Abstract
Paclitaxel (Taxol®) is an anti-cancer drug that induces mitotic arrest via microtubule hyperstabilization, but causes side effects due to its hydrophobicity and cellular promiscuity. The targeted cytotoxicity of hydrophilic paclitaxel-conjugated polyamidoamine (PAMAM) dendrimers has been demonstrated in cultured cancer cells. Mechanisms of action responsible for this cytotoxicity are unknown—i.e., whether the cytotoxicity is due to paclitaxel stabilization of microtubules — as is whether paclitaxel is released intracellularly from the dendrimer. To determine whether the conjugated paclitaxel can bind microtubules, we used a combination of ensemble and single microtubule imaging techniques in vitro. We demonstrate that these conjugates adversely affect microtubules by: (1) promoting the polymerization and stabilization of microtubules in a paclitaxel-dependent manner; and (2) bundling pre-formed microtubules in a paclitaxel-independent manner, potentially due to protonation of tertiary amines in the dendrimer interior. Our results provide mechanistic insights into the cytotoxicity of paclitaxel-conjugated PAMAM dendrimers and uncover unexpected risks of using such conjugates therapeutically.
Keywords: paclitaxel, PAMAM dendrimers, microtubules, microtubule bundling, targeted cancer drug delivery
Introduction
Paclitaxel is a small organic molecule that is marketed as an anti-cancer drug (Taxol®, Bristol-Myers Squibb, New York, NY; Figure 1) because of its cytotoxic properties that derive from its ability to hyperstabilize microtubules against depolymerization and, consequently, arrest cell division, particularly of rapidly dividing cells.1 Since paclitaxel is poorly water soluble, it is currently solubilized in a mixture of polyethoxylated castor oil and ethanol prior to injection into the patient.2 These toxic solubilizing agents, the promiscuous cytotoxicity of paclitaxel, and the fact that paclitaxel has a high binding affinity for plasma proteins, lead to detrimental side effects and decreased drug efficiency.2 Nevertheless, paclitaxel has proven to be a successful cancer drug,1 making the design of a targeted delivery strategy using a water-soluble form of paclitaxel very desirable.
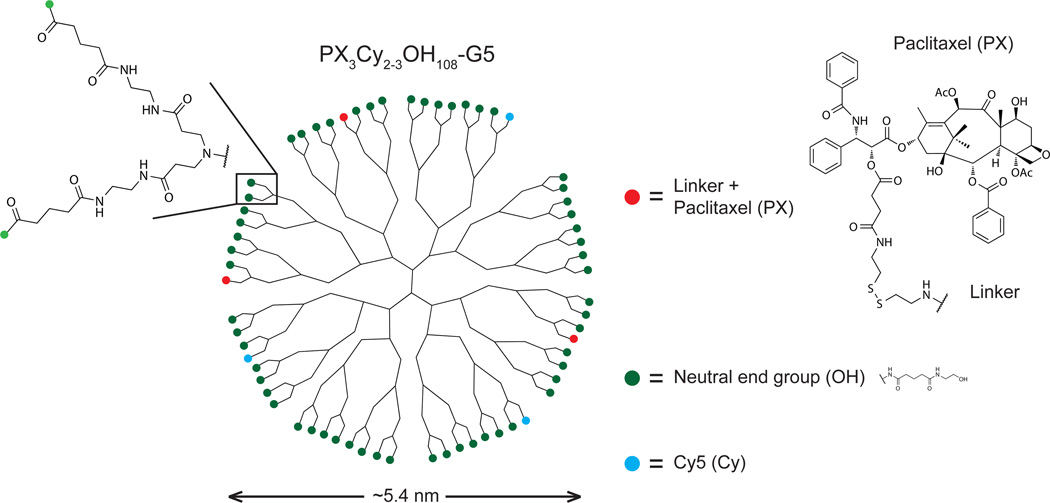
Figure 1.
Two-dimensional schematic of the fifth generation (G5), paclitaxel (PX)-conjugated PAMAM dendrimer used in this study (systematically termed PX3Cy2–3OH108-G5 by its chemical composition). The theoretical number of terminal branches per G5 dendrimer is 128, but only a fraction of the branches (64) are shown for clarity. The batch used in this study had 144 end groups on average, as determined by potentiometric titration. The modification of all end groups from the original primary amines is depicted by colored circles, showing covalent attachment of 3 paclitaxel molecules (PX: red) and 3 Cy5 molecules (Cy: blue), on average. The remaining surface groups were neutralized by the addition of 108 hydroxyl groups, on average (OH: dark green). The chemical structures of the hydroxyl end groups (OH) and paclitaxel (PX)—covalently linked to the dendrimer at the 2’OH (red) by a linker containing an ester, disulfide bridge, and amide bond—are shown on the right.
Numerous targeted delivery strategies for paclitaxel that aim to overcome its limitations are currently being explored experimentally and in clinical trials.2–4 One potential strategy is to use a water-soluble drug carrier such as a polyamidoamine (PAMAM) dendrimer. PAMAM dendrimers are branched, organic nanoparticles that are highly multivalent, allowing for the attachment of many (different) ligands (Figure 1).5, 6 PAMAM dendrimers have shown promise as a drug delivery platform in the treatment of many diseases, including cancer.4, 7–10 Notably, paclitaxel-conjugated generation 5 (G5)11 PAMAM dendrimers have demonstrated cytotoxicity against cancerous cells.12 The mechanism(s) of cytotoxicity of these paclitaxel-conjugated dendrimers, however, was not investigated. That is, it is not known if the observed cytotoxicity is due to paclitaxel stabilization of microtubules, or to some other mechanism(s). The dendrimers were designed so that the paclitaxel load can be cleaved from the dendrimer carrier inside the cell, via the hydrolysis of the ester bond at the C2’-OH of paclitaxel (Figure 1), and/or a second ester bond that tethers the drug-linker to the dendrimer carrier. This mechanism has not been experimentally confirmed, although numerous studies suggest that such paclitaxel prodrugs similarly employing a C2’-ester bond have no in vitro activity, but gain activity in vivo, suggesting that this ester bond may be cleaved intracellularly to release paclitaxel.13–15 We have now replaced the second ester bond within the linker with a disulfide bridge and amide bond (Figure 1)16, 17 to provide better chemical stability of the linker during synthesis. In addition, a disulfide bond may provide a second mechanism for the intracellular release of the paclitaxel load, as it can be cleaved in the reductive environment of endosomes or lysosomes.16, 17 However, the questions remain whether paclitaxel is cleaved from the dendrimer carrier in the cell (and if so, how rapidly), and whether the still paclitaxel-coupled or paclitaxel-free dendrimer may contribute to cytotoxicity by binding to microtubules.
To address the question of whether PAMAM dendrimers, with or without paclitaxel, bind to microtubules, we here have used a turbidity assay as well as two single microtubule microscopy techniques, total internal reflection fluorescence microscopy (TIRFM) and transmission electron microscopy (TEM), to investigate the effects of paclitaxel-conjugated or unconjugated dendrimers on purified microtubules in vitro. We performed these assays in vitro in order to directly observe the effects of the dendrimers on microtubules, while removing the possibility of the conjugates causing cytoxicity through non-microtubule interactions. First, we find that the paclitaxel-conjugated G5 PAMAM dendrimers stabilize polymerizing microtubules against depolymerization, in a paclitaxel-dependent manner, although much less efficiently than free paclitaxel. Second, and more surprisingly, we find that G5 PAMAM dendrimers are able to bind and bundle preformed microtubules, even after removing both paclitaxel and any surface charges (by addition of hydroxyl groups to the terminal primary amine groups). This paclitaxel-independent mode of action is probably induced, at least in part, by electrostatic interactions, involving protonated tertiary amine functionalities of the interior dendrimer core (Figure 1). Both modes of action result in microtubules stabilized against depolymerization, which is expected to impair cellular mitosis. Therefore, at least at the concentrations used in this study, our results call to attention the need to carefully consider potentially harmful molecular interactions mediated even by surface-neutralized drug carriers during the design of novel targeted drug delivery strategies.
Materials and Methods
Materials
2-(N-morpholino)ethanesulfonic acid (MES), MgCl2, and guanosine triphosphate (GTP) were all purchased from Fisher Scientific (Waltham, MA). EGTA, paclitaxel, protocatechuate acid (PCA), and protocatechuate-3,4-dioxygenase (PCD) from Sigma-Aldrich (St. Louis, MO); Trolox from Acros Organics (Geel, Belgium); GpCpp (GMPCPP) from Jena Biosciences (Jena, Germany); 5-(6)-TAMRA succimidyl ester (TMR) from Molecular Probes (Eugene, OR); and Cy5 succimidyl ester (Cy) from GE Healthcare Life Sciences (Piscataway Township, NJ).
Tubulin purification and polymerization
Tubulin was purified from bovine brain and labeled with TMR, as described before.18, 19 For all experiments, microtubules were polymerized by incubating 2 mg/mL (≈ 20 µM) α/β-tubulin dimers (using a mix of TMR-labeled and unlabeled tubulin dimers to achieve a final ratio of 1 TMR dye per 20 dimers, as determined by UV-Vis, where noted) with 4 mM MgCl2 and 1 mM GTP, or GMPCPP where noted, in MEM806:8 buffer (80 mM MES-KOH, pH 6.8, 1 mM EGTA, 2 mM MgCl2) at 37°C for 30 min. After polymerization, the microtubules were stabilized with 10 µM paclitaxel, unless otherwise noted.
Synthesis and characterization of paclitaxel-conjugated G5 PAMAM dendrimers
G5 PAMAM dendrimers ((NH2)114-G5) were purchased from Dendritech, Inc. (Midland, MI). The dendrimers were first purified using a 10 kDa molecular weight cut off (MWCO) dialysis membrane, achieving a relatively monodisperse population (polydispersity index (PDI) = 1.01–1.05, determined by gel permeation chromatography (GPC)).20 The average number of primary amine end groups was determined to be 114 by potentiometric titration after purification by dialysis, membrane filtration, and lyophilization.21 These dendrimers were first reacted with 5 molar equivalents of Cy5 NHS-ester to achieve an average of 2–3 Cy5 molecules per dendrimer, as determined by 1H NMR and UV-Vis. The Cy5 labeled PAMAM amine-terminated dendrimer was then fully capped with carboxylic acid groups by reacting glutaric anhydride with the primary amine end groups on the dendrimer. Finally, the Cy5-carboxylated dendrimer was reacted with 10 molar equivalents of paclitaxel, previously conjugated to the ester-disulfide-amide linker (see Supplementary Materials), to achieve an average of 3.2 paclitaxel molecules per dendrimer, as determined by 1H NMR. The residual active groups were neutralized by addition of hydroxyl groups. More details concerning the synthesis, purification, and characterization of the paclitaxel-conjugated G5 dendrimer, as well as of the paclitaxel-conjugated G3 dendrimer and the paclitaxel linkers used for each dendrimer, are available in the Supplementary Materials.
Turbidity Assays
Turbidity assays were conducted as described previously.22 Briefly, 2.0 mg/mL of unlabeled tubulin (≈ 20 µM) was mixed with 4 mM MgCl2 in MEM806:8 and incubated on ice for 30 min. Meanwhile, the following components were pre-warmed at 37° in cuvettes: (1) no additional components; (2) 1 mM GTP; (3) 1 mM GTP and 10 µM paclitaxel; (4) 1 mM GTP and 3.33 µM PX3Cy2–3OH108-G5; or (5) 1 mM GTP and 3.33 µM Cy2–3OH111-G5. After 30 min, the tubulin mixtures were added to the cuvettes and the readings were begun immediately. The turbidity (polymerization) of the reaction mixtures was monitored at 340 nm once every minute for 30 min at 37°C on a Beckman DU-640 UV-Vis Spectrometer utilizing a High Performance Peltier Temperature Controller (Beckman Coulter, Brea, CA).
TIRFM
Imaging chambers were prepared by affixing a cover glass (No. 1.5, 24×30 mm, VWR, Radnor, PA) to a glass slide (Fischer Scientific, Waltham, MA) with double-sided sticky tape. After the imaging solution was flown into the chamber, the chamber was sealed with candle wax. Images were taken on an inverted fluorescence microscope (model IX81, Olympus, Center Valley, PA) using a 60x objective lens. Samples were illuminated at either 532 nm (for TMR; type Compass 315M, Coherent Inc., Santa Clara, CA) or 635 nm (for Cy5; type Cube 640–100C, Coherent Inc., Santa Clara, CA) at the critical angle, using a cellˆ TIRF™ Illuminator (Olympus, Center Valley, PA). Fluorescent emissions were split into four separate channels using a QV2 Quad View Imaging System (Photometrics, Tuscon, AZ) and projected onto an EMCCD camera (model Evolve 512, Photometrics, Tuscon, AZ). Fluorescent images were viewed using MetaMorph software (Molecular Devices, Sunnyvale, CA).
TIRFM Polymerization Assays
2.0 mg/mL of a mix of unlabeled and TMR-labeled (see above) tubulin (≈ 20 µM) was mixed with 4 mM MgCl2 and 1 mM GTP in MEM806:8 and either (1) no additional components; (2) 10 µM paclitaxel; (3) 3.33 µM PX3Cy2–3OH108-G5; or (4) 3.33 µM Cy2–3OH111-G5 at 37°C for 30 min. The resulting tubulin mixtures were incubated at room temperature for another 180 min to allow unstable microtubules to depolymerize it was first determined that no microtubules from sample (1) could be detected by TIRFM after 180 min incubation at room temperature). Prior to visualization by TIRFM, the samples were either diluted to 4 (sample 2) or 15 (samples 1, 3, 4) µM tubulin in MEM806:8supplemented with an oxygen scavenging system (OSS; 5 mM PCA, 50 nM PCD, 2 mM Trolox).
Co-localization of Dendrimers and Pre-formed Microtubules Observed by TIRFM
TMR-labeled, paclitaxel- or GMPCPP-stabilized microtubules were polymerized and stabilized as described above (see section Tubulin purification and polymerization) and incubated with either (1) PX3Cy2–3OH108-G5; or (2) Cy2–3OH111-G5 and OSS at a 7:1 ratio of dendrimers:tubulin dimers (2.1:0.29 µM) in MEM806:8 for 30, 105, or 180 min at room temperature prior to visualization by TIRFM. To determine the proportion of microtubules bound by dendrimers for each sample, a coverslip area of 0.5–2 mm2 was scanned, the bound and unbound microtubules were counted, and the number of microtubules counted were normalized to the total coverslip area scanned. Statistical significance of the difference of the proportion of dendrimer-bound microtubules counted between samples was determined using the Fisher’s exact test at significance level α = 0.05.
TEM
Carbon-coated copper mesh TEM grids were glow-discharged using a Solarus 950 (Gatan, Inc., Pleasanton, CA). Samples were placed on the carbon-coated side of the grid and negatively stained with a 0.75 % solution of uranyl formate.23 It was assumed that the acidic pH of the stain would not significantly alter the pH of the sample as the stain would fix the sample in ≤ 10 ms.24 Samples were imaged using a Morgagni 268 transmission electron microscope (FEI, Hillsboro, OR).
Microtubule Bundling Observed by TEM
TMR-labeled microtubules were polymerized as described above (see section Tubulin purification and polymerization) and pelleted from unpolymerized tubulin by centrifugation (Airfuge, rotor A-100, Beckman, Fullerton, CA; 30 s at 30 psi). After centrifugation, microtubules were resuspended in MEM806:8 to an estimated 3.2 µM, based on the microtubule recovery efficiencies we determined previously (data not shown). Microtubules (at 0.64 µM tubulin) were then incubated with either (1) 10 µM paclitaxel; (2) 3.33 µM PX3Cy2–3OH108-G5 (which equates to 10 µM of conjugated paclitaxel); or (3) 3.33 µMCy2–3OH111-G5 to achieve a 1:1 ratio of dendrimer:tubulin dimers in MEM80 at variable pH, adjusted with KOH, for 30 min at room temperature prior to visualization by TEM.
Quantitative Analysis of Microtubule Bundling in TEM Images
The diameter of microtubule (bundles) in TEM images taken at 3,095× magnification was measured at 10 pixel intervals and the corresponding microtubule length was weighted according to the number of bundled microtubules determined to be in that length. The diameter of a single, unbundled microtubule, within two standard deviations from the mean, was assigned to the weight of 1, and this was used as a basis to assign diameter ranges to higher weights for bundled microtubules. The fraction of bundled microtubules in a sample was calculated as the fraction of bundled microtubule length per total measured microtubule length. This analytical process was automated using a MATLAB (MathWorks, Natick, MA) script written in-house.
Statistical Tests for Significance in Bundling
Statistical significance of the difference of mean bundled microtubule length between samples was tested using the Student’s t-test, assuming equal variances, at significance level α = 0.05. Prior to this test, it was verified that the two samples being compared had equal variances using the F-test at significance level α = 0.05. All statistical tests were performed using the Statistics Toolbox in MATLAB (MathWorks, Natick, MA).
Results and Discussion
Probing the Interactions of Modified PAMAM Dendrimers with Microtubules
The design of the paclitaxel-conjugated, generation 5 (G5) or 3 (G3), PAMAM dendrimers used in this study, hereafter referred to by their stoichiometry as PX3Cy2–3OH108-G5 (Figure 1) and PX3Cy2–3OH26-G3, respectively, was based on the G5 paclitaxel-conjugate used in our previous cytotoxicity study.12 As the purpose of the previously designed conjugate was for targeted cancer drug delivery, the conjugate bore the therapeutic drug paclitaxel, the tumor targeting molecule folic acid, and the fluorescent molecule FITC. In the current study, we did not include folic acid on the conjugates for simplicity since we were not testing them on cells, and replaced the fluorophore FITC with Cy5 to achieve the most sensitive detection by objective-type TIRFM.25
For increased stability during chemical conjugation, a new paclitaxel-dendrimer linker was developed that contained an ester-disulfide-amide linker instead of the double-ester linker used in the previous study (Figure 1).12 An additional benefit to this change is that the disulfide bond may be cleaved in the reductive environment of the endosome or lysosome, if the conjugate enters the cell via receptor-mediated endocytosis. This mechanism has shown promise for the delivery of drug therapeutics linked to carriers with disulfide bonds,26 including the specific case using paclitaxel,16, 17 although the supporting results are still debated.27
Generally, the synthesis, purification, and analysis of the conjugates were performed following standard methods described elsewhere (Materials and Methods and Supplementary Materials).28 Cy5 was first conjugated to the G5, or G3, dendrimer at a mean stoichiometric ratio of 2–3 fluorophores per dendrimer, as determined by 1H NMR and UV-Vis spectroscopy. Next, the paclitaxel-linker was conjugated to the G5 and G3 dendrimer core at a mean stoichiometric ratio of 3.2 and 2.8, respectively, drug molecules per dendrimer, as determined by 1H NMR spectroscopy. Prior to conjugation to the dendrimer, the integrity of the paclitaxel-linker was confirmed by high resolution mass spectroscopy and 1H NMR spectroscopy. Finally, the remaining surface groups on the dendrimer, initially primary amines, were neutralized by the addition of hydroxyl groups, as previous studies have found that highly cationic PAMAM dendrimers disrupt cellular membranes.29, 30 UPLC was used to confirm that no detectable level of free paclitaxel was present in the conjugate preparations (see Supplementary Materials) and that the paclitaxel conjugate was stable in water for up to 20 h, which is much greater than the time period of an experiment (≤ 6 h). As negative controls, G5 dendrimers without paclitaxel were synthesized in parallel with PX3Cy2–3OH108-G5, with the exemption of the paclitaxel conjugation steps. Therefore, the only chemical difference between these dendrimers and PX3Cy2–3OH108-G5 is the presence of paclitaxel. Hereafter this dendrimer will be referred to as Cy2–3OH111-G5.
Microtubule polymerization and all experiments were carried out in a standard, near-physiological buffer termed “MEM806:8”, composed of 80 mM MES-KOH, pH 6.8, 1 mM ethylene glycol tetraacetic acid (EGTA), and 2 mM MgCl2 (Materials and Methods). This buffer will be referred to as only “MEM80” when the pH is varied from 6.8. MEM806:8 is identical in composition to BRB80, a buffer commonly used for polymerizing microtubules in vitro,31 except the buffering agent PIPES is replaced by MES.32 For observing dendrimer binding by TIRFM co-localization, we used sparsely tetramethylrhodamine (TMR)-labeled tubulin purified from bovine brain, as described before,19 and a dendrimer:tubulin dimer ratio of 7:1. No binding events were observed by TIRFM when the dendrimer concentration was significantly less than a 7-fold excess of the tubulin concentration. Since there are on average three paclitaxel molecules conjugated to each dendrimer, this ratio would equate to achieving a cellular paclitaxel concentration of 300–400 µM, assuming an average cellular tubulin concentration of 15–20 µM.33, 34 To decrease background signal from unbound dendrimers, we performed TEM assays at a slightly lesser dendrimer:tubulin dimer ratio of 1:1, which corresponds to a cellular paclitaxel concentration of 15–20 µM. Note that although the paclitaxel concentrations used in our experiments seem very high in this context, these concentrations are standard for in vitro stabilization of microtubules,35 which we are focusing on in this study.
G5 PAMAM Dendrimers Induce Turbidities in Tubulin Solutions Comparable to Paclitaxel
Turbidity assays are standard ensemble microtubule polymerization assays as the turbidity of a tubulin solution is a reliable measure of the mass of microtubules present.36 To determine the ability of PX3Cy2–3OH108-G5 to induce microtubule polymerization and stabilization compared to paclitaxel, we mixed tubulin together with these components and monitored the turbidity of the mixtures at 37°C for 30 min, as described in the Materials and Methods section and elsewhere.22 The following controls were also tested for comparison: (1) tubulin in the absence of GTP, paclitaxel, or dendrimers; and (2) tubulin, GTP, and Cy2–3OH111-G5.
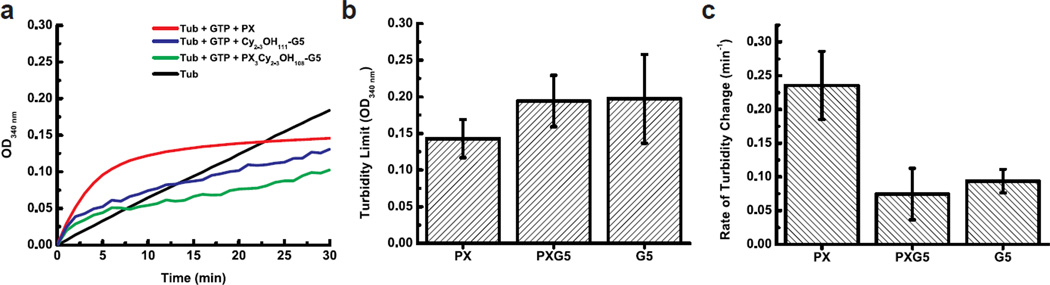
The results of these turbidity assays are summarized in Figure 2. During the time course of the experiment, the turbidity of all tubulin mixtures increased in an exponential fashion (R2 = 0.96–0.99), except for the tubulin mixture not containing GTP, paclitaxel, or dendrimers, which increased in a linear fashion (R2 = 0.9995) (Figure 2a). Notably, the turbidity limits reached by the tubulin mixtures in the presence of paclitaxel, PX3Cy2–3OH108-G5, or Cy2–3OH111-G5 were not significantly different (p > 0.05, Student’s t-test), on average (Figure 2b). However, the rate of turbidity change was about 3-fold greater in the presence of paclitaxel, compared to that measured in the presence of PX3Cy2–3OH108-G5, or Cy2–3OH111-G5 (which were not significantly different from each other; p > 0.05, Student’s t-test) (Figure 2c). The exponential increase in turbidity of the tubulin mixtures in the presence of dendrimers while at 37°C suggests the formation of microtubules or alternatively, non-microtubule tubulin aggregates. To distinguish between these alternatives, we next turned to TIRFM, which should allow visualization of single microtubules.
Figure 2.
Turbidity polymerization assays. (a) The average turbidities of 2 mg/mL tubulin (Tub) solutions in the presence of (1) no extra components (black; n = 2); (2) GTP and paclitaxel (PX) (red; n = 3); (3) GTP and PX3Cy2–3OH108-G5 (green; n = 3); or (4) GTP and Cy2–3OH111-G5 (blue; n = 4) was monitored at 340 nm once every minute for 30 min at 37°C. Error bars are not shown for clarity. All turbidity trends, with the exception of tubulin alone (which was fit with a linear equation), were fit with the exponential growth equation y = A(1-e−kt) at R2 = 0.96–0.99. (b) The average turbidity limits (parameter A from fit) from the tubulin mixtures in the presence of GTP and (1) paclitaxel (PX); (2) PX3Cy2–3OH108-G5 (PXG5); or (3) Cy2–3OH111-G5 (G5). (c) The average rate of turbidity changes (parameter k from fit) from the tubulin mixtures.
PX3Cy2–3OH108-G5 Stabilize Polymerizing Microtubules
To determine if the dendrimers are able to induce to the formation of microtubule-like structures, we followed the same general procedure as with the turbidity assays, except mixing in a small fraction of TMR-labeled tubulin (5% of all tubulin dimers carry one TMR dye, see Materials and Methods) to allow visualization by TIRFM. To further test the ability of PX3Cy2–3OH108-G5 to not only promote polymerization of microtubules, but to stabilize them, we incubated the tubulin mixtures at room temperature following the 30 min incubation at 37°C. Prior to the experiment, we determined how long a tubulin mixture in the presence of only GTP has to be incubated at room temperature (180 min) before the concentration of unpolymerized tubulin in solution becomes so great that the fluorescence from this tubulin obscures the fluorescence of any remaining microtubules on the slide surface.
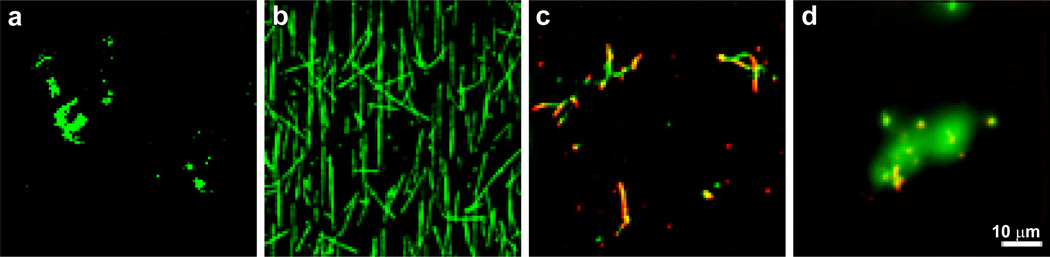
The results of these TIRFM experiments are summarized in Figure 3. No microtubules were observed in the absence of paclitaxel after 180 min (Figure 3a); but instead a fluorescent haze (not shown) due to the high concentration of unpolymerized tubulin in solution. Contrast adjustment of these images revealed unpolymerized tubulin on the surface (Figure 3a). By contrast, typical microtubules, identified as green rods based on their TMR fluorescence, were polymerized and stabilized by free paclitaxel, in the absence of dendrimers (Figure 3b). In the presence of PX3Cy2–3OH108-G5, microtubule-like green rods (Figure 3c), similar to those observed in Figure 3a, were co-localized with PX3Cy2–3OH108-G5, which are found through their red Cy5 fluorescence. With a few exceptions, PX3Cy2–3OH108-G5 dendrimers were often observed to colocalize at the ends of the microtubule-like rods. The apparent microtubules observed in Figure 3c were likely stabilized by PX3Cy2–3OH108-G5, since microtubules polymerized at the tubulin concentrations used in this experiment completely depolymerize after 180 min (the incubation time used in this experiment) in the absence of dendrimers and paclitaxel (Figure 3a). However, stabilization by PX3Cy2–3OH108-G5 was very inefficient compared to free paclitaxel, as evidenced by the difference in microtubule density between Figures 3b and c, made even greater by the fact that the tubulin imaging concentration in Figure 3c was about 4-fold greater than the concentration in Figure 3b. This reduction in tubulin imaging concentration for Figure 3b was implemented in order to get a clear image, as the microtubules were initially too dense on the slide at the imaging concentration of Figure 3a; c–d. In the presence of paclitaxel-unconjugated Cy2–3OH111-G5, only non-cylindrical tubulin aggregates were observed, which were seemingly bound by dendrimers, as evidenced by the co-localization of green TMR and red Cy5 fluorescence in some areas (Figure 3d).
Figure 3.
Polymerization assays imaged by TIRFM. Images from the TMR channel (green: microtubules) are overlaid onto images from the Cy5 channel (red: dendrimers). Scale bar shown is 10 µm. In these assays, 2 mg/mL α/β-tubulin heterodimers are incubated with either (a) no stabilizers; (b) free paclitaxel; (c) PX3Cy2–3OH108-G5; or (d) Cy2–3OH111-G5 in MEM806:8 supplemented with 1 mM GTP at 37°C for 30 min, then at room temperature for 180 min. To obtain the best image in the presence of a high density of microtubules, the microtubule-paclitaxel sample (b) was diluted 4-fold (as compared to the imaging concentration in the other three samples, which were equal) immediately prior to imaging.
Together, these results suggest that PX3Cy2–3OH108-G5 is able to bind microtubules specifically through the conjugated drug, resulting in the stabilization of microtubules polymerized in the presence of the paclitaxel-dendrimer conjugate. This stabilization is, however, very inefficient compared to free paclitaxel. This apparent decrease in binding affinity of the dendrimer-conjugated paclitaxel compared to free paclitaxel may result from its conjugation to the dendrimer, as previous studies have shown that paclitaxel prodrugs modified at the same paclitaxel functional group, the 2’ OH (see Figure 1), have reduced affinity for microtubules.13–15 Additionally, or alternatively, the size of the dendrimer (≈ 5.4 nm) may hinder the binding of a sufficient number of paclitaxel-conjugated dendrimers to accomplish microtubule stabilization. Notably, while some microtubules were bound along their entire length by PX3Cy2–3OH108-G5, many were only bound at the ends (Figure 3c). It is possible in these cases that tubulin dimers bound by PX3Cy2–3OH108-G5 are added to the (+) end of an existing microtubule, stabilizing the (+) end and slowing depolymerization. It is also important to note that Cy2–3OH111-G5—and presumably PX3Cy2–3OH108-G5 as well—binds tubulin through an interaction independent of paclitaxel, and that this interaction does not promote stabilization of microtubules.
Interpreting these results together with the turbidity results (Figure 2), PX3Cy2–3OH108-G5 is able to induce a very low level of polymerization via the conjugated paclitaxel. In addition, as the TIRFM image in Figure 3d suggests, even in the absence of conjugated paclitaxel, Cy2–3OH111-G5 is able to induce tubulin aggregation, which may also lead to an increase in turbidity. Presumably, since PX3Cy2–3OH108-G5 is identical in structure to Cy2–3OH111-G5 with the exception of three molecules of paclitaxel, PX3Cy2–3OH108-G5 can also induce tubulin aggregation. These interpretations may explain the observation that the dendrimers increase the turbidity of a tubulin solution to a level comparable with paclitaxel (Figure 2).
PX3Cy2–3OH108-G5 Binds Paclitaxel-but Not GMPCPP-stabilized Microtubules
We next aimed to determine if PX3Cy2–3OH108-G5 could bind and stabilize pre-assembled microtubules, which it would also encounter in the cell. To test this idea, we probed for co-localization of PX3Cy2–3OH108-G5 with pre-assembled microtubules by TIRFM. Tubulin dimers were preassembled into microtubules and these microtubules were stabilized with an equimolar concentration of free paclitaxel (Materials and Methods), the concentration of which was subsequently reduced by 4-fold dilution upon incubation for 30, 105, or 180 min at room temperature with PX3Cy2–3OH108-G5 or Cy2–3OH111-G5 in MEM806:8which did not contain any additional free paclitaxel, at a 7:1 ratio of dendrimers:tubulin dimers. The number of microtubules bound or unbound by dendrimers was counted for each sample and normalized by the total coverslip surface area scanned.
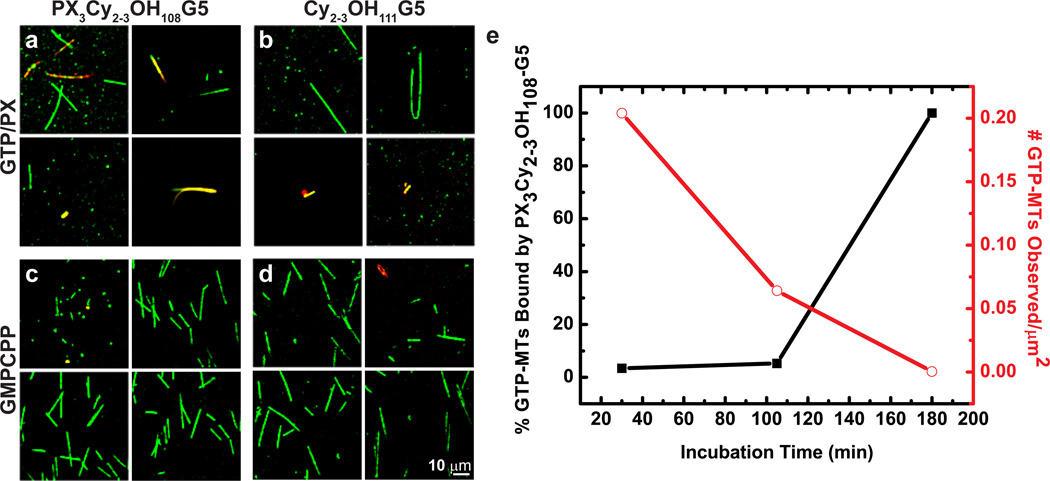
When incubated with PX3Cy2–3OH108-G5, two distinct populations of microtubules were observed (Figure 4a): (1) those that only fluoresced in the TMR emission channel (green in images, corresponding to tubulin); and (2) those that fluoresced in the Cy5 channel (corresponding to PX3Cy2–3OH108-G5 fluorescence) only (red in images) or in addition to the TMR channel (yellow in images). Population 1 appears entirely free of PX3Cy2–3OH108-G5 whereas population 2 is bound throughout by PX3Cy2–3OH108-G5, sometimes so extensively that the TMR signal is not observed, likely due to fluorescence resonance energy transfer (FRET) from TMR on the tubulin to Cy5 on the dendrimer (top left image in Figure 4a). We note that no microtubules that are only bound by dendrimers at the microtubules ends are observed in the present experiment as they were in Figure 3c, and that few uniformly bound microtubules are observed in Figure 3c compared to the present experiment. This apparent discrepancy may be due to limited sampling.
Figure 4.
Co-localization assays between dendrimers and pre-formed microtubules visualized by TIRFM. Images from the TMR channel (green: microtubules) and Cy5 channel (red: dendrimers) are overlaid. Scale bar shown is 10 µm. Microtubules were polymerized, stabilized with either free paclitaxel (a–b) or GMPCPP (c–d), then incubated with either PX3Cy2–3OH108-G5 (a,c) or Cy2–3OH111-G5 (b,d) for 30, 105, or 180 min at room temperature. All images shown are from the 30 min time point, and each 2 × 2 grid of images are from the same sample. The number of microtubules bound or unbound by dendrimers was counted for each sample and normalized by the total coverslip surface area scanned. The plot in (e) shows these normalized data for the PX-stabilized microtubules and the PX3Cy2–3OH108-G5 samples over time.
Different structural populations of PX3Cy2–3OH108-G5-bound microtubules were also observed. Some appeared similar to unbound microtubules (top two images in Figure 4a), but some had a distinctly larger diameter (bottom left image in Figure 4a) whereas others seemed to be bundled microtubules, as represented by the microtubule(s) in the bottom right image of Figure 4a, which appears to have a split tail.
Overall, the fraction of the total microtubule population that was bound by PX3Cy2–3OH108-G5 was initially very small—only 3.4% at the 30 min time point. The bound fraction increased over time, but this partially coincided with a drastic decrease in the total microtubule population (Figure 4e). This population decrease was likely due to inadequate stabilization by PX3Cy2–3-OH108-G5. The very few microtubules that remained at the end of the experiment were bound, and therefore presumably stabilized, by PX3Cy2–3OH108-G5. Recall that PX3Cy2–3OH108-G5 was observed to stabilize microtubules in Figure 3c, and that this stabilization seemed much less efficient than that induced by free paclitaxel. However, we cannot quantitatively compare the extent of stabilization in this latter experiment with the current experiment as the former provided only qualitative observations. The negative control dendrimer, Cy2–3OH111-G5, showed an even smaller incidence of binding: only 2.2% of paclitaxel-microtubules after 105 min. No binding was observed at the 30 and 180 min time points. As found with PX3Cy2–3OH108-G5, all bound microtubules observed were bound by Cy2–3OH111-G5 along their entire length (Figure 4b).
Because we observed so few microtubules bound by PX3Cy2–3OH108-G5, we next asked whether PX3Cy2–3OH108-G5 may still be competing for paclitaxel binding sites with the free paclitaxel used to initially stabilize the microtubules immediately after polymerization, even though its concentration was reduced 20-fold by buffer exchange upon incubation with PX3Cy2–3OH108-G5. To this end, we polymerized stable microtubules in the presence of GMPCPP (a slowly hydrolyzable analogue of GTP)37 instead of GTP and paclitaxel. Hereafter, we will refer to these populations as GMPCPP-microtubules and GTP-microtubules, respectively. The results of this experiment are shown in Figure 4c–d. The fraction of PX3Cy2–3OH108-G5-bound GTP-microtubules observed was significantly greater (p < 0.05) than PX3Cy2–3OH108-G5-bound GMPCPP-microtubules at all time points. With Cy2–3OH111-G5, this difference was significantly greater only at the 105 min time point. These observations suggest that PX3Cy2–3OH108-G5 can either associate more readily with GTP-microtubules or dissociate more readily from GMPCPP-microtubules, and that this observation is specifically caused by the paclitaxel on the dendrimer. As these observations directly oppose those obtained with free paclitaxel,38 the paclitaxel-dependent binding mechanism of PX3Cy2–3OH108-G5 does not likely include access of the interior paclitaxel binding site through the microtubule walls.38 Since the dendrimers are always binding the entire microtubule length, and it is unlikely that PX3Cy2–3OH108G5 is able to diffuse along the microtubule length while inside the microtubule,39 PX3Cy2–3OH108-G5 is also not likely binding the microtubule by entering through its ends. Alternatively it is possible that PX3Cy2–3OH108-G5 is not binding pre-assembled microtubules, but assembling microtubules that have their luminal paclitaxel binding sites exposed to solution, and remaining associated with these microtubules as they complete assembly. This notion supported by the fact that GTP-microtubules depolymerize faster than GMPCPP-microtubules,37 consistent with the observation of more dendrimer bound GTP-microtubules than GMPCPP-microtubules. Furthermore, this may be the binding mechanism utilized by PX3Cy2–3OH108-G5 in the experiments shown in Figure 3. We therefore conclude that the low binding incidence of PX3Cy2–3OH108-G5 is unlikely due to competition with the free paclitaxel used to stabilize the microtubules in the previous experiment.
In addition to G5 paclitaxel-conjugated dendrimers, we also tested the ability of a G3 paclitaxel-conjugated dendrimer to bind microtubules. G3 PAMAM dendrimers are smaller than G5 PAMAM dendrimers, ≈3.6 nm vs. 5.4 nm.5, 40 Our G3 conjugate (see Supplementary Materials for structural details), named PX3Cy2–3OH26-G3 according to its stoichiometry, has the same number of conjugated paclitaxel and Cy5 molecules per dendrimer as our G5 conjugates, and the same neutral hydroxyl structure on the remaining end groups. When PX3Cy2–3OH26-G3 was incubated with either paclitaxel- or GMPCPP-stabilized microtubules for 30, 105, or 180 min, under the same conditions used for the G5 conjugates (see above), no binding events were observed (data not shown). The fact that our G3 paclitaxel-conjugates cannot bind microtubules, but our G5 paclitaxel-conjugates can, may be due to the lower density of protonatable amines in the G3 vs. G5 dendrimer core (1041 vs. 40 at pH 6.8; see results in subsequent sections).
Taking these observations together, we propose that PX3Cy2–3OH108-G5 is able to bind microtubules by two mechanisms: (1) binding and stabilizing polymerizing microtubules in a paclitaxel-dependent manner; and (2) binding pre-formed microtubules in a paclitaxel-independent manner, potentially mediating bundling of these microtubules. The first mechanism is supported by the polymerization experiments described above (see Figure 3). The second mechanism is supported by the fact that high concentrations of paclitaxel (≥33 nM),42 and certain microtubule binding proteins,43 are known to induce bundling of microtubules, as well as by the two lower images of Figure 4a.
It is important to note that while the experiments described thus far suggest these two mechanisms, the ability of PX3Cy2–3OH108-G5 to bind and stabilize microtubules appears by TIRFM to be very low and inefficient, as compared to free paclitaxel. This may be due to a decreased ability, compared to free paclitaxel, of the conjugated paclitaxel to access its binding site in the microtubule lumen. It has been proposed that in vivo small molecules such as paclitaxel are able to access the microtubule lumen through ≈ 1 nm pores in the microtubule wall,38, 44 whereas large molecules, such as the enzyme tubulin acetyltransferase, are able to access the microtubule lumen either through large transient openings in the microtubule wall or by copolymerization.45, 46 The size of PX3Cy2–3OH108-G5 is on the order of tubulin acetyltransferase, and indeed we have observed that PX3Cy2–3OH108-G5 is able to bind and stabilize polymerizing microtubules to some degree, however, it is possible that the entry mechanism utilized by large molecules such as tubulin acetyltransferase in vivo is not readily available to PX3Cy2–3OH108-G5 in vitro. It is also possible that PX3Cy2–3OH108-G5 may be able to access the microtubule lumen via the ≈ 17 nm diameter openings at the microtubule ends (Figure 3c). However, modeling studies have predicted that paclitaxel would take days to reach half equilibrium concentration in the center of a 40 µm microtubule (typical microtubule lengths are 1–100 µm), and an antibody would take years.39 The paclitaxel-conjugated dendrimer, which has a molecular weight far greater than that of paclitaxel alone, but lower than that of an antibody, would also likely take days to years. Therefore, it is unlikely that PX3Cy2–3OH108-G5 is accessing its luminal binding site via this mechanism on the microtubules that we observe to be bound by PX3Cy2–3OH108-G5 along their entire length (Figure 4), and may be binding the outside wall of the microtubule in these instances.
Surface Neutralized G5 PAMAM Dendrimers Bundle Microtubules in a Paclitaxel-Independent Manner
The bundling mechanism discussed above is indirectly supported by the observation that some dendrimer-bound microtubules seem to have a greater diameter than unbound microtubules. Figures 4a–b (microtubules in the presence of PX3Cy2–3OH108-G5 or Cy2–3OH111-G5, respectively) show this qualitatively. In addition, the results of Figure 2 in conjunction with those of Figure 3 suggest that the dendrimers induce the formation of large tubulin aggregates. In order to directly observe the structure of these PX3Cy2–3OH108-G5-bound microtubules and determine whether they are bundled, we next turned to an imaging technique with greater spatial resolution than TIRFM, electron microscopy.
Electron microscopy is routinely used to measure the diameter of microtubules.47–50 Employing the same reagents that were used for the TIRFM experiments, and the same protocol—with the additional step of separating polymerized microtubules from unpolymerized tubulin via centrifugation to decrease tubulin background (Materials and Methods)—we sought to examine dendrimerbound microtubules via negative-stain TEM. Negative staining enables single microtubules to be observed, which is advantageous since dendrimers may have multiple effects on microtubules. Using TEM, microtubule diameter cannot be precisely measured due to the requirement that the sample be dried, thereby collapsing the microtubules. Nonetheless, bundled microtubules will be easily distinguished from unbundled microtubules.
To determine if PX3Cy2–3OH108-G5 induces microtubule bundling, we preformed the same assays as were performed for the TIRFM imaging above (Figure 4). That is, we incubated preassembled microtubules with either paclitaxel, PX3Cy2–3OH108-G5, or Cy2–3OH111-G5 for 30 min at room temperature, and then imaged the samples via TEM. To obtain a rough estimate of the degree of bundling in each sample (Figure 5g), the diameter of a microtubule (bundle) was measured at 10 pixel intervals and weighted according to the estimated number of bundled microtubules present in that interval, where the mean diameter of a single, unbundled microtubule within two standard deviations was assigned a weight equal to one. This measurement results in a set of bundled or unbundled microtubule lengths, which are then summed and used to calculate the fraction of bundled microtubule lengths for each sample. This analytic method only provides a rough estimate of the degree of bundling, as we cannot measure the thickness of the microtubule bundle in the z-direction from the TEM images. However, using the planar thickness of the bundles to estimate the number of microtubules present should be sufficient to compare the degree of bundling among the samples tested.
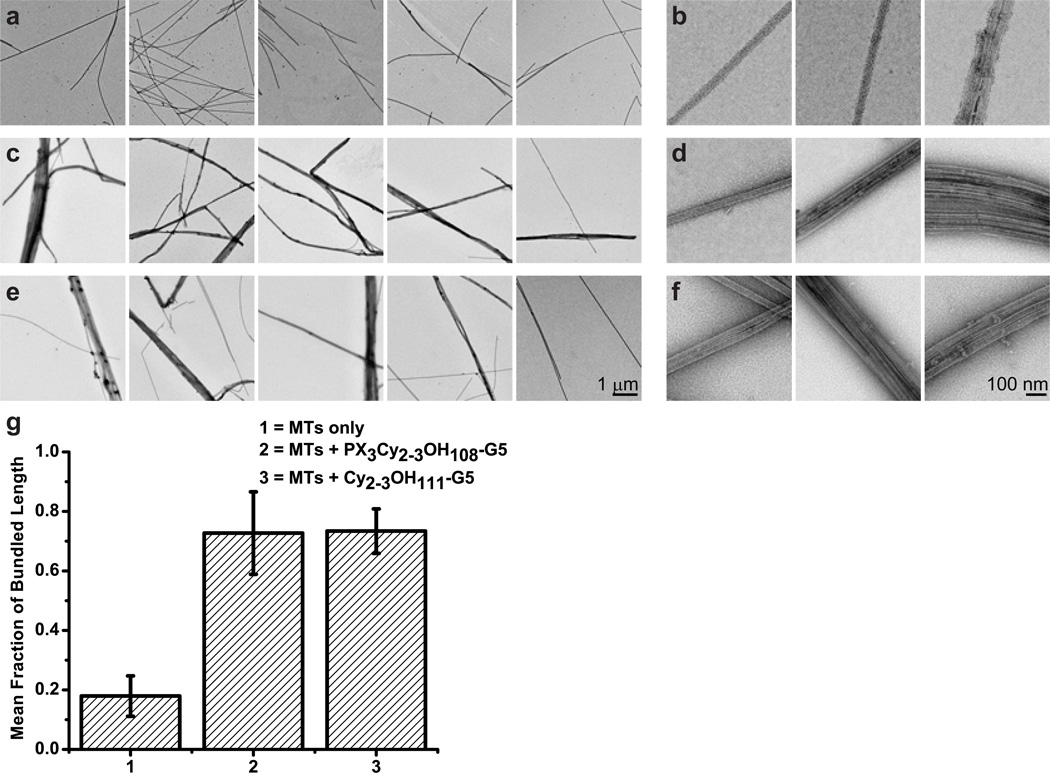
Figure 5.
Negative-stain TEM images obtained at 3,095× (a, c, e; scale bar = 1 µm) or 24,628× magnification (b, d, f; scale bar = 100 nm). Microtubules are incubated in MEM806:8 at room temperature for 30 min with either: paclitaxel (a–b); PX3Cy2–3OH108-G5 (c–d); or Cy2–3OH111-G5 (e–f). The amount of bundling in a population is represented as the fraction of bundled microtubule length (g).
Figure 5a–b shows the typical structure of paclitaxel-stabilized microtubules in the absence of dendrimers at both low and high magnification. By comparison, Figure 5c–d shows microtubules in the presence of PX3Cy2–3OH108-G5, revealing that PX3Cy2–3OH108-G5 induces formation of microtubule bundles of various sizes. The PAMAM dendrimers are not easily resolved in these images due to their small size, low electron mass compared to the carbon imaging grid, and high background from dendrimers and unpolymerized tubulin. Small bundles, containing 2–3 microtubules, were observed in the absence of dendrimers, but the fraction of the total population represented by these bundles was significantly smaller (p < 0.05) than the fraction observed in the presence of PX3Cy2–3OH108-G5, as quantified in Figure 5g. Intriguingly, a large amount of microtubule bundles was also observed in the presence of Cy2–3OH111-G5 (Figure 5e–f), and this bundled fraction was not significantly different from the bundling fraction measured in the presence of PX3Cy2–3OH108-G5 (p > 0.05) (Figure 5g).
Surface Neutralized G5 PAMAM Dendrimers Bundle Microtubules via Electrostatic Interactions
We next sought to identify the properties of PX3Cy2–3OH108-G5 and Cy2–3OH108-G5 that induce microtubule bundling. Microtubule bundles have been observed both in vitro and in vivo, induced by paclitaxel,42 microtubule-associated proteins (MAPs),43 or polyamines.51 The mechanism of action for these microtubule bundling agents has been postulated to be cross-linking and/or charge shielding (i.e., binding the C-terminal tails of the microtubules where the majority of the negative charge is located, thereby reducing electrostatic repulsion between microtubules and allowing them to associate laterally). The fact that there is no significant difference in the amount of bundling induced by PX3Cy2–3OH108-G5 as compared to Cy2–3OH111-G5 (see Figure 5) precludes the contribution of paclitaxel. We therefore considered the commonalities in the chemical structures of PX3Cy2–3OH108-G5 and Cy2–3OH111-G5 (Figure 1)—the fluorescent dye Cy5 and the dendrimer core itself—to determine if either of these components contribute to the observed bundling.
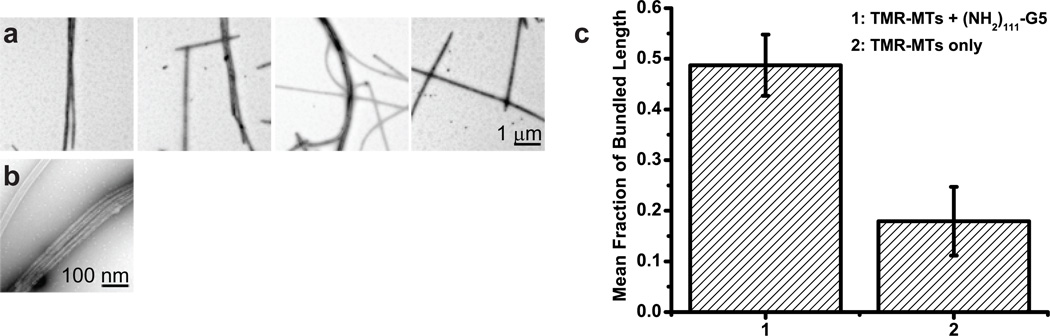
First, considering the dendrimer core structure, we realized that it contains interior tertiary amines with pKa equal to 6.5 ± 0.2 (derived from potentiometric titration; see Supplementary Materials), and therefore a fraction of these will be positively charged at neutral pH. It is feasible that the interior amines of the dendrimer core could encounter the microtubule surface, as the branched structure of the core is highly flexible and has been predicted by molecular dynamics simulations, and demonstrated by atomic force microscopy, to flatten into a disc-like structure upon encountering a surface.52 We therefore hypothesize that these protonated tertiary amines contact the microtubule surface and induce microtubule bundling through cross-linking and/or charge shielding. In support of this hypothesis, we find that unmodified G5 PAMAM dendrimers, hereafter referred to as (NH2)114-G5, carrying 114 protonatable primary amines on the surface, but no paclitaxel or Cy5, are also able to bundle microtubules to a significant degree compared to microtubules alone (p < 0.05) (Figure 6), although its bundling ability is seemingly lesser than that of PX3Cy2–3OH108-G5 (or by extension, Cy2–3OH111-G5). This difference is possibly attributed to the fact that (NH2)114-G5 dendrimers would be highly electrostatically repulsed from each other as they are highly cationic, and therefore fewer (NH2)114-G5 might bind a microtubule than the surface neutralized PX3Cy2–3OH108-G5 or Cy2–3OH111-G5. Because the cationic (NH2)114-G5 was able to bundle microtubules, we therefore sought to test the impact of the positive charges from the interior tertiary amines of PX3Cy2–3OH108-G5 on the ability of these dendrimers to bundle microtubules.
Figure 6.
Microtubules are incubated with (NH2)114-G5 for 30 min in MEM806:8 and visualized by negative-stain TEM at 3,095× (a; scale bar = 1 µm) or 24,628× (b; scale bar = 100 nm). The amount of bundling in each sample was quantified as before (c), from ten 25.38 µm2 images from each of three trials. The data from microtubules in the absence of dendrimers (sample 2 in c) was introduced previously (Figure 5), but is shown here for reference.
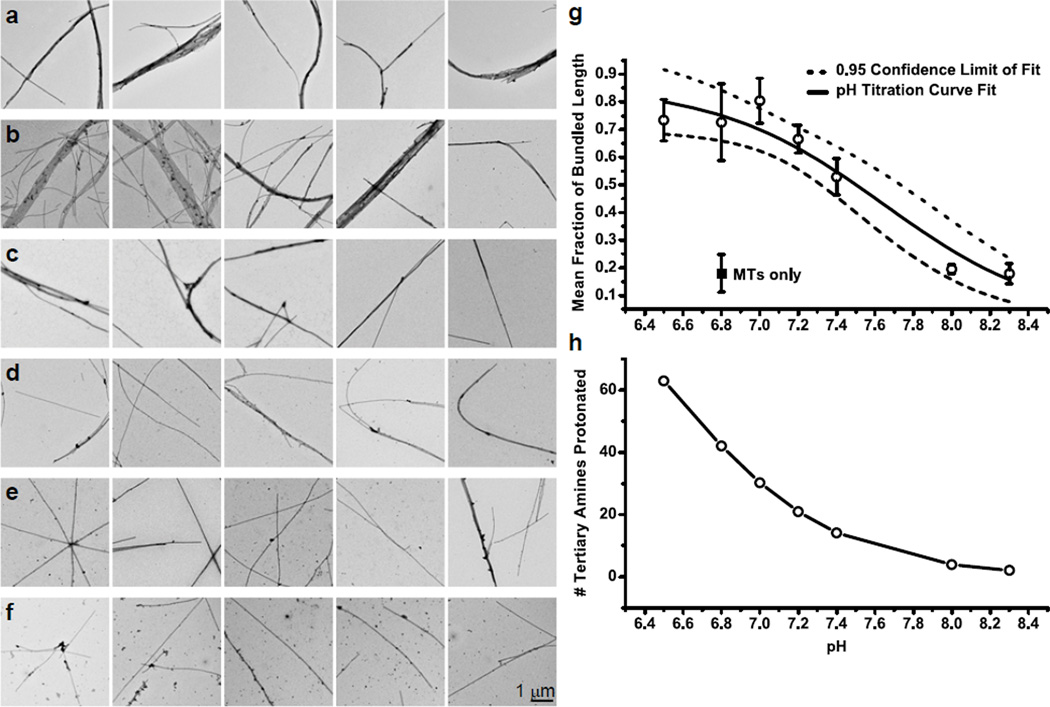
To test the hypothesis that the cationic amines of the dendrimer core interior contribute to the microtubule bundling observed by TIRFM and TEM, we incrementally increased the pH of the MEM80 incubation buffer from 6.5 to 8.3 using KOH, and visualized the resulting dendrimer-microtubule complexes by TEM. As the pH increases, the fraction of tertiary amines in the dendrimer core that are protonated, and therefore positively charged, decreases (Figure 7g). If the observed dendrimer-induced bundling is mediated by electrostatics, microtubule bundling will decrease with increasing pH. Only PX3Cy2–3OH108-G5 dendrimers were used for these experiments.
Figure 7.
Negative stain TEM images obtained at 3,095× magnification (scale bar = 1 µm). Microtubules are incubated at room temperature with PX3Cy2–3OH108-G5 for 30 min in MEM80, in which the pH has been adjusted with KOH to: (a) 6.5; (b) 7.0; (c) 7.2; (d) 7.4; (e) 8.0; or (f) 8.3. Bundling in each sample was estimated as before (g), from ten 25.38 µm2 images from each of three trials, and a pH titration curve () was fit to the data (R2 = 0.94). The amount of bundling in the absence of dendrimers is shown for reference (solid square labeled ‘MTs only’). The Henderson-Hasselbalch equation () is used to determine the theoretical number of protonated amines in the dendrimer core at the tested pH values (h).
The images of Figure 7a–f show qualitatively that the amount of bundling does indeed decrease as the pH increases. The degree of bundling in each sample is roughly estimated as before (Figure 7g), and a pH titration curve is fit to the data with an excellent correlation coefficient, R2 = 0.94, suggesting that the observed bundling is indeed pH dependent. From the titration curve, a pKa of bundling equal to 7.6 ± 0.1 is extracted.
The source of the bundling pH dependence could be one or both of the following: (1) electrostatic interactions between the cationic amines in the dendrimer core and the anionic C-terminal tubulin tails on the microtubule surface; or (2) the instability of microtubules at alkaline pH.53 Considering the first mechanism, the pH increase would cause both a decrease in the number of protonated, cationic amines in the dendrimer core, as well as an increase in the number of deprotonated, anionic C-terminal tubulin tails on the microtubule surface. These changes would decrease any electrostatic attraction between the dendrimers and the microtubules, as well as increase the electrostatic repulsion between microtubules that do not have the negative charges on their C-terminal tubulin tails shielded by bound dendrimers. Considering the second mechanism, it is known that microtubule disassembly increases with increasing pH.53 Therefore, we may be observing a decrease in microtubule bundling with increasing pH simply due to increased microtubule disassembly. To distinguish between these two possibilities, additional experiments, such as quantification of microtubule bundling in the presence of PAMAM dendrimers after cleavage of the anionic C-terminal tubulin tails using subtilisin,54 must be performed.
Implications of Dendrimer-Induced Microtubule Structural Changes for Cancer Treatment
Both dendrimer-induced microtubule structural changes observed in this paper—stabilization of polymerizing microtubules and bundling of pre-formed microtubules—have the potential to stall mitosis in a dividing cell. In this respect, paclitaxel-conjugated dendrimers show promise for use as cancer therapeutics, even without, or upon slow, cleavage of the drug from the carrier. However, one of these structural changes was not specific to the cancer drug paclitaxel, but rather the dendrimer core itself.
Our previous study demonstrated that untargeted, surface neutralized G5 PAMAM dendrimers are not cytotoxic,12, 55 implying that paclitaxel, and less the dendrimer core, induces cytotoxicity. While differences in dendrimer concentration between the current study utilizing purified proteins and the prior study utilizing cultured cells may explain this difference, or the possibility that paclitaxel was cleaved from the dendrimer carrier intracellularly in the prior study, the current study underscores the need for exercising caution when designing dendrimer-conjugates for the treatment of diseases both responsive and non-responsive to paclitaxel, as the dendrimers may have unintended cytotoxic effects not specific to paclitaxel due to their potential direct electrostatic interactions with microtubules. More broadly, the current study highlights the need for careful toxicity studies to be performed and target cell specificity to be achieved to control for and avoid such side effects; not only when using PAMAM dendrimers as a carrier, but any other carrier molecules as well.
Nanoparticle toxicity is a topic that is very important to everyday life, as nanomaterials are being incorporated into a wide array of consumer products in addition to therapeutics.56–58 Although it is recognized that nanoparticle toxicity stems from the ability of nanoparticles to cross cellular membranes, and that this ability derives from their small size59 and surface charge,29, 60 it is not completely understood what the molecular basis of the toxicity is, or what physicochemical properties of nanoparticles contribute to this toxicity.58–60 The present study further emphasizes the need for additional studies addressing these questions, and advances a mechanism of cytotoxicity for flexible cationic nanoparticles involving interior, cationic tertiary amines.
Conclusions
In the present study, we find that paclitaxel-conjugated, fifth generation, PAMAM dendrimers are able to affect microtubule structure via two mechanisms: (1) by stabilizing polymerizing microtubules; and (2) by bundling pre-formed microtubules. The latter mode of action is not specific to the activity of paclitaxel, as surface neutralized G5 PAMAM dendrimers that are not conjugated with paclitaxel are also able to bundle pre-formed microtubules to the same degree as paclitaxel-conjugated dendrimers. The mechanism for this bundling is at least partially electrostatically driven, mediated by tertiary amines located in the interior of the polyvalent dendrimer core that cannot be neutralized through chemical modifications. The results of this study, therefore, demonstrate both the promise of paclitaxel-conjugated dendrimers in the treatment of cancer and the necessity for further careful toxicity studies of these and other cationic nanoparticles.
Supplementary Material
Acknowledgement
We gratefully acknowledge the support of the National Science Foundation (NSF), under grant EFRI-BSBA 0938019. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation (NSF). E. Cline was also supported in part by training grant NIH T-32-GM007315.
Footnotes
Supporting Information Available
Detailed synthesis, purification, and characterization of paclitaxel-dendrimer conjugates. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Wang TH, Wang HS, Soong YK. Cancer. 2000;88:2619–2628. doi: 10.1002/1097-0142(20000601)88:11<2619::aid-cncr26>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 2.Marupudi NI, Han JE, Li KW, Renard VM, Tyler BM, Brem H. Expert Opin. Drug Saf. 2007;6:609–621. doi: 10.1517/14740338.6.5.609. [DOI] [PubMed] [Google Scholar]
- 3.Liu YJ, Zhang B, Yan B. Int. J. Mol. Sci. 2011;12:4395–4413. doi: 10.3390/ijms12074395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali I, Rahis U, Salim K, Rather MA, Wani WA, Haque A. Curr. Cancer Drug Targets. 2011;11:135–146. doi: 10.2174/156800911794328493. [DOI] [PubMed] [Google Scholar]
- 5.Esfand R, Tomalia DA. Drug Discovery Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 6.Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P. Polym. J. 1985;17:117–132. [Google Scholar]
- 7.Baker JR. Hematology Am. Soc. Hematol. Educ. Program. 2009:708–719. doi: 10.1182/asheducation-2009.1.708. [DOI] [PubMed] [Google Scholar]
- 8.Yellepeddi VK, Kumar A, Palakurthi S. Expert Opin. Drug Delivery. 2009;6:835–850. doi: 10.1517/17425240903061251. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y, Wang J, Rao T, He X, Xu T. Front. Biosci. 2008;13:1447–1471. doi: 10.2741/2774. [DOI] [PubMed] [Google Scholar]
- 10.Tekade RK, Kumar PV, Jain NK. Chem. Rev. 2009;109:49–87. doi: 10.1021/cr068212n. [DOI] [PubMed] [Google Scholar]
- 11.Nourse A, Millar DB, Minton AP. Biopolymers. 2000;53:316–328. doi: 10.1002/(SICI)1097-0282(20000405)53:4<316::AID-BIP4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 12.Majoros IJ, Myc A, Thomas T, Mehta CB, Baker JR. Biomacromolecules. 2006;7:572–579. doi: 10.1021/bm0506142. [DOI] [PubMed] [Google Scholar]
- 13.Mellado W, Magri NF, Kingston DG, Garcia-Arenas R, Orr GA, Horwitz SB. Biochem. Biophys. Res. Commun. 1984;124:329–336. doi: 10.1016/0006-291x(84)91557-2. [DOI] [PubMed] [Google Scholar]
- 14.Gueritte-Voegelein F, Guenard D, Lavelle F, Le Goff MT, Mangatal L, Potier PJ. Med. Chem. 1991;34:992–998. doi: 10.1021/jm00107a017. [DOI] [PubMed] [Google Scholar]
- 15.Mathew AE, Mejillano MR, Nath JP, Himes RH, Stella VJ. J. Med. Chem. 1992;35:145–151. doi: 10.1021/jm00079a019. [DOI] [PubMed] [Google Scholar]
- 16.Lim J, Chouai A, Lo ST, Liu W, Sun X, Simanek EE. Bioconjugate Chem. 2009;20:2154–2161. doi: 10.1021/bc900324z. [DOI] [PubMed] [Google Scholar]
- 17.Vrudhula VM, MacMaster JF, Li Z, Kerr DE, Senter PD. Bioorg. Med. Chem. Lett. 2002;12:3591–3594. doi: 10.1016/s0960-894x(02)00784-9. [DOI] [PubMed] [Google Scholar]
- 18.Castoldi M, Popov AV. Protein Expression Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 19.Kim T, Kao MT, Hasselbrink EF, Meyhofer E. Nano Lett. 2007;7:211–217. doi: 10.1021/nl061474k. [DOI] [PubMed] [Google Scholar]
- 20.Mullen DG, Desai A, van Dongen MA, Barash M, Baker JJR, Banaszak Holl MM. Macromolecules. 2012;45:5316–5320. doi: 10.1021/ma300485p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majoros IJ, Thomas TP, Mehta CB, Baker JR. J. Med. Chem. 2005;48:5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 22.Davis A, Martinez S, Nelson D, Middleton K. Methods Cell Biol. 2010;95:331–351. doi: 10.1016/S0091-679X(10)95018-8. [DOI] [PubMed] [Google Scholar]
- 23.Ohi M, Li Y, Cheng Y, Walz T. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao FQ, Craig RJ. Struct. Biol. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 25.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Austin CD, Wen X, Gazzard L, Nelson C, Scheller RH, Scales SJ. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17987–17992. doi: 10.1073/pnas.0509035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MH, Choi SK, Thomas TP, Desai A, Lee KH, Kotlyar A, Banaszak Holl MM, Baker JR. Eur. J. Med. Chem. 2012;47:560–572. doi: 10.1016/j.ejmech.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Holl MM. Nano Lett. 2008;8:420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- 30.Jain K, Kesharwani P, Gupta U, Jain NK. Int. J. Pharm. 2010;394:122–142. doi: 10.1016/j.ijpharm.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 31.Lakamper S, Kallipolitou A, Woehlke G, Schliwa M, Meyhofer E. Biophys. J. 2003;84:1833–1843. doi: 10.1016/S0006-3495(03)74991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minoura I, Katayama E, Sekimoto K, Muto E. Biophys. J. 2010;98:1589–1597. doi: 10.1016/j.bpj.2009.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiller G, Weber K. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 34.Van de Water rL, Olmsted JB. J. Biol. Chem. 1980;255:10744–10751. [PubMed] [Google Scholar]
- 35.Parness J, Horwitz SB. J. Cell Biol. 1981;91:479–487. doi: 10.1083/jcb.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaskin F, Cantor CR, Shelanski ML. J. Mol. Biol. 1974;89:737–755. doi: 10.1016/0022-2836(74)90048-5. [DOI] [PubMed] [Google Scholar]
- 37.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Mol. Biol. Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross JL, Fygenson DK. Biophys. J. 2003;84:3959–3967. doi: 10.1016/S0006-3495(03)75123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odde D. Eur. Biophys. J. 1998;27:514–520. doi: 10.1007/s002490050161. [DOI] [PubMed] [Google Scholar]
- 40.Tomalia DA, Naylor AM, Goddard rWA. Angew. Chem. Int. Ed. Engl. 1990;29:138–175. [Google Scholar]
- 41.Diallo MS, Christie S, Swaminathan P, Balogh L, Shi X, Um W, Papelis C, Goddard rWA, Johnson JJH. Langmuir. 2004;20:2640–2651. doi: 10.1021/la036108k. [DOI] [PubMed] [Google Scholar]
- 42.Jordan MA, Wendell K, Gardiner S, Derry WB, Copp H, Wilson L. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 43.Walczak CE, Shaw SL. Cell. 2010;142:364–367. doi: 10.1016/j.cell.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 44.Nogales E, Whittaker M, Milligan RA, Downing KH. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 45.Gaertig J, Wloga D. Curr. Biol. 2012;22:R483–R485. doi: 10.1016/j.cub.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 46.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chretien D, Metoz F, Verde F, Karsenti E, Wade RH. J. Cell Biol. 1992;117:1031–1040. doi: 10.1083/jcb.117.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chretien D, Wade RH. Biol. Cell. 1991;71:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 49.Diaz JF, Valpuesta JM, Chacon P, Diakun G, Andreu JM. J. Biol. Chem. 1998;273:33803–33810. doi: 10.1074/jbc.273.50.33803. [DOI] [PubMed] [Google Scholar]
- 50.Ray S, Meyhofer E, Milligan RA, Howard JJ. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamon L, Savarin P, Curmi PA, Pastre D. Biophys. J. 2011;101:205–216. doi: 10.1016/j.bpj.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mecke A, Lee I, Baker JR, Holl MMB, Orr BG. Eur. Phys. J. E: Soft Matter Biol. Phys. 2004;14:7–16. doi: 10.1140/epje/i2003-10087-5. [DOI] [PubMed] [Google Scholar]
- 53.Regula CS, Pfeiffer JR, Berlin RD. J. Cell Biol. 1981;89:45–53. doi: 10.1083/jcb.89.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skiniotis G, Cochran JC, Muller J, Mandelkow E, Gilbert SP, Hoenger A. EMBO J. 2004;23:989–999. doi: 10.1038/sj.emboj.7600118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas TP, Majoros I, Kotlyar A, Mullen D, Holl MMB, Baker JR. Biomacromolecules. 2009;10:3207–3214. doi: 10.1021/bm900683r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Card JW, Jonaitis TS, Tafazoli S, Magnuson BA. Crit. Rev. Toxicol. 2011;41:20–49. doi: 10.3109/10408444.2010.524636. [DOI] [PubMed] [Google Scholar]
- 57.Clift MJD, Gehr P, Rothen-Rutishauser B. Arch. Toxicol. 2011;85:723–731. doi: 10.1007/s00204-010-0560-6. [DOI] [PubMed] [Google Scholar]
- 58.Hubbs AF, Mercer RR, Benkovic SA, Harkema J, Sriram K, Schwegler-Berry D, Goravanahally MP, Nurkiewicz TR, Castranova V, Sargent LM. Toxicol. Pathol. 2011;39:301–324. doi: 10.1177/0192623310390705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberdorster G, Stone V, Donaldson K. Nanotoxicology. 2007;1:2–25. [Google Scholar]
- 60.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Chem. Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.