Abstract
A long-standing question in evolutionary biology concerns the effect of recombination in shaping the genomic architecture of organisms and, in particular, how this impacts the speciation process. Despite efforts employed in the last decade, the role of chromosomal reorganizations in the human–chimpanzee speciation process remains unresolved. Through whole-genome comparisons, we have analyzed the genome-wide impact of genomic shuffling in the distribution of human recombination rates during the human–chimpanzee speciation process. We have constructed a highly refined map of the reorganizations and evolutionary breakpoint regions in the human and chimpanzee genomes based on orthologous genes and genome sequence alignments. The analysis of the most recent human and chimpanzee recombination maps inferred from genome-wide single-nucleotide polymorphism data revealed that the standardized recombination rate was significantly lower in rearranged than in collinear chromosomes. In fact, rearranged chromosomes presented significantly lower recombination rates than chromosomes that have been maintained since the ancestor of great apes, and this was related with the lineage in which they become fixed. Importantly, inverted regions had lower recombination rates than collinear and noninverted regions, independently of the effect of centromeres. Our observations have implications for the chromosomal speciation theory, providing new evidences for the contribution of inversions in suppressing recombination in mammals.
Keywords: recombination, speciation, human, hemiplasy, chimpanzee, inversions, reorganizations
Introduction
Reorganization (shuffling) of the genomic landscape plays an important role in the evolutionary processes as well as in the development of inherited diseases and carcinogenesis. Traditionally, it has been argued that chromosomal reorganizations may contribute to speciation due to the underdominant fitness effects associated with meiotic abnormalities, and the creation of unbalanced gametes in heterozygotes (White et al. 1978). But this model has important limitations given that “underdominance” is likely to occur in small, inbred populations, or when rearrangements are weakly underdominant individually, but strongly underdominant in combination (White et al. 1978; King 1993), and are difficult to test in natural populations. More recently, a number of related studies have proposed an alternative explanation by which chromosomal rearrangements could reduce gene flow and potentially contribute to speciation by the suppression of recombination (Noor et al. 2001; Rieseberg 2001). According to this “suppressed recombination” model, chromosome rearrangements could have a minimal influence on fitness, but would suppress recombination leading to the reduction of gene flow across genomic regions and to the accumulation of incompatibilities. Recombination provides physical connections between homologs during the first meiotic division, contributing to correct chromosomal segregation. Although recombination can occur at the somatic level (such as V(D)J recombination produced in the immune system), only those recombination events occurring in the germ line are relevant for the speciation process. Recombination introduces inheritable new chromosomal variants that can become fixed with a probability that depends on various population genetic parameters (i.e., frequency, effective population size, among others), contributing, in the long term, to the formation of new species. Few empirical data are available that address the mechanisms by which new chromosomal variants are fixed in populations of mammalian species, and how recombination influences chromosomal speciation and vice versa. In this regard, two models [Kirkpatrick–Barton model (Kirkpatrick and Barton 2006) and Navarro–Barton model (Navarro and Barton 2003)] have been formulated to explain 1) how, under divergent selection, chromosomal rearrangements can be fixed in two parapatric populations (in the presence of gene flow) and 2) by which mechanisms these contribute to speciation (revised in Faria and Navarro 2010). In both case, these formulations require a reduction of recombination between heterokaryotyes (chromosomes displaying alternate forms for rearrangements) as a crucial factor for speciation in parapatry.
Direct and indirect evidences of suppressed recombination within rearranged segments have been reported in the literature (Brown and O'Neill 2010; Faria and Navarro 2010). Direct evidence includes the analysis of recombination in the gametes and/or the offspring when reorganization (inversions and/or translocations) occurs. Data supporting recombination suppression by inversions has been provided by early cytogenetic studies in mammals, especially rodents (Ashley et al. 1981; Greenbaum and Reed 1984; Hale 1986) and Drosophila (Navarro and Ruiz 1997; Navarro et al. 1997). Hale (1986) described heterosynapsis (asynapsis) and, therefore, suppression of chiasmata (chromosomal configurations resulted from meiotic crossovers [COs]) formation within heterozygous pericentric inversions in the Sitka deer mouse (Peromyscus sitkenssi) as a mechanism for the maintenance of pericentric inversion polymorphisms in wild populations. Borodin et al. (Borodin et al. 2008) detected a reduction in MLH1 foci (a meiotic protein that marks COs) in translocated chromosomes of the common shrew (Sorex araneus), whereas two independent studies (Castiglia and Capanna 2002; Dumas and Britton-Davidian 2002) have reported a reduction in chiasmata number in house mice (Mus musculus domesticus) with Robertsonian (Rb) translocations.
On the other hand, indirect evidence for the suppression of recombination has included the analysis of rates of genetic divergence between rearranged and collinear chromosomes (Navarro and Barton 2003). High rates of sequence divergence detected in genes located at, or near, chromosomal rearrangements have been interpreted as indirect evidence of chromosomal speciation through suppressed recombination (Marques-Bonet et al. 2007). This latter approach has been used in several studies on Drosophila (Brown et al. 2004; Kulathinal et al. 2008), Helianthus sp. (sunflower) (Rieseberg et al. 1995, 1999), Solanaceae (Rieseberg and Willis 2007), and Anopheles (Besansky et al. 2003; Michel et al. 2006). In mammals, two studies (Yannic et al. 2009; Franchini et al. 2010) detected reduced gene flow within the reorganized regions (Rb translocations in heterokaryotypes) in house mouse and shrew populations, respectively, probably as a consequence of a fall-off in recombination around the centromeric regions. The study of the human and chimpanzee, on the other hand, (Navarro and Barton 2003) has provided contradictory results so far (Vallender and Lahn 2004; Zhang et al. 2004; Marques-Bonet and Navarro 2005; Marques-Bonet et al. 2007) and demonstration of recombination suppression in these species remains elusive.
Moreover, if genomic shuffling is affecting evolutionary and speciation processes, through the mechanical shearing at evolutionary breakpoints, how does this reorganization impact meiotic recombination? The assumption that some chromosomal regions have been reused during the mammalian chromosomal evolution (Murphy et al. 2005; Ruiz-Herrera et al. 2006; Larkin et al. 2009; Robinson and Ruiz-Herrera 2010) has lead evolutionary biologists to investigate whether there is any particular DNA configuration or composition underpinning genomic instability. In this sense, it has been reported that breakpoint regions co-localize with fragile sites (Ruiz-Herrera et al. 2005) and are enriched in repetitive elements, such as tandem repeats (Ruiz-Herrera et al. 2006; Farre et al. 2011), segmental duplications (Bailey and Eichler 2006; Kehrer-Sawatzki and Cooper 2008), and transposable elements (Caceres et al. 1999; Bourque 2009; Carbone et al. 2009; Delprat et al. 2009; Longo et al. 2009; Farre et al. 2011). However, few empirical data focus on the relationship between evolutionary breakpoint regions (EBRs) and recombination rates. Initial studies in Drosophila have described a strong reduction of recombination around inversion breakpoints and within the reorganization itself (Navarro et al. 1997). But the question whether this pattern also holds for mammals (in our study human and chimpanzee) remains unanswered despite the efforts in the last decade (Navarro and Barton 2003; Vallender and Lahn 2004; Zhang et al. 2004; Marques-Bonet and Navarro 2005; Marques-Bonet et al. 2007). Here, we analyze the recombination rate in homologous synteny blocks ([HSBs] i.e., regions where the gene order has been conserved among species) and EBRs (i.e., regions where the synteny has been disrupted due to genome shuffling—see Ruiz-Herrera et al. 2006) in the human and chimpanzee genomes by taking advantage of the most recent human and chimpanzee recombination map inferred from genome-wide single-nucleotide polymorphism (SNP) data (Kong et al. 2010; Auton et al. 2012). Moreover, we determine whether chromosomal reorganizations (i.e., inversions) may have had a genome-wide impact in the distribution of human and chimpanzee recombination rates. Overall, our data provide compelling evidence for the existence of low recombination rates within genomic regions that have been rearranged in the chromosomal evolution of human and chimpanzee.
Results
Whole-Genome Comparisons between Human and Chimpanzee Genomes
A total of 17,360 orthologous genes between the human and chimpanzee genomes and 16,409 orthologous genes between the human and orangutan were used in our estimations of EBRs. We identified 43 HSBs and 37 EBRs in the human genome, ranging from 5 bp to 171 kb with a median length of 9.8 kb (supplementary table S1, Supplementary Material online). Overall, we confirmed and refined the breakpoints involved in nine large inversions detected by previous studies (Kehrer-Sawatzki and Cooper 2008), affecting homologous chromosomes 1, 4, 5, 9, 12, 15, 16, 17, and 18, in addition to the fusion responsible for human chromosome 2. Additionally, we detected four indels (insertion/deletions) (three of them in chromosome 2 and one in chromosome 10) and 8 microinversions (less than 4.3 Mb) affecting chromosomes 1, 7, 10, 19, X and Y. As a whole, macrorearrangements encompassed 318 Mb of the whole human genome (ranging from 12.3 to 77.36 Mb, with a median size of 40 Mb), whereas the microrearrangements spanned 10.8 Mb, ranging from 12.5 kb to 4.3 Mb. Finally, we divided the rearranged chromosomes into regions considered as inverted and noninverted (i.e., if they were included or excluded in the reorganization). Overall, inverted regions encompassed 328.57 Mb of the human genome, while noninverted regions represented 1.67 Gb of the human genome.
Using the chimpanzee genome as a reference, we detected 38 EBRs when compared with the human genome, ranging from 2 pb to 756 kb and a median length of 42.8 kb (supplementary table S1, Supplementary Material online). Largely, macrorearrangements affected 298.29 Mb of the chimpanzee genome, whereas microrearrangements spanned 10.61 Mb. Once we divided the rearranged chromosomes into inverted and noninverted regions, we found that, as a whole, inverted regions occupied 309.63 Mb and noninverted regions encompassed 1.31 Gb of the chimpanzee genome.
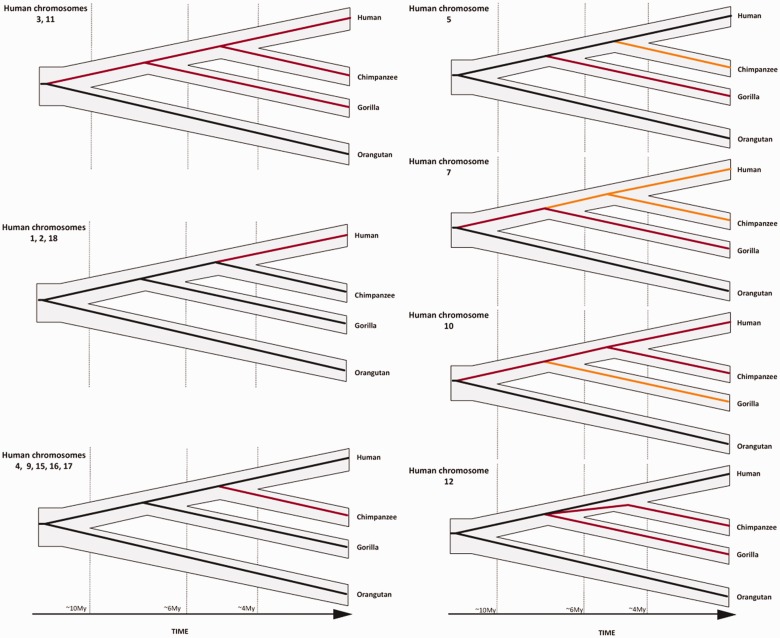
Finally, we proceeded to date the large inversions detected based on previous reports (Yunis and Prakash 1982; Kehrer-Sawatzki and Cooper 2008) (fig. 1). It is known that the ancestral form of orthologous chromosomes 3 and 11 are conserved in orangutan, but suffered an inversion that has been fixed in the human–chimpanzee–gorilla ancestor. The orangutan also presents the ancestral forms for orthologous chromosomes 7 and 10, each of which suffered different inversions at different speciation nodes (fig. 1). Human chromosomes 1, 2, 4, 5, 9, 12, 15, 16, 17, and 18 are rearranged between human and chimpanzee; inversions affecting human chromosome 1 and 18 have occurred only in the human lineage (i.e., are autapomorphies), whereas the rest (on chimpanzee chromosomes 4, 5, 9, 12, 15, 16, and 17) have become fixed in the lineage leading to chimpanzee (Kehrer-Sawatzki and Cooper 2008). Finally, human chromosomes 6, 8, 13, 14, 19, 20, 21, and 22 have been maintained as collinear orthologous in the great apes since common ancestry.
Fig. 1.
Evolutionary history of human chromosomes superimposed on the phylogeny of great apes. Black lines within the phylogenetic tree represent the ancestral state of the chromosomes, whereas red and orange lines represent the rearranged forms. Orangutan maintains the ancestral form for orthologous chromosomes 3 and 11, whereas human, chimpanzee, and gorilla forms are derived. Orthologous chromosomes 1, 2, and 18 have been rearranged in the lineage leading to humans, whereas orthologous chromosomes 4, 9, 15, 16, and 17 are rearranged in the lineage leading to chimpanzee. Ancestral chromosome 5 has been maintained in orangutan and human but has suffered two independent inversions in chimpanzee and gorilla, respectively. Chromosome 7 has suffered one inversion, which has been fixed in gorilla, and another inversion has been fixed in the lineage leading to human and chimpanzee. Chromosome 10 underwent one inversion that was fixed in human and chimpanzee, and a new inversion fixed in gorilla. Finally, chromosome 12 has maintained the ancestral form in humans and orangutans but has undergone an inversion that has been fixed in chimpanzee and gorilla, therefore, the polymorphic state has persisted across multiple speciation nodes (gorilla–human–chimpanzee and human–chimp).
Recombination Rates and Chromosomal Reorganizations
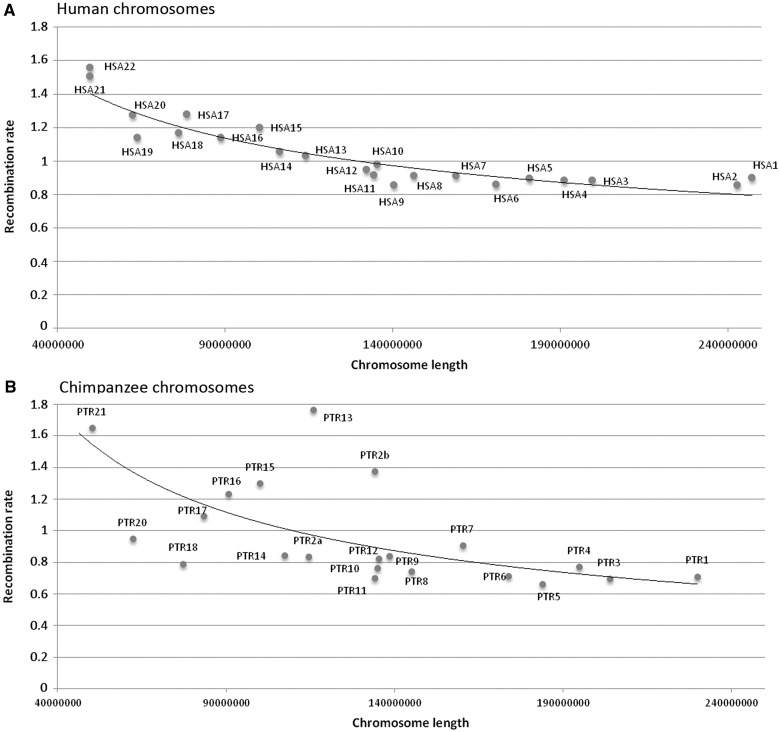
We analyzed the standardized human recombination maps described by Kong et al. (2010) to study the association between recombination rate and genomic shuffling in the human genome. Comparable data for the chimpanzee whole genome sequence was also used (Auton et al. 2012). For the analysis of the human genome, we used the sex-averaged (female and male) recombination map, which provides a total of 4,006 “hotspots” with standardized recombination rate (SRR) ≥10. A first analysis showed that SRR is not homogeneously distributed among human chromosomes (Kruskal–Wallis test, P < 0.0001). The lowest average SRR (0.856) was on human chromosome 9, whereas the highest (1.559) was on human chromosome 22 (fig. 2A). We found a strong correlation between recombination rate and chromosome size (R2 = 0.867, P value < 0.0001) indicating that smaller chromosomes have a higher recombination rates than do larger chromosomes (fig. 2A). For the chimpanzee genome, we calculated the average recombination rate in windows of 10 kb to compare it with the human data. We found the same pattern as in human chromosomes, given that smaller chromosomes showed higher recombination rate than larger chromosomes (R2 = 0.380, P value = 0.0016) (fig. 2B). These results corroborate previous observations in mammals that show how larger chromosomes tend to accumulate larger numbers of COs, and that each chromosome generally presents at least one CO (Sun et al. 2005). Small chromosomes, on the other hand, are expected to have higher recombination rates than large chromosomes to ensure the resolution of, at least, one CO, and therefore guarantee a correct disjunction during meiosis. Moreover, we observed that recombination rate was not uniformly distributed across each human chromosome, which presented clusters of “hotspots” and “coldspots” along chromosomal regions (fig. 3A and supplementary fig. S1, Supplementary Material online). This follows the nonrandom distribution of COs described in other species (Petes 2001).
Fig. 2.
Correlation between chromosomal length and recombination rates. SRRs for each human (A) and chimpanzee (B) chromosomes are shown.
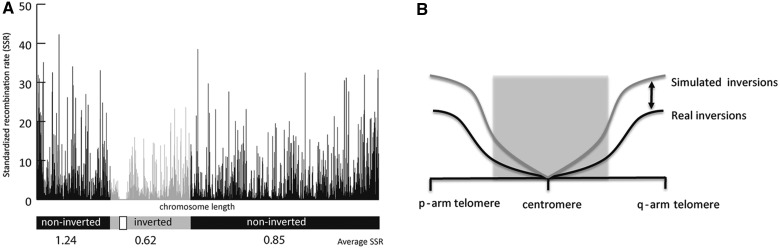
Fig. 3.
Distribution of recombination rates. (A) SRR along the human chromosome 4. SRR (y axis) are shown as needles across nonoverlapping windows of 10 kb in the whole chromosomal length (x axis). The genomic region affected by an inversion is depicted in gray, whereas noninverted regions are showed in black. The white rectangle indicates the centromere. Average recombination rate for each region is shown in numbers in the x axis. (B) Schematic representation of how recombination rates (SRR) varies along observed and simulated inversions. Average SRR across inverted and noninverted regions in observed (black) and simulated (dark gray) chromosomes. The light gray square shows the inverted region around the centromere. Differences between simulated and observed inversions are significantly different (t test, P value < 0.0001) (see text for details).
We compared SRR between collinear and rearranged chromosomes to assess whether the distribution of recombination rate is affected by chromosomal reorganizations. First, the average recombination rate was estimated by considering all the 10 kb windows for each chromosome as a whole. In the human genome, this was found to be significantly higher in collinear (0.975) than in rearranged (0.944) chromosomes (Mann–Whitney’s U test, P < 0.0001) (table 1). We then classified rearranged chromosomes into genomic regions affected (inverted) or not affected (noninverted) by rearrangement. Importantly, inverted regions present significantly lower recombination rates (0.715) than do collinear (0.975) and noninverted regions (1.001) (Kruskal–Wallis test, P < 0.0001), showing a possible effect of suppression of recombination within inverted regions (table 1). To evaluate the effect of chromosomal length in the results, we performed a regression analysis in rearranged and collinear chromosomes and compared the resulting slopes (−0.38 for collinear and −0.29 for rearranged) by means of a t test. Differences among slopes between collinear and rearranged chromosomes were not statistically significant (t test, P = 0.217), suggesting that in both collinear and rearranged chromosomes, SRR correlate with the chromosomal size (R2 = 0.93 for collinear chromosomes and R2 = 0.72 for rearranged).
Table 1.
Comparison of Means Recombination Rates in Each Type of Chromosome and Region (inverted, noninverted, or collinear).
| Type of Region | Mean SRR | Standard Error |
|---|---|---|
| Collinear | 0.975 | 0.009 |
| Rearranged | 0.944 | 0.006 |
| Collinear | 0.975 | 0.009 |
| Noninverted | 1.001 | 0.007 |
| Inverted | 0.715 | 0.012 |
Note.—Rearranged chromosomes exhibited a lower recombination rate than do collinear chromosomes (Mann–Whitney’s U test, P < 0.0001). Recombination rate is significantly lower in inverted regions compared with collinear and noninverted regions (Kruskal–Wallis test, P < 0.0001).
Finally, we considered the length of the region involved in rearrangements by grouping each inverted region into micro- or macrorearrangements. In doing so, we detected a significantly lower recombination rates in genomic regions within macro- (0.713) rather than within microrearrangements (0.976) and nonrearranged regions (0.996) (Kruskal–Wallis test, P < 0.0001), suggesting that macrorearrangements have a stronger impact on reducing recombination rate than do microrearrangements (table 2). When analyzing each rearranged human chromosome separately, a striking pattern emerged whereby regions affected by inversions showed lower SRR than did noninverted regions (fig. 3A and supplementary fig. S1, Supplementary Material online). Moreover, when the size of the inverted genomic region is considered, macroreorganizations have lower recombination rates than microrearrangements (supplementary fig. S1, Supplementary Material online), observations consistent with data obtained by the whole genome analysis. Mirroring these results, we observed the same pattern in the chimpanzee genome; we found that collinear chromosomes have higher recombination rate (1.081) than reorganized chromosomes (0.871) (Mann–Whitney’s U test, P < 0.0001). However, when focusing on the different genomic regions, we observed that SRR at the chimpanzee centromeres were higher (1.795) than expected (almost zero) due to the centromeric interference (Kong et al. 2010). It is likely that these results are due to inaccurate positioning of centromeres in the chimpanzee assembly or lack of power in assigning accurately recombination rates in these regions.
Table 2.
Comparison of Means of Recombination Rate in Regions that Have Suffered Macro- and Microrearrangements.
| Type of Rearrangement | Mean SRR | Standard Error |
|---|---|---|
| Nonrearranged | 0.996 | 0.005 |
| Microrearrangement | 0.976 | 0.108 |
| Macrorearrangement | 0.713 | 0.012 |
Note.—Recombination rate is lower in regions affected by macrorearrangements compared with those affected by microrearrangements or those that are not rearranged (Kruskal–Wallis test, P < 0.0001).
Interestingly, we observed the same pattern for sex-specific human recombination maps (male and female, Kong et al. 2010). In both sexes, the observed recombination rate was greater in collinear than in rearranged chromosomes, which, in turn, exhibited lower recombination rate in inverted regions than in noninverted regions. In addition, macrorearrangements showed lower recombination rates than microrearrangements and collinear chromosomes in both sexes (0.788, 0.958, and 0.986 for the female recombination map and 0.771, 0.931, and 0.989 for the male recombination map).
Given that all the macroreorganizations described involved the centromere (pericentric inversions), we tested whether the suppression of recombination that we observed within reorganized areas was due to the low recombination rate characteristic of pericentromeric regions (Kong et al. 2010). To do so, we simulated all types of inversions in collinear chromosomes (3, 6, 8, 11, 13, 14, 20, 21, and 22) of the same variety of sizes (absolute and relative) than the observed ones. We performed two different tests (based on 10,000 permutations with randomization) to finally obtain an average SRR for simulated and observed inverted and noninverted regions. In the first permutation test (from now on called the Mb test), we simulated the observed size in Mb of each inversion and its position regarding the centromere in collinear chromosomes. In the second test (referred as the % test), we simulated the relative size, in percentage, of the chromosome involved in each observed inversion and the position from the centromere in collinear chromosomes. We found that, in the human genome, simulated macroinverted regions have lower SRR (0.837 for the Mb test and 0.700 for the % test, P value < 0.0001) than simulated noninverted regions (1.025 for the Mb test and 1.058 for the % test, P value < 0.0001) (supplementary table S2, Supplementary Material online). More importantly, the observed inverted regions have lower SRR (0.717) than simulated inverted regions (0.837) in the Mb test (P value < 0.0001) (fig. 3B and supplementary table S2, Supplementary Material online). When considering the relative size of inversions (% test), this signal was not detected most likely due to the fact that simulations of inversions based on percentage performed in collinear chromosomes could include small regions next to the centromere, especially in the case of small chromosomes. Regarding the chimpanzee genome, we confirmed the same trend, because observed inverted regions have lower SRR (0.909) than simulated inverted regions (1.280 for the Mb test, P value = 0.0001 and 1.234 for the % test, P value = 0.0001) (supplementary table S2, Supplementary Material online). Overall, our data suggest that centromere position would have an influence in the reduction of the recombination rates, but the effect is not strong enough to explain the reduction of recombination rate observed in reorganized chromosomes.
It has been described that EBRs tend to have higher divergence rates than other regions in the genome (Navarro et al. 1997; Marques-Bonet and Navarro 2005), and divergence rate strongly correlates with recombination (Hellmann et al. 2003). Therefore, we decided to compare recombination rates between EBRs and HSBs in the human genome. Although not statistically significant (Mann–Whitney’s U test, P = 0.078), we nonetheless found a lower recombination rate in EBRs (0.492) than in HSBs (0.962). Remarkably, none of the evolutionary breakpoints detected show recombination “hotspots” (regions with SRR higher than 10) but they contained significantly less “coldspots” than did the HSBs (Fisher’s exact test, P < 0.0001), suggesting that this tendency may be of relevance. We also analyzed the recombination rate in the genomic regions surrounding the breakpoints. To do so, we utilized a “breakpoint-edge” (BP-edge) that spanned a region 100 kb upstream or downstream from the breakpoint coordinates. This showed that EBR are surrounded by regions of high recombination (SRR = 0.877), although the differences between EBRs and BP-edge were not statistically significant (Mann–Whitney’s U test, P = 0.634).
We did not expect to find any effect on recombination rates for inversions that have been fixed in the chimpanzee lineage (i.e., chromosomes 4, 5, 9, 12, 15, 16, and 17) but, surprisingly, we detected lower SRR not only in macrorearrangements affecting human chromosomes 1 and 18 (human-specific inversions), but also in human chromosomes 2, 4, 5, 9, 12, 16, and 17 (supplementary fig. S1, Supplementary Material online). To further investigate this striking pattern, we studied the SRR in human and chimpanzee chromosomes taking into account their evolutionary history. We classified chromosomes 1, 2, and 18 as human specific and chromosomes 4, 5, 9, 12, 15, 16, and 17 as chimpanzee specific because these chromosomes have been fixed in each lineage. Chromosomes 3, 7, 10, and 11 were considered as orangutan-ancestral because human and chimpanzee forms are derivative; and finally, chromosomes 6, 8, 9, 13, 14, 19, 20, 21, and 22 as great apes ancestral because they have been maintained collinear during great apes evolutionary history. When analyzing the SRR in the human genome, human-specific chromosomes have the lowest SRR (0.913) followed by orangutan ancestral chromosomes (0.917) and chimpanzee-specific chromosomes (0.998) (Kruskal–Wallis test, P < 0.0001). Moreover, we found the highest SRR (0.999) in great apes ancestral chromosomes (Kruskal–Wallis test, P < 0.0001). Regression analysis considering the influence of chromosomal size in SRR indicated that differences among slopes between human-, chimp-, and orangutan-specific chromosomal forms were not statistically significant (t test, P = 0.198). These results suggest that the observed differences in SRR are most probably due to the evolutionary history of each chromosome and independent of the chromosomal size. Meanwhile, when analyzing the SRR in the chimpanzee genome, chimpanzee-specific chromosomes (4, 5, 9, 12, 15, 16, and 17) have lower SRR (0.752) than the rest (Kruskal–Wallis test, P < 0.0001). These data suggest that those chromosomes that have been maintained collinear during evolutionary history retained higher recombination rates than those that have been altered during evolution in each particular lineage.
Distribution of Hotspot Motifs
Prdm9 is the only known speciation-associated gene described in mammals (Mihola et al. 2009) and codifies for a meiotic-specific histone H3 methyltransferase that is thought to locate at recombination sites by the recognition of a specific 13 bp DNA motif (CCNCCNTNNCCNC in humans, according to Myers et al. 2005). Specifically, studies in mice have revealed that the Prdm9 protein determines the position where recombination takes place, being directly involved in the recruitment of the recombination initiation machinery during meiosis (Brick et al. 2012). Although just approximately 40% of the human recombination hotspots contain the specific motif (Myers et al. 2005, 2010), it has been described that hotspots containing the consensus sequence have a stronger activity than those that do not (Smagulova et al. 2011). Therefore, we analyzed the distribution of the human consensus Prdm9 DNA motif in searching for a mechanistic explanation for the low recombination rates observed within inverted regions. Out of the 83,091 different loci found in the human genome sequence, 17,254 loci were excluded because they were located at telomeric and subtelomeric regions, leaving us with a total of 65,837 loci. After merging these data with the different chromosome types (collinear vs. rearranged), we found that rearranged chromosomes have higher density of hotspot loci (27.16 loci/Mb) than collinear chromosomes (23.11 loci/Mb) (Mann–Whitney U test, P < 0.0001). When analyzing the different genomic regions within reorganized chromosomes (inverted vs. noninverted), we observed that inverted regions have lower density of hotspot loci (26.49 loci/Mb) than noninverted regions (27.28 loci/Mb) (Kruskal–Wallis test, P < 0.0001).
Discussion
Our own study represents a departure from those conducted previously in that it relies on the use of a recent and high-resolution (10 kb) genome-wide map of recombination rates in the human and chimpanzee to refine genome reshuffling between both species. These recombination rates are not directly quantified from gametes, but inferred from genome-wide SNP data from a human population of 38,167 individuals in the case of humans (Kong et al. 2010) and 10 nonrelated individuals in the case of chimpanzees (Auton et al. 2012). These maps estimate the location of recombination events in the progeny (see Lynn et al. 2004, for a review) and reflect the integration of population-level processes over several generations. Although we are aware that these maps estimate the recombination of the extant species, they provide an historical view of recombination events, incorporating data on population growth and natural selection among others (Clark et al. 2010). Using this approach, we provide evidences of a reduction of recombination within genomic regions that have been implicated in the chromosomal evolution between human and chimpanzee.
Reduced Recombination Rates within Reorganized Genomic Regions
When initially proposed, the “suppressed recombination” model was considered as a compelling hypothesis to explain the contribution of large genome reshuffling in the formation of new species (Noor et al. 2001; Rieseberg 2001). Under this assumption, chromosome rearrangements in heterokaryotypes would suppress recombination thus contributing to a reduction of gene flow across genomic regions and the accumulation of genetic incompatibilities. Most subsequent studies have used sequence divergence (patterns of nucleotide differentiation) between species as an indirect estimation of recombination. But this approximation has its limitations, and the interpretation of amino acid divergence, as an effect of recombination, can be problematic (reviewed in Noor and Bennett 2009). As an example, Bullaughey et al. (2008) found no correlation between either broad- or fine-scale rates of recombination and rates of protein evolution (measured by dN/dS ratios) between human, chimpanzee, and rhesus macaque, suggesting that additional parameters should be considered.
To circumvent this limitation, we studied the most recent recombination maps for human and chimpanzee based on SNP data. When considering the human genome as a whole, we found that recombination rate was significantly higher in collinear than in rearranged chromosomes (Mann–Whitney’s U test, P < 0.0001 and permutation tests). Moreover, those genomic regions within the macroreorganizations (historically detected by cytogenetic studies) have a significantly lower recombination rate than both microrearrangements and nonrearranged regions (Kruskal–Wallis test, P < 0.0001 and permutation tests). This pattern was also observed in the chimpanzee. These data support the existence of a possible suppression of recombination effect associated with reorganized chromosomal regions—this being more substantial in large inversions. Previous studies by Navarro and Barton (2003) compared the recombination rates (cM/Mb) in collinear and rearranged chromosomes between human and chimpanzees using an earlier version of the human recombination map (Kong et al. 2002). Although no statistical differences were found, they noted a tendency for rearranged chromosomes to show a reduced recombination rate compared with collinear chromosomes. Later studies, however, have shown different trends (Zhang et al. 2004; Marques-Bonet et al. 2007; Szamalek et al. 2007), underscoring the uncertainty surrounding recombination rates. Here, we have shown that the effects of genome reshuffling on the distribution of recombination rates can be assessed when combining an accurate delineation of the chromosomal reorganizations and a high-resolution standardized recombination map. We detected not only a lower recombination rate within rearranged genomic regions, but also found that regions not affected by the reorganization in rearranged chromosomes (noninverted regions) presented significantly higher recombination rates than do collinear chromosomes in the human genome (Kruskal–Wallis test, P < 0.0001 and permutation tests). This pattern is not unexpected given that at least one CO per pair of homologous chromosomes is necessary to ensure proper disjunction. Therefore, chromosomes affected by rearrangements showed that the noninverted regions accumulate recombination events that are absent within the inverted region—the so-called “inter- and intrachromosomal effect”— thus explaining the global increase of recombination rate in regions outside inversions (Sturtevant 1919; Schultz and Redfiled 1951).
More importantly, our observations validate the relevance of chromosomal reorganizations in modeling the recombination landscape. Our model follows two lines of evidence. Kirkpatrick and Barton (2006) have suggested that selection could favor reorganizations (i.e., inversions) that reduce recombination of alleles involved in local adaptation. This situation would, eventually, contribute to the fixation of chromosomal reorganizations in different subpopulations in parapatry (connected by gene flow). On the other hand, it has been proposed that chromosomal reorganizations can occasionally survive as polymorphic states for considerable lengths of time, although this would depend on historical variables including effective population size and spatial population structure. Termed hemiplasy (Avise and Robinson 2008), this hypothesis suggests that derived chromosomal rearrangements may have persisted as polymorphisms across multiple speciation nodes (Robinson et al. 2008; Robinson and Ropiquet 2011). This has been the case for chiropteran and afrotherian species (Robinson et al. 2008), Perissodactyla (Trifonov et al. 2008), Rodentia (Badenhorst et al. 2011), Bovidae (Robinson and Ropiquet 2011), and this is also probably true for Primates (Dutrillaux and Couturier 1981; Rumpler et al. 2008). Incomplete lineage sorting (when a gene tree is topologically inconsistent with the species tree) has been detected in the human–chimpanzee–gorilla species phylogeny in genome-wide studies (Chen and Li 2001; Patterson et al. 2006; Hobolth et al. 2011; Scally et al. 2012). In fact, chimpanzee and gorilla chromosomes 12 represent a clear example because they share the same derivative form, whereas human chromosome 12 has maintained the ancestral state from the human–chimpanzee–gorilla ancestor (fig. 1). Under such conditions, it is plausible that inversions could have been maintained as heterokaryotypes in the human–chimp ancestral population (between 6 and 4 Ma according to Hobolth et al. 2011 or 5.5 and 7 Ma according to Scally et al. 2012). This would result in recombination suppression within the reorganized genomic regions involved and this suppression would persist up to the present population gradually returning to the same levels observed in ancestral collinear chromosomes.
This interpretation is supported by our data given that those genomic regions contained within the inversions characterizing the human–chimpanzee–orangutan speciation node presented lower recombination rates (0.917) than ancestral collinear chromosomes (0.999) but higher than the case of inversions that have been fixed after the human–chimpanzee divergence (0.913). Therefore, chromosomal forms would have different recombination rates according to the speciation node where they had become fixed. We detect lower SRR in recently rearranged chromosomes (human–chimpanzee node, ∼4 Ma), intermediate SRR in those fixed in the human–chimpanzee–orangutan node (∼10 Ma) and higher SRR in those chromosomes that maintained the ancestral form for great apes. Then, from an ancestral human–chimp population characterized with persisting floating heterokaryotypes, seven inversions (affecting chromosomes 4, 5, 9, 12, 16, and 17) have become fixed in the lineage leading to chimpanzees, whereas two inversions (affecting chromosomes 1 and 18) have been fixed in the lineage leading to humans. We found lower recombination rates in human chromosomes that have been rearranged and fixed in human lineage, as well as lower recombination rates in chimpanzee chromosomes fixed in chimpanzee lineage. Therefore, the reduction of suppression within inversions that took place while ancestral human–chimpanzee population was polymorphic is still traceable in both the human and chimpanzee genomes. These data fit with previous results on gene-expression divergence between human and chimpanzee (Marques-Bonet et al. 2004). The maintenance of the polymorphic state could increase the time of suppressed recombination, which, in turns, could explain gene-expression divergence in both lineages. It is also possible that the persistence of polymorphism in the ancestral population vary for each rearrangement, so they could exhibit quite different divergence times (Noor and Bennett 2009), and thus explaining the contradictory results obtained in previous studies (Navarro and Barton 2003; Vallender and Lahn 2004; Zhang et al. 2004; Marques-Bonet and Navarro 2005; Marques-Bonet et al. 2007).
Additional, mechanistic forces, however, might have also played a role in maintaining low recombination rates within reorganized regions. Is in this scenario where meiotic hotspot density can influence the distribution of recombination rates. In fact, our results on the distribution of hotspots motifs associated to PRDM9 in the human genome revealed that inverted regions have lower density of hotspot loci than noninverted regions within reorganized chromosomes. The persistence of heterokaryotypes in the human and chimpanzee speciation node could be responsible of these results; and, altogether with the lower hotspot density distribution observed, might suggest a mechanistic explanation for the low recombination rates that characterize inverted regions.
Materials and Methods
Whole-Genome Comparisons and Evolutionary Breakpoint Definition
We obtained human (hg18), chimpanzee (pantro2), and orangutan (PPYG2) orthologous genes from Biomart and downloaded the masked genome sequences from Ensembl v64 database. To detect the EBRs and HSBs between human and chimpanzee whole-genome sequences, we applied two recently described algorithms: SyntenyTracker (Donthu et al. 2009) and Cassis (Baudet et al. 2010). The former approach relies on the detection of HSBs among different species genomes. Based on the genomic positions of orthologous genes, this algorithm establishes temporary synteny blocks and merges neighboring blocks spaced less than a given threshold and having the same orientation. We used the default parameters proposed by the authors (Donthu et al. 2009) and set different thresholds in the distance between blocks parameter (250 kb, 500 kb, and 1 Mb), obtaining the best performance regarding number and size of the HSBs at 1 Mb. Then, we defined the EBRs by sequence coordinates that delimit the start and end of contiguous HSBs. Cassis (Baudet et al. 2010), on the other hand, is specially designed to define breakpoint regions. The algorithm establishes the putative location of EBRs using the position of orthologous genes as markers and then by means of sequence alignment, more accurately defines the EBR coordinates. The Cassis algorithm was run using default parameters.
We then compared the EBRs detected by our approach with previously published comparisons of human and chimpanzee genomes (Feuk et al. 2005; Kehrer-Sawatzki and Cooper 2008) to establish a reliable EBRs data set. All the macroreorganizations initially described by Kehrer-Sawatzki and Cooper (2008) affecting human chromosomes 1, 4, 5, 9, 12, 15, 16, and 17 were refined by our in silico analysis, except for the inversion breakpoints of human chromosome 18, which proved to be in regions rich in repetitive sequences. In the case of this chromosome, we used the coordinates defined by cytogenetic studies (Kehrer-Sawatzki and Cooper 2008). Finally, we filtered our estimated EBRs data set retaining EBRs identified using Cassis that met any of the following criteria: 1) they were identified using Synteny Tracker, 2) they were part of the macrorearrangements experimentally validated by fluorescent in situ hybridization (Kehrer-Sawatzki and Cooper 2008), or 3) they were experimentally validated by Feuk et al. (2005).
Once the EBRs were defined in both the human and chimpanzee genomes, we grouped the chromosomes into rearranged (those affected by reorganizations) or collinear (if they were conserved), using the orangutan as an outgroup. We divided the rearranged chromosomes into two additional genomic regions: 1) inverted or 2) noninverted, if they were affected by the reorganization or not, respectively. Then, we recognized macrorearrangements as those where the inverted regions spanned >4.3 Mb and microrearrangements if they spanned <4.3 Mb.
Finally, we defined the genomic positions of centromeres and telomeres as those described by Ensembl v64 for human genome (hg18) and ±40 kb from those described by UCSC for chimpanzee genome (pantro2).
Recombination Rates
Genetic maps for the human and chimpanzee genomes were extracted from Kong et al. (2010) and Auton et al. (2012), respectively. In the case of the human genetic map, the authors inferred genomic recombination rates from 289,658 SNP data from 38,167 individuals using linkage disequilibrium patterns with a resolution of nonoverlapping windows of 10 kb. Recombination rate data are estimated for 2.4 Gb of the human genome, excluding chromosome X and the 5 Mb at the ends of each autosomal chromosome. For each window, the SRR is calculated as a fraction of the genetic distance divided by the overall average distance (Kong et al. 2010). Genomic regions with a recombination rate ≥10 were considered as “hotspots,” whereas regions with a recombination rate equal to 0 were considered “coldspots.” Regarding the chimpanzee recombination map, Auton et al. (2012) inferred genomic recombination rates for autosomes from 5.3 million SNPs identified in 10 nonrelated Western chimpanzees. By this way, the authors identified 6,290 “hotspots” using a coalescent simulation test after the genomic recombination rate estimation. To perform the same analysis as in human, we calculated the average recombination rate in windows of 10 kb along the chimpanzee autosomes.
We finally merged the coordinates of recombination rate windows with the positions of EBRs and the different types of regions detected (collinear, noninverted, and inverted) in both species using in-house Perl scripts.
Statistical Analysis
Statistical analyses were performed using the JMP v7 package. Given that the genomic distribution of recombination rates did not follow a normal distribution, we applied nonparametric analysis (Mann–Whitney U test or Kruskal–Wallis test) to assess the differential recombination rates between EBRs/HSBs, inverted/noninverted/collinear regions, and collinear/rearranged chromosomes. We applied the Bonferroni correction when necessary. Fisher’s exact test was applied to compare the distribution of recombination “hotspots” and “coldspots” in EBRs and HSBs. For the evaluations of the relationship between chromosome lengths and recombination rates, we performed regression analysis and applied a t test to test slopes and analyze residuals.
Permutation Test
To test the effect of the inversions on the recombination rate, we performed two series of permutation tests. In the first test, we simulated inversions in collinear chromosomes of the same variety of absolute size (in Mb) than the observed ones, whereas in the second test we simulated inversions in collinear chromosomes of the same relative size (in percentage of chromosome) than the observed ones. In the two series of tests each inversion was simulated in the collinear chromosome maintaining not only the size (relative or absolute) but also the distribution around the centromere to reproduce the properties of the centromeres. Both tests were performed using in-house python scripts and were based on 10,000 permutations with randomization. At each round, we compared the mean SRR of the permuted data set with the mean of the observed inversion for each chromosome. The significance level was determined counting, at each permutation, how many times the mean SRR of the simulated inversion is larger than the mean SRR in the observed inversion. For each test and inversion, we considered three possible scenarios: 1) Hypothesis 1—the average SRR of simulated inverted regions is higher than those in simulated noninverted regions; 2) Hypothesis 2—the average SRR of observed inversions is higher than those of simulated inversions; and 3) Hypothesis 3—the average SRR of observed noninverted regions is higher than simulated noninverted regions. The threshold was fixed in the 5%, therefore P values < 0.05 indicate that the hypothesis tested can be rejected. Moreover, we applied a t test with a significance level of 0.05 to compare 1) the overall mean SRR of all observed inverted regions against the overall mean SRR of all simulated inverted regions, and 2) the overall mean of observed macroinversions with the overall mean of simulated macroinversions.
Hotspot Motif Identification
To identify the DNA motif associated with the PRDM9 hotspot activity in the human genome (CCNCCNTNNCCNC, according to Myers et al. 2005), we used the FIMO algorithm (Grant et al. 2011) as part of the MEME suite. We created the matrix input file assigning a probability of 1 or 0.25 for each nitrogenated base in each position and ran the algorithm using a P value output threshold of 0.0001 and the rest of parameters as default. Then, we merged the positions of the motifs with the different types of regions (inverted, noninverted, and collinear) in the human genome.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge T.J. Robinson, C. Gilbert, M. Garcia-Caldés, and M. Ponsà for insightful discussions and comments on the manuscript. They are also thankful to the anonymous reviewers whose suggestions improved the manuscript. This work was supported by the Ministerio de Ciencia y Tecnologia (CGL-2010-20170) and the Universitat Autònoma de Barcelona (PhD fellowship to M.F.).
References
- Ashley T, Moses MJ, Solari AJ. Fine structure and behaviour of a pericentric inversion in the sand rat, Psammomys obesus. J Cell Sci. 1981;50:105–119. doi: 10.1242/jcs.50.1.105. [DOI] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, et al. (23 co-authors) A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Robinson TJ. Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol. 2008;57:503–507. doi: 10.1080/10635150802164587. [DOI] [PubMed] [Google Scholar]
- Badenhorst D, Dobigny G, Adega F, Chaves R, O'Brien PC, Ferguson-Smith MA, Waters PD, Robinson TJ. Chromosomal evolution in Rattini (Muridae, Rodentia) Chromosome Res. 2011;19:709–727. doi: 10.1007/s10577-011-9227-2. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Baudet C, Lemaitre C, Dias Z, Gautier C, Tannier E, Sagot MF. Cassis: detection of genomic rearrangement breakpoints. Bioinformatics. 2010;26:1897–1898. doi: 10.1093/bioinformatics/btq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Toure Y, Sagnon N. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci U S A. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodin PM, Karamysheva TV, Belonogova NM, Torgasheva AA, Rubtsov NB, Searle JB. Recombination map of the common shrew, Sorex araneus (eulipotyphla, mammalia) Genetics. 2008;178:621–632. doi: 10.1534/genetics.107.079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, O’Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Brown KM, Burk LM, Henagan LM, Noor MA. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution. 2004;58:1856–1860. doi: 10.1111/j.0014-3820.2004.tb00469.x. [DOI] [PubMed] [Google Scholar]
- Bullaughey K, Przeworski M, Coop G. No effect of recombination on the efficacy of natural selection in primates. Genome Res. 2008;18:544–554. doi: 10.1101/gr.071548.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, et al. (12 co-authors) Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:e1000538. doi: 10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia R, Capanna E. Chiasma repatterning across a chromosomal hybrid zone between chromosomal races of Mus musculus domesticus. Genetica. 2002;114:35–40. doi: 10.1023/a:1014626330022. [DOI] [PubMed] [Google Scholar]
- Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Wang X, Matise T. Contrasting methods of quantifying fine structure of human recombination. Annu Rev Genomics Hum Genet. 2010;11:45–64. doi: 10.1146/annurev-genom-082908-150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprat A, Negre B, Puig M, Ruiz A. The transposon Galileo generates natural chromosomal inversions in Drosophila by ectopic recombination. PLoS One. 2009;4:e7883. doi: 10.1371/journal.pone.0007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donthu R, Lewin HA, Larkin DM. SyntenyTracker: a tool for defining homologous synteny blocks using radiation hybrid maps and whole-genome sequence. BMC Res Notes. 2009;2:148. doi: 10.1186/1756-0500-2-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas D, Britton-Davidian J. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics. 2002;162:1355–1366. doi: 10.1093/genetics/162.3.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrillaux B, Couturier J. The ancestral karyotype of platyrrhine monkeys. Cytogenet Cell Genet. 1981;30:232–242. doi: 10.1159/000131614. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25:660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Farre M, Bosch M, Lopez-Giraldez F, Ponsa M, Ruiz-Herrera A. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS One. 2011;6:e27239. doi: 10.1371/journal.pone.0027239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, MacDonald JR, Tang T, Carson AR, Li M, Rao G, Khaja R, Scherer SW. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini P, Colangelo P, Solano E, Capanna E, Verheyen E, Castiglia R. Reduced gene flow at pericentromeric loci in a hybrid zone involving chromosomal races of the house mouse Mus musculus domesticus. Evolution. 2010;64:2020–2032. doi: 10.1111/j.1558-5646.2010.00964.x. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum IF, Reed MJ. Evidence for heterosynaptic pairing of the inverted segment in pericentric inversion heterozygotes of the deer mouse (Peromyscus maniculatus) Cytogenet Cell Genet. 1984;38:106–111. doi: 10.1159/000132040. [DOI] [PubMed] [Google Scholar]
- Hale DW. Heterosynapsis and suppression of chiasmata within heterozygous pericentric inversions of the sitka deer mouse. Chromosoma. 1986;94:425–432. doi: 10.1007/BF00292751. [DOI] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Paabo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am J Hum Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 2011;21:349–356. doi: 10.1101/gr.114751.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res. 2008;16:41–56. doi: 10.1007/s10577-007-1207-1. [DOI] [PubMed] [Google Scholar]
- King M. Cambridge: Cambridge University Press; 1993. Species evolution. The role of chromosome change. [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, et al. (16 co-authors) A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, et al. (15 co-authors) Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MA. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci U S A. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19:770–777. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, Carone DM, NISC Comparative Sequencing Program. Green ED, O'Neill MJ, O'Neill RJ. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics. 2009;10:334. doi: 10.1186/1471-2164-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Ashley T, Hassold T. Variation in human meiotic recombination. Annu Rev Genomics Hum Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Caceres M, Bertranpetit J, Preuss TM, Thomas JW, Navarro A. Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet. 2004;20:524–529. doi: 10.1016/j.tig.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Navarro A. Chromosomal rearrangements are associated with higher rates of molecular evolution in mammals. Gene. 2005;353:147–154. doi: 10.1016/j.gene.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Sanchez-Ruiz J, Armengol L, Khaja R, Bertranpetit J, Lopez-Bigas N, Rocchi M, Gazave E, Navarro A. On the association between chromosomal rearrangements and genic evolution in humans and chimpanzees. Genome Biol. 2007;8:R230. doi: 10.1186/gb-2007-8-10-r230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AP, Grushko O, Guelbeogo WM, Lobo NF, Sagnon N, Costantini C, Besansky NJ. Divergence with gene flow in Anopheles funestus from the Sudan savanna of Burkina Faso, West Africa. Genetics. 2006;173:1389–1395. doi: 10.1534/genetics.106.059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, et al. (25 co-authors) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betran E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Ruiz A. On the fertility effects of pericentric inversions. Genetics. 1997;147:931–933. doi: 10.1093/genetics/147.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Linder CR, Seiler GJ. Chromosomal and genic barriers to introgression in helianthus. Genetics. 1995;141:1163–1171. doi: 10.1093/genetics/141.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TJ, Ropiquet A. Examination of hemiplasy, homoplasy and phylogenetic discordance in chromosomal evolution of the bovidae. Syst Biol. 2011;60:439–450. doi: 10.1093/sysbio/syr045. [DOI] [PubMed] [Google Scholar]
- Robinson TJ, Ruiz-Herrera A. Mammalian chromosomal evolution: from ancestral states to evolutionary regions. In: Pontarotti P, editor. Evolutionary biology—concepts, molecular and morphological evolution. Berlin (Germany): Springer; 2010. pp. 143–158. [Google Scholar]
- Robinson TJ, Ruiz-Herrera A, Avise JC. Hemiplasy and homoplasy in the karyotypic phylogenies of mammals. Proc Natl Acad Sci U S A. 2008;105:14477–14481. doi: 10.1073/pnas.0807433105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Castresana J, Robinson TJ. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Garcia F, Giulotto E, Attolini C, Egozcue J, Ponsa M, Garcia M. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet Genome Res. 2005;108:234–247. doi: 10.1159/000080822. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Warter S, Hauwy M, Fausser JL, Roos C, Zinner D. Comparing chromosomal and mitochondrial phylogenies of sportive lemurs (genus Lepilemur, Primates) Chromosome Res. 2008;16:1143–1158. doi: 10.1007/s10577-008-1265-z. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, et al. (72 co-authors) Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Redfiled H. Interchromosomal effects on crossing over in Drosophila. Cold Spring Harb Symp Quant Biol. 1951;16:175–197. doi: 10.1101/sqb.1951.016.01.015. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. Contributions to genetics of Drosophila melanogaster. Washington (DC): Carnegie Institution; 1919. Inherited linkage variation in the second chromosome; pp. 305–341. [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Trpkov K, Rademaker A, Ko E, Martin RH. Variation in meiotic recombination frequencies among human males. Hum Genet. 2005;116:172–178. doi: 10.1007/s00439-004-1215-6. [DOI] [PubMed] [Google Scholar]
- Szamalek JM, Cooper DN, Hoegel J, Hameister H, Kehrer-Sawatzki H. Chromosomal speciation of humans and chimpanzees revisited: studies of DNA divergence within inverted regions. Cytogenet Genome Res. 2007;116:53–60. doi: 10.1159/000097417. [DOI] [PubMed] [Google Scholar]
- Trifonov VA, Stanyon R, Nesterenko AI, et al. (14 co-authors) Multidirectional cross-species painting illuminates the history of karyotypic evolution in perissodactyla. Chromosome Res. 2008;16:89–107. doi: 10.1007/s10577-007-1201-7. [DOI] [PubMed] [Google Scholar]
- Vallender EJ, Lahn BT. Effects of chromosomal rearrangements on human-chimpanzee molecular evolution. Genomics. 2004;84:757–761. doi: 10.1016/j.ygeno.2004.07.005. [DOI] [PubMed] [Google Scholar]
- White BJ, Crandall C, Raveche ES, Hjio JH. Laboratory mice carrying three pairs of Robertsonian translocations: establishment of a strain and analysis of meiotic segregation. Cytogenet Cell Genet. 1978;21:113–138. doi: 10.1159/000130886. [DOI] [PubMed] [Google Scholar]
- Yannic G, Basset P, Hausser J. Chromosomal rearrangements and gene flow over time in an inter-specific hybrid zone of the Sorex araneus group. Heredity. 2009;102:616–625. doi: 10.1038/hdy.2009.19. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Podlaha O. Testing the chromosomal speciation hypothesis for humans and chimpanzees. Genome Res. 2004;14:845–851. doi: 10.1101/gr.1891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.