Abstract
The Opisthokonta clade includes Metazoa, Fungi, and several unicellular lineages, such as choanoflagellates, filastereans, ichthyosporeans, and nucleariids. To date, studies of the evolutionary diversity of opisthokonts have focused exclusively on metazoans, fungi, and, very recently, choanoflagellates. Thus, very little is known about diversity among the filastereans, ichthyosporeans, and nucleariids. To better understand the evolutionary diversity and ecology of the opisthokonts, here we analyze published environmental data from nonfungal unicellular opisthokonts and report 18S ribosomal DNA phylogenetic analyses. Our data reveal extensive diversity among all unicellular opisthokonts, except for the filastereans. We identify several clades that consist exclusively of environmental sequences, especially among ichthyosporeans and choanoflagellates. Moreover, we show that the ichthyosporeans represent a significant percentage of overall unicellular opisthokont diversity, with a greater ecological role in marine environments than previously believed. Our results provide a useful phylogenetic framework for future ecological and evolutionary studies of these poorly known lineages.
Keywords: unicellular opisthokonts, diversity, distribution, choanoflagellates, ichthyosporeans
The opisthokonts include two of the most well-studied kingdoms of life: the metazoans and the fungi. Recent phylogenetic and phylogenomic analyses have shown that the opisthokonts also include several unicellular lineages (Steenkamp et al. 2006). These include the nucleariids (Amaral-Zettler et al. 2001), Fonticula alba (Brown et al. 2009), the filastereans (Shalchian-Tabrizi et al. 2008), the ichthyosporeans (also known as DRIPs or mesomycetozoeans) (Ragan et al. 1996), Corallochytrium limacisporum (Raghu-Kumar 1987), and the choanoflagellates (Carr et al. 2008) (for a review see Paps and Ruiz-Trillo 2010). Although we have some knowledge of the diversity of metazoans, fungi, and choanoflagellates, our current understanding of the evolutionary diversity of the other opisthokont lineages is very poor. To date, most phylogenetic studies of the opisthokonts have focused on data from cultured organisms (same references as above). A handful of studies take environmental sequences into account, but they focus on a single clade and/or use a limited number of environmental sequences (Marshall and Berbee 2010; del Campo and Massana 2011). To fill this gap and obtain an exhaustive description of the diversity (i.e., species richness and abundance) of all nonfungal unicellular opisthokonts, we screened published 18S ribosomal gene clone libraries and performed phylogenetic analyses. In terms of the environmental data provided, the 18S ribosomal gene is, by far, the most sequenced gene (Pawlowski et al. 2012), and therefore, it is the ideal candidate for searching for environmental diversity.
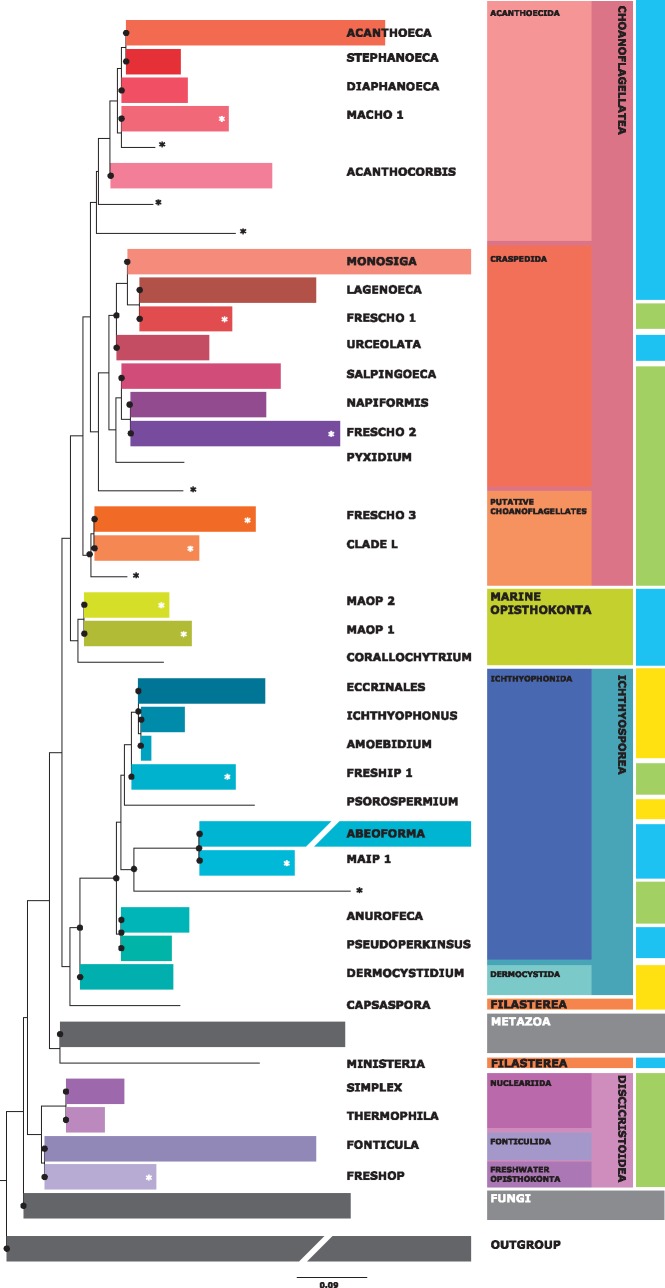
Our final phylogenetic tree, based on complete 18S ribosomal DNA (rDNA) sequences, is shown in figure 1. Our topology confirms the division of the choanoflagellates into two main clades: Acanthoecida and Craspedida (Carr et al. 2008; Nitsche et al. 2011), with some internal clades being formed exclusively of environmental sequences (del Campo and Massana 2011). We also recover three environmental clades (Clade L from Weber et al. 2012, FRESCHO 3, and a singleton) of mainly freshwater choanoflagellates that branch as a sister group to the Acanthoecida and Craspedida clades. Furthermore, branching as a sister group to all the choanoflagellates, there is a group constituted of Corallochytrium and two clades of marine environmental sequences: the novel MAOP 1 and the previously described MAOP 2 (Uncultured Opisthokonts 1 in Marshall and Berbee 2010). These two marine clades may well represent a completely novel lineage of marine opisthokonts. It is worth mentioning that the Corallochytrium clade, for which so far only one single species has been described (Cavalier-Smith and Allsopp 1996), is fairly well populated with environmental sequences and represents an important percentage of the unicellular opisthokonts in their entirety (5%, table 1). Our tree also recovers all previously described clades and families within the Ichthyosporea. There are two well-defined exclusively environmental clades among the Ichthyosporea: one freshwater (FRESHIP 1) and the other marine (MAIP 1). The latter (Marine Ichthyosporean 1) is fairly abundant in the environmental clone libraries, with 34 sequences used in this analysis, which represent 6% of the total of unicellular opisthokonts. Unexpectedly, we did not retrieve any filasterean environmental sequences, either from the symbiont Capsaspora or from the free-living marine Ministeria, which both appear as singletons. Among the Holomycota, we retrieved Nuclearia, Fonticula, and the environmental clade FRESHOP (previously described by Marshall and Berbee 2010) as a monophyletic group but with no statistical support. This clade was previously described as class Discicristoidea (Cavalier-Smith 2009), and our data show that it is a mainly a freshwater group.
Fig. 1.
Maximum likelihood (ML) phylogenetic tree of the opisthokonts constructed with 227 complete 18S rDNA sequences (1,436 informative positions). Exclusively environmental clades are indicated with *. The last bar on the right indicates the main environmental origin of the different clades: blue (marine), green (freshwater), and yellow (symbiont). Clades indicated as symbiont are those in which the species are known to be symbiont and that had almost no environmental sequences (maximum one sequence). Environmental clades consisting of a single sequence have not been named. Because of the increasing number of new clades described and for the sake of clarity, the names have been changed from del Campo and Massana (2011) (supplementary information S1, Supplementary Material online). 1,000 pseudoreplicate ML bootstrap values over 50% are indicated with a black dot for the defined clades. The scale bar indicates 0.09 substitutions per position. The complete tree is shown in the supplementary information S2, Supplementary Material online. The order and class names given are based on Cavalier-Smith and Chao (2003), Shalchian-Tabrizi et al. (2008), and Cavalier-Smith (2009). MACHO, marine choanoflagellates; FRESCHO, freshwater choanoflagellates; MAOP, marine opisthokonts; FRESHIP, freshwater ichthyosporeans; MAIP, marine ichthyosporeans; FRESHOP, freshwater opisthokonts.
Table 1.
Assignation of Sequences from Both the Cultures and the Environment to the Different Defined Clades (fig. 1).
| Clade | #seq | %env | %env opisto | Environments |
|||
|---|---|---|---|---|---|---|---|
| M | SL | F | SB | ||||
| Acanthoeca | 48 | 79 | 6 | 94 | 6 | — | — |
| Stephanoeca | 17 | 71 | 2 | 100 | — | — | — |
| Diaphanoeca | 55 | 91 | 9 | 100 | — | — | — |
| MACHO 1 | 16 | 88 | 2 | 100 | — | — | — |
| Acanthocorbis | 71 | 90 | 11 | 100 | — | — | — |
| Monosiga | 35 | 71 | 4 | 100 | — | — | — |
| Langenoeca | 17 | 88 | 3 | 82 | 6 | 12 | — |
| FRESCHO 1 | 22 | 100 | 4 | — | 5 | 95 | — |
| Urceolata | 12 | 75 | 2 | 100 | — | — | — |
| Salpingoeca | 31 | 87 | 5 | — | 6 | 94 | — |
| Napiformis | 15 | 93 | 2 | — | — | 100 | — |
| FRESCHO2 | 5 | 100 | 1 | — | — | 100 | — |
| Pyxidium | 2 | 50 | NA | — | — | 100 | — |
| FRESCHO 4a | 18 | 100 | 3 | — | — | 100 | — |
| FRESCHO 3 | 33 | 100 | 6 | — | — | 100 | — |
| Clade L | 18 | 100 | 3 | 44 | — | 56 | —— |
| Other MACHOa | 9 | 100 | 2 | 100 | — | — | — |
| Other FRESCHOa | 5 | 100 | 1 | — | 20 | 80 | — |
| Choanoflagellatea | 429 | 88 | 66 | 66 | 2 | 32 | — |
| MAOP 2 | 15 | 100 | 3 | 100 | — | — | — |
| MAOP 1 | 9 | 100 | 2 | 100 | — | — | — |
| Corallochytrium | 29 | 97 | 5 | 100 | — | — | — |
| Marine Opisthokonts | 42 | 98 | 9 | 100 | — | — | — |
| Eccrinales | 15 | — | — | — | — | — | 100 |
| Ichthyophonus | 38 | — | — | — | — | — | 100 |
| Amoebidium | 7 | 57 | 1 | — | — | 57 | 43 |
| Psorospermium | 1 | — | — | — | — | — | 100 |
| FRESHIP 1 | 4 | 100 | 1 | — | — | 100 | — |
| Abeoforma | 48 | 33 | 3 | 31 | 2 | — | 67 |
| MAIP 1 | 34 | 100 | 6 | 94 | 3 | 3 | — |
| FRESHIP 2a | 3 | 100 | 1 | — | — | 100 | — |
| Anurofeca | 11 | 82 | 2 | — | — | 82 | 18 |
| Pseudoperkinsus | 82 | 82 | 11 | 82 | — | — | 18 |
| Dermocystidium | 29 | 7 | NA | — | 3 | 3 | 93 |
| Other MAIPa | 5 | 100 | 1 | 100 | — | — | — |
| Ichthyosporea | 277 | 52 | 24 | 43 | 1 | 8 | 48 |
| Capsaspora | 5 | — | — | — | — | — | 100 |
| Ministeria | 2 | — | — | 100 | — | — | — |
| Filasterea | 7 | — | — | 29 | — | 71 | |
| Simplex | 11 | 64 | 1 | — | 9 | 91 | — |
| Termophila | 8 | 38 | 1 | — | 13 | 88 | — |
| Fonticula | 2 | 50 | NA | 50 | — | 50 | — |
| FRESHOP | 2 | 100 | NA | — | — | 100 | — |
| Discicristoidea | 23 | 57 | 2 | 4 | 9 | 87 | — |
| Total | 789 | 75 | 100 | 58 | 2 | 23 | 17 |
Note.—#seq, total number of sequences retrieved from GenBank; %env, percentage of environmental sequences; %env opisto, percentage of environmental sequences within the unicellular opisthokonts; M, marine; SL, salt lake; F, freshwater; SB, symbiont. NA, not appicable. Sequences were assigned using two different maximum likelihood phylogenetic trees: one with only complete sequences (fig. 1) and one inferred from both complete and partial 18S rDNA sequences (442 sequences in total, 1,286 informative positions, shown in supplementary information S3, Supplementary Material online).
aClades that appear as an unnamed singletons in figure 1 or that are only recovered from partial sequences.
Besides finding new environmental clades within the opisthokonts, our data suggest that the ichthyosporeans could play a more significant ecological role in both marine and freshwater environments than previously thought. To date, all the described ichthyosporeans have been isolated from metazoans. Therefore, it was assumed that all ichthyosporeans were either parasites or commensals of vertebrates and invertebrates (Ragan et al. 1996; Mendoza et al. 2002). However, our data show that some ichthyosporean groups, such as the Abeoforma, Pseudoperkinsus, and Anurofeca clades, contain an important proportion (up to 82%) of environmental sequences. These are similar percentages of environmental sequences as some choanoflagellate clades (table 1). In contrast, clearly parasitic ichthyosporean clades, such as Ichthyophonus and Dermocystidium, have much smaller percentages of environmental sequences (0–7%). Moreover, we identified two ichthyosporean clades, MAIP 1 and FRESHIP 1, which are exclusively environmental. Moreover, the ichthyosporeans represent an important percentage (24%) of the overall unicellular opisthokont sequences, with some clades (such as MAIP1 and Pseudoperkinsus) representing 6% and 11% of the total of unicellular opisthokont sequences, respectively; similar values to those of some choanoflagellate groups (table 1). A possible explanation for this large percentage of environmental sequences among the ichthyosporeans is that some clades may have representatives with a free-living stage, at least. However, we cannot rule out the possibility that they are infecting small organisms (such as Syndiniales infecting Dinoflagellates, see Guillou et al. 2008) that can be recovered in the 0.2–3 µm fraction used in most of the studies (Massana and Pedrós-Alió 2008).
Overall, our data show a large and hidden diversity of nonfungal unicellular opisthokonts, except for the filastereans. This hidden diversity, unveiled only after analyzing the complete set of 18S rDNA sequences from all published environmental studies based on clone libraries, indicates an important bias in the representation previously assigned to cultures, which constitute only a very small fraction of the real diversity. As expected, choanoflagellates are the most abundant and important unicellular opisthokont group in the environment, but both the ichthyosporeans and Corallochytrium clade represent as well an important fraction of the unicellular opisthokont diversity and have the potential to be prominent ecological players. Thus, it is crucial to increase our efforts in culturing the little-known unicellular opisthokonts to better understand their ecological role. Our phylogenetic tree provides an important phylogenetic framework for future evolutionary studies of the ichthyosporeans, nucleariids, Corallochytrium, filastereans, and choanoflagellates. Moreover, new environmental data, single-cell genomics, and the application of fluorescence in situ hybridization to environmental samples are also needed to further increase our knowledge of the real unicellular opisthokont diversity.
Materials and Methods
Environmental 18S rDNA sequences of opisthokonts were obtained from GenBank following a two-step screening process. First, we retrieved sequences using the NCBI taxonomy tool. The sequences were further checked by blastn to confirm their phylogenetic assignment. Second, we used these and other published sequences from cultures or environmental surveys that belong to the target groups (but are not labeled as such in GenBank) to retrieve additional sequences using blastn. Putative chimeric sequences were further checked with KeyDNATools (Guillou et al. 2013) as described elsewhere (Guillou et al. 2008). A total of 789 sequences were finally retrieved from GenBank. Maximum likelihood phylogenetic trees were constructed using RAxML with the GTRCATI model of evolution and with wide taxon coverage, to establish whether ambiguous divergent sequences belonged to a given group. Repeated runs on distinct starting trees were carried out to select the tree with the best topology (the most likely of 1,000 alternative trees). Details on the phylogenetic methods are explained elsewhere (del Campo and Massana 2011).
Supplementary Material
Supplementary information S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Diego Mallo for bioinformatic support and to Ramon Massana for helpful discussions. This work was supported by an Institució Catalana per a la Recerca i Estudis Avançats contract, a European Research Council Starting Grant (ERC-2007-StG-206883), and a grant (BFU2011-23434) from Ministerio de Economía y Competitividad (MINECO) to I.R.-T.
References
- Amaral-Zettler LA, Nerad TA, O’Kelly CJ, Sogin ML. The nucleariid amoebae: more protists at the animal-fungal boundary. J Eukaryot Microbiol. 2001;48:293–297. doi: 10.1111/j.1550-7408.2001.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Carr M, Leadbeater BSC, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Megaphylogeny, cell body plans, adaptive zones: causes and timing of eukaryote basal radiations. J Eukaryot Microbiol. 2009;56:26–33. doi: 10.1111/j.1550-7408.2008.00373.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Paula Allsopp MTE. Corallochytrium, an enigmatic non-flagellate protozoan related to choanoflagellates. Eur J Protistol. 1996;32:306–310. [Google Scholar]
- del Campo J, Massana R. Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist. 2011;162:435–448. doi: 10.1016/j.protis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S, et al. (32 co-authors) The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, Scanlan DJ, Worden AZ. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- Marshall WL, Berbee ML. Facing unknowns: living cultures (Pirum gemmata gen. nov., sp. nov., and Abeoforma whisleri, gen. nov., sp. nov.) from invertebrate digestive tracts represent an undescribed clade within the unicellular Opisthokont lineage Ichthyosporea (Mesomycetozoea) Protist. 2010;162:33–57. doi: 10.1016/j.protis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Massana R, Pedrós-Alió C. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Taylor JW, Ajello L. The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Ann Rev Microbiol. 2002;56:315–344. doi: 10.1146/annurev.micro.56.012302.160950. [DOI] [PubMed] [Google Scholar]
- Nitsche F, Carr M, Arndt H, Leadbeater BS. Higher level taxonomy and molecular phylogenetics of the choanoflagellatea. J Eukaryot Microbiol. 2011;58:452–462. doi: 10.1111/j.1550-7408.2011.00572.x. [DOI] [PubMed] [Google Scholar]
- Paps J, Ruiz-Trillo I. 2010. Animals and their unicellular ancestors. In: Encyclopedia of life sciences (ELS). John Wiley & Sons, Ltd: Chichester, UK. doi:10.1002/9780470015902.a0022853. [Google Scholar]
- Pawlowski J, Audic S, Adl SM, et al. (33 co-authors) CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA, Goggin CL, Cawthorn RJ, Cerenius L, Jamieson AV, Plourde SM, Rand TG, Söderhall K, Gutell RR. A novel clade of protistan parasites near the animal-fungal divergence. Proc Natl Acad Sci U S A. 1996;93:11907–11912. doi: 10.1073/pnas.93.21.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu-kumar S. Occurrence of the traustochytrid Corallochytrium limacisporum gen. et sp. nov. in the coral reef lagoons of the Lakshadweep Islands in the Arabian Sea. Bot Mar. 1987;30:83–89. [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr RJS, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS One. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Weber F, del Campo J, Wylezich C, Massana R, Jürgens K. Unveiling trophic functions of uncultured protist taxa by incubation experiments in the brackish Baltic Sea. PLoS One. 2012;7:e41970. doi: 10.1371/journal.pone.0041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.