Automatically map data values that are associated with probes and probe-sets for many common platforms and chips, such as those from Affymetrix, Agilent and Illumina, based on files published by the vendors. By default, IGV maps the data values to the loci of individual probe sets, which typically cover a small portion of a gene, but users can choose to have values mapped to the entire gene locus instead. (iii) Perform mappings provided by the user in the input gene expression file.
Genomic annotations
IGV supports a number of formats for genomic annotations, including BED, GFF, GTF2 [20] and PSL [21]. Visual representation of annotations follows many of the conventions introduced by the UCSC Genome Browser. For example, gene exonic regions are displayed as solid blocks connected by thin lines representing introns. By default, annotations are drawn on a single row in ‘collapsed’ mode. Tracks that contain overlapping features, such as multiple isoforms for a gene, can be expanded to reveal all features.
Sample attributes
Tracks can be annotated with metadata by loading a tab-delimited sample information file. The metadata might include, for example, clinical, experimental or computational data such as patient identifier, pedigree, phenotype, outcome, cluster membership, etc. Metadata is displayed as a color-coded matrix in the attribute panel. Each column in the matrix corresponds to a specific attribute, and colors are used to distinguish different values of that attribute. Colors are assigned explicitly by the user or chosen automatically by IGV. Numeric attributes can have a one- or two-color continuous color scale in lieu of specific colors for each value.
Interacting with the view
IGV provides a variety of ways to interact with views of the data. To select the genomic region to view and the zoom level, an investigator can enter genomic coordinates, search for genomic features by name, zoom in or out by clicking on the railroad track control in the upper right of the window or by using mouse shortcuts, and pan by click-dragging in the data panel. Tracks can be grouped and filtered, based on one or more sample attribute values. They also can be sorted based on attribute values or on data values in a genomic subregion. IGV selects default display parameters, such as graph type and colors, for each track, based on the data type and any preferences the user may have set. A right click on any track will bring up a menu with display options specific to the type of data displayed in the track. A preferences window also provides many data type-specific options.
Viewing multiple genomic regions
With the IGV 2.0 release, we introduced a flexible, ‘multilocus’ mode to support viewing multiple genomic regions, side by side. In this view, the data panel in the IGV window is subdivided into a series of vertical panels, one for each region, all displaying the same set of tracks. All track manipulation features, e.g. grouping, overlaying and sorting functions, are available and applied simultaneously to all panels. Panning and zooming within each panel are supported and the user can change the order of the panels by dragging them to different locations in the window. Currently, IGV supports two different types of multilocus views.
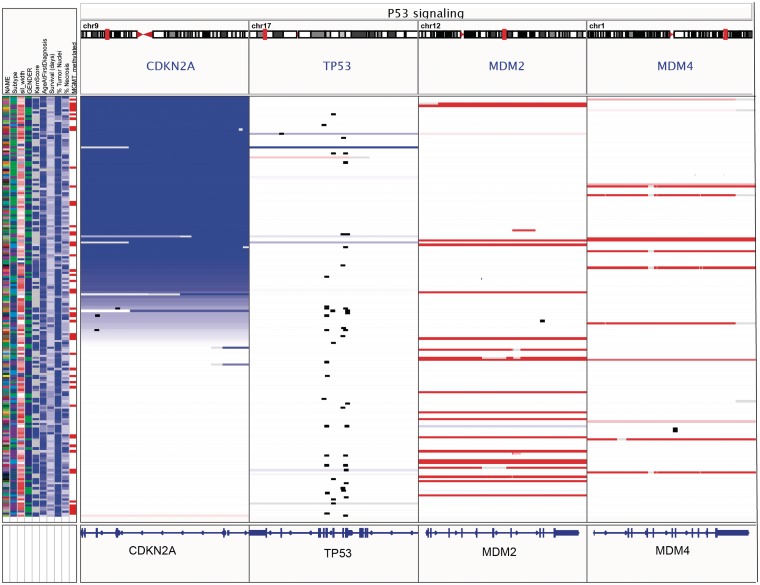
The first type of multilocus view is invoked by specifying regions, by genomic locus or gene name, either by entering them in the search box or using the ‘Gene Lists’ option from the IGV menu. IGV provides a number of predefined gene lists representing pathways from public databases, and users can also create and save their own lists. This is illustrated in Figure 7, with a view of copy number, mutation and clinical data from 202 glioblastoma samples from the TCGA project [22]. The window has been split into panels corresponding to four genes from the p53 signaling pathway.
Figure 7:
Gene-list view of copy number, mutation and clinical data from 202 glioblastoma samples from the TCGA project. The IGV window has been split into panels corresponding to four genes from the p53 signaling pathway. Copy number is indicated by color, with blue denoting deletion and red amplification. Mutations are overlaid as small black rectangles. The samples have been sorted by copy number of CDKN2A. In this view it is apparent that deletion of CDKN2A and mutation of TP53 tend to be mutually exclusive.
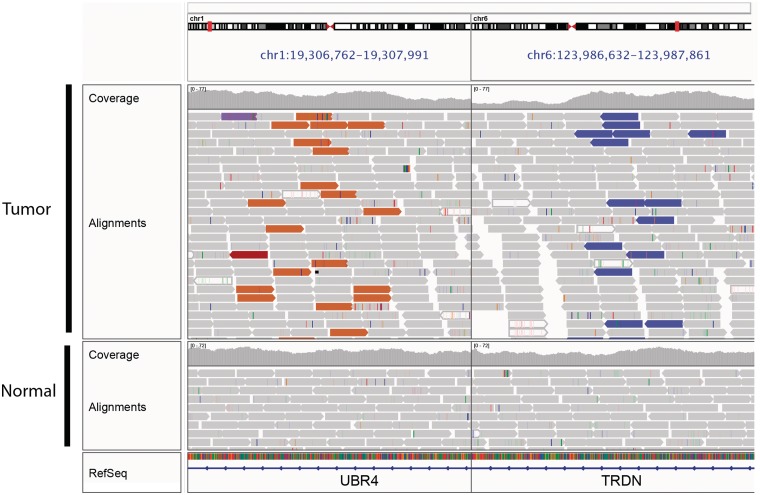
The second type of multilocus view is useful for viewing paired-end sequence read alignments when the mates are aligned to distant regions of the genome. Selecting a read and choosing ‘Show Mate Region’ from the options menu will split the view and display the genomic regions of both mates. Figure 8 illustrates viewing both sides of a balanced translocation in a glioblastoma tumor sample.
Figure 8:
Split-screen view of read alignments from a glioblastoma multiforme tumor sample and matched normal, displaying regions of chromosomes 1 and 6. In this example, alignments whose mate pairs are mapped to unexpected locations are color-coded by the chromosome of the mate; other alignments are displayed in light gray. The brown alignments on the left panel and purple alignments on the right are matepairs, indicating a fusion between these loci. There is no evidence of this rearrangement in the matched normal.
Saving and sharing sessions
Users can save the current state of an IGV session to a file. This file stores information on which data sets are loaded (the data sets themselves are not stored), how they have been grouped and sorted, and the current view and zoom level. Saved session files can be sent to collaborators to replicate the same view, as long as they have access to the same data files. It is also possible to share sessions with remote users by putting the session file, along with the associated data files, in a web-accessible location. Others can then reproduce the session in IGV by using an HTML link of the form: http://www.broadinstitute.org/igv/projects/current/igv.php?sessionURL=URL&locus=locus.
For example, the following link opens IGV on the ‘gbm_subtypes_session.xml’ session file hosted at the Broad Institute and goes to the specified locus on chromosome 7. http://www.broadinstitute.org/igv/projects/current/igv.php?sessionURL=http://www.broadinstitute.org/igvdata/tcga/gbmsubtypes/gbm_subtypes_session.xml&locus=chr7:55054218-55206232.
Controlling IGV
As a desktop application, the most common mode of interaction with IGV is through a graphical user interface. However, external programs can also control IGV using the following interfaces: (i) a batch script interpreter, (ii) a socket port interface and (iii) an HTTP interface:
Scripting allows many IGV actions to be automated, such as loading data, navigation, sorting and image generation. This enables visiting a large number of genomic sites quickly and producing image snapshots that can later be viewed offline. Often, this is used to visually validate a large number of variant sites and flag those needed for follow-up inspection.
The port interface can support similar use cases, but also makes it possible to control IGV from any language or tool that can write to a socket. For example, MATLAB users have used this capability to tie IGV to interactive analyses, loading files and jumping to loci in response to commands from MATLAB functions.
The HTTP interface supports creating links to launch IGV on a specific data set or send data to an already running IGV. Users can easily embed these links in their own pages and documents. This feature has been used to launch IGV from Excel spreadsheets, Word documents and to view data presented by web portals, including the Tumorscape Portal [23] and the cBio Cancer Genomics Portal [24].
Utilities
The ‘igvtools’ provide a set of utilities for preprocessing data files. These include utilities for: (i) converting files to the Binary Tiled Data (TDF) format for faster loading and retrieval of large data sets, (ii) computing read alignment coverage, (iii) computing feature density and (iv) creating an index file for feature files and text SAM alignment files. The ‘igvtools’ utilities can be run from the IGV user interface, or downloaded as a separate package from the web site and run from the command line.
FUTURE DIRECTIONS
While originally developed for use in cancer genome characterization studies, IGV is now used extensively in a broad range of basic biology and biomedical studies, and will continue to evolve with the changing needs of the biomedical research community. Here, we name a few of the opportunities and challenges on the immediate horizon. First, the increasing scale of NGS data sets will continue to challenge the capabilities of existing visualization tools, including the IGV. Even now, large studies can comprise hundreds to thousands of whole-genome and whole-exome sequencing experiments. Visualization of these data will require new approaches for aggregating data intelligently to reveal trends, while continuing to provide access to lower-level details on demand. We also anticipate the need for augmenting IGV with auxiliary nongenomic views, such as network views to highlight functional relationships in a pathway. Finally, we plan to couple the IGV with external tools to enable intelligent data-driven search and navigation.
Key Points.
The IGV is a high-performance desktop viewer that efficiently handles large heterogeneous data sets, while providing a smooth and intuitive user experience at all levels of genome resolution.
IGV allows researchers to visualize many different types of genomic data together, including NGS data, variant calls, microarray data and genome annotations. Importantly, it also supports integrating metadata, such as clinical, phenotypic and other attribute information.
IGV provides flexible and fast loading of local and remote data sets. For indexed files, IGV loads data as needed for regions in view, thereby minimizing memory usage and data transfer of remote files.
IGV has a flexible ‘multilocus’ mode that supports viewing multiple genomic regions, side by side.
IGV is freely available at http://www.broadinstitute.org/igv.
FUNDING
National Institute of General Medical Sciences (R01GM074024); National Cancer Institute (R21CA135827); National Human Genome Research Institute (U54HG003067) and Starr Cancer Consortium (I5-A500).
Acknowledgements
We thank the following collaborators for their contributions to components described in this manuscript: Damon May, Fred Hutchinson Cancer Research Center, for the RNA-seq splice junction viewer; Fabien Campagne, Campagne Laboratory, Institute for Computational Biomedicine, Weill Cornell Medical College, for the Goby alignment format modules and Benjamin Berman of the USC Epigenome Center for the bisulfite sequencing components.
Biographies
Helga Thorvaldsdóttir is a senior software project manager in the Cancer Program at the Broad Institute of MIT and Harvard.
James T. Robinson is a principle software engineer in the Cancer Program at the Broad Institute of MIT and Harvard, where he has worked on omics visualization software since 2006.
Jill P. Mesirov is Chief Informatics Officer of the Broad Institute of MIT and Harvard, where she directs the Computational Biology and Bioinformatics Organization, and a member of the Koch Institute for Integrative Cancer Research at MIT.
References
- 1.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milne I, Bayer M, Cardle L, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carver T, Bohme U, Otto TD, et al. BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformatics. 2010;26:676–7. doi: 10.1093/bioinformatics/btq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiume M, Williams V, Brook A, et al. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–44. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutherford K, Parkhill J, Crook J, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanger Institute. GFF: an Exchange Format for Feature Description. http://www.sanger.ac.uk/resources/software/gff/ (21 December 2011, date last accessed)
- 12.UCSC Genome Bioinformatics. BED Format. http://genome.ucsc.edu/FAQ/FAQformat.html#format1 (21 December 2011, date last accessed)
- 13.UCSC Genome Bioinformatics. Wiggle Track Format (WIG) http://genome.ucsc.edu/goldenPath/help/wiggle (21 December 2011, date last accessed)
- 14.Campagne Laboratory, Institute for Computational Biology, Weill Cornell Medical School. Goby. http://campagnelab.org/software/goby/ (21 December 2011, date last accessed)
- 15.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–9. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent WJ, Zweig AS, Barber G, et al. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–7. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 18.Dowell RD, Jokerst RM, Day A, et al. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40–6. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brent Laboratory, Washington University in St. Louis. GTF2 Format (Revised Ensembl GTF). http://mblab.wustl.edu/GTF2.html (21 December 2011, date last accessed)
- 21.UCSC Genome Bioinformatics. PSL Format. http://genome.ucsc.edu/FAQ/FAQformat.html#format2 (21 December 2011, date last accessed)
- 22.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Memorial Sloan-Kettering Cancer Center. cBio Cancer Genomics Portal. http://www.cbioportal.org/ (21 December 2011, date last accessed)