Abstract
Rapid and sensitive methods of discriminating between healthy tissue and metastases are critical for predicting disease course and designing therapeutic strategies. We report here the use of an array of gold nanoparticle–green fluorescent protein elements to rapidly detect metastatic cancer cells (in minutes), as well as to discriminate between organ-specific metastases and their corresponding normal tissues through their overall intracellular proteome signatures. Spontaneous metastases established in a new pre-clinical non-small cell lung cancer metastasis model in athymic mice were used to provide a challenging and realistic testbed for clinical cancer diagnosis. Full differentiation between the analyte cell/tissue was achieved with as little as 200 ng of intracellular protein (~1000 cells) for each nanoparticle, indicating high sensitivity of this sensor array. Notably, the sensor created a distinct fingerprint pattern for the normal and metastatic tumor tissues. Moreover, this array-based approach is unbiased, precluding the requirement of a priori knowledge of the disease biomarkers. Taken together, these studies demonstrate the utility of this sensor for creating fingerprints of cells and tissues in different states, and present a generalizable platform for rapid screening amenable to microbiopsy samples.
Keywords: Array-based sensing, metastasis, tissue, lysates, gold nanoparticle
Metastasis is frequently the terminal process in the progression of tumors.1 As an example, non-small cell lung cancer (NSCLC) is aggressively metastatic, making it a leading cause of cancer related deaths worldwide.2,3 NSCLC can metastasize to any organ in the body, with adrenal gland, brain, bone, lymph nodes, and liver being most commonly affected. Different sub-types of NSCLC also differ in terms of their evolution, morbidity, mortality, and treatment.4 Rapid and sensitive methods to molecularly stratify metastases such as those arising from NSCLC would provide important information for predicting disease course and indicate therapeutic strategies.
Currently available protein-based methods for phenotyping tumor or metastatic cells rely on extracellular (cell surface-bound or secreted) and intracellular biomarkers.5,6 Cell detection based on cell surface protein biomarkers generally involves the use of specific antibodies.7,8 Intracellular protein biomarkers9 have been explored using emerging proteomic techniques, such as 2-D gel electrophoresis (2D-SDS-PAGE)10 and mass spectrometry.11 While these proteomic methods provide potential approaches for cancer phenotyping, they generally require prior knowledge of the biomarkers being detected.12 However, cells do not always express unique biomarkers that allow inter-individual differentiation between tumors of a given type or their metastases.13 Detection methods are further complicated by the requirement of identifying several protein variants expressed from a single gene, with each variant being potentially subject to additional post-translational modifications that regulate activity and conformation.14 An alternative means of cell discrimination is to profile the phenotypic signatures of cells by means of differential display analysis that reveals genomic, proteomic, metabolomic, or phenotypic alterations.15 Key methods such as RT-PCR,16 electrophoresis,17 and Raman spectroscopy18 have been employed in this respect, but sophisticated instrumentation and complex operational steps, low throughput, and prolonged assay times restrict their applicability in the clinic.
Array-based sensing approaches that discriminate between analytes based on their overall signatures have emerged as a potential alternative for point-of-care diagnosis.19-21 In this strategy, a unique global diagnostic pattern is derived from the responses acquired from differential binding interactions of the analytes with a sensor array featuring selective receptors. Then, comparing the detected profile of an unknown case to the global database can allow us to predict its class.19,22 This differential sensing method analogous to mammalian olfaction presents a powerful tool for discriminating subtly different analytes and their complex mixtures23-25 even in bio-matrices.26,27 This selective array-based approach is unbiased, precluding the need to pre-identify specific biomarkers.19,21 To date, this strategy has been successfully applied to cell surface-based identification of bacteria28,29 and to determine cell type, and state of cultured mammalian cells.30-32 However, intracellular proteome signatures have not been exploited for identification of cells, nor has this strategy been applied to in vivo systems. More importantly, the utilization of array-based sensor to discriminate between tissues, a much more complex matrix, would minimize the influence of phenotypic changes upon in vitro cell cultures,33 and provide a robust and generic tool for clinical diagnostics.
We demonstrate here efficient discrimination between site-specific metastases and healthy state using cell and tissue lysates through a sensor array comprised of gold nanoparticle (NP)-green fluorescent protein (GFP) complexes as sensor elements.26 We hypothesized that site-specific metastasis could be effectively identified and discriminated from their normal states using the unique proteomic profiles10,34,35 of the cells and tissues. To test this hypothesis, we first established an in vivo experimental metastasis model by inoculating NCI-H1299 non-small lung cancer cells that developed spontaneous metastases in multiple organs. Lysates from the intact tumor samples, as well as tumor cells isolated from these metastatic lesions were then used in the sensing experiments. These studies demonstrate that the sensor was able to completely differentiate between site-specific metastatic cells with high sensitivity. Notably, the sensor could distinctly profile normal tissues and organ-specific metastases using their lysate protein signatures. Overall, we demonstrate for the first time the utility of array-based sensing using gold naoparticles for sensing tissues, providing a promising strategy for cancer diagnostics.
RESULTS AND DISCUSSION
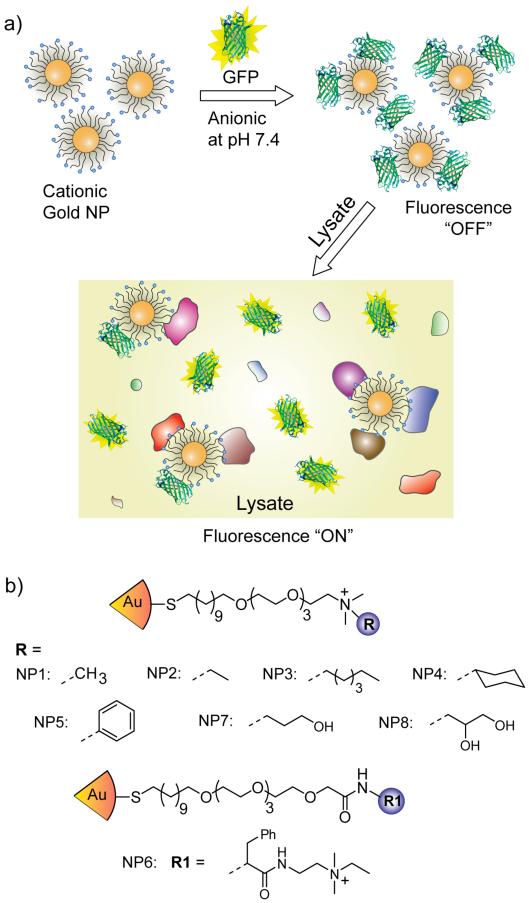
Our present sensing strategy is based on non-covalent conjugates between functionalized gold NPs (core diameter ~2nm) and GFP bearing complimentary charges (Figure 1a). Gold NPs provide appropriate scaffold for the selective sensor array owing to the features such as size commensurate with proteins, tunability of the structure and functionality required for selectivity, high loading of recognition elements leading to high sensitivity, excellent stability, and fluorescence quenching ability.36 In the present sensor, negatively-charged GFP binds efficiently with positively-charged gold NPs, with concomitant quenching of GFP fluorescence by the particle cores. Upon incubation with lysates, the cellular proteins compete with GFP for binding to the particle surface, resulting in the displacement of GFP from the particle surface with concomitant restoration of fluorescence (Figure 1a). The differential interactions between the cationic NPs and various cellular proteins in the lysates generate fluorescence patterns characteristic of the cell/tissue type, thus enabling us to discern between the cell/tissue states based on the lysate composition.
Figure 1.
(a) Schematic illustration of fluorescence modulation by the competitive binding between the quenched NP-GFP complex and the lysate proteins. (b) Ligand structure of the NPs used in this study.
We hypothesized that the selectivity required for sensing would be provided by ligand headgroups on the NPs that present different non-covalent interactions with the analytes. To this end, we prepared particles capable of hydrophobic (NP1 – 4, NP6), aromatic stacking (NP5, NP6) and hydrogen bonding (NP7, NP8) interactions (Figure 1b). These NPs are expected to generate differential fluorescence responses by tuning the NP-GFP and NP-analyte interactions, a prediction validated by variation in NP-GFP avidities as quantified through fluorescence titrations between GFP and NPs (Figure S2, Table S1 in the Supporting Information). Significantly, the relatively simple structures of the ligands streamlines sensor fabrication while ensuring unbiased screening.37-39
The sensor array was generated by incubating the NPs and GFP followed by loading into a 96-well microplate. These complexes were then incubated (for 30 min) with analyte lysates from cells and tissues to determine the changes in fluorescence of the NP-GFP solutions. The protein contents of lysates were quantified by bicinchoninic acid (BCA) assays and sensing studies were carried out using a constant amount of proteins, thus reducing cell-to-cell and batch-to-batch variation. Furthermore, sensing with a particular amount of lysate proteins essentially relies on the differential protein expression patterns of different cells/tissues. Following quantification, titrations of NP-GFP complexes with different amounts of lysate proteins were performed providing differentiation with 200 ng, the cellular protein content of ~1000 cells. The ability of the sensor array to identify cells using such small sample sizes, pairs this strategy well with clinical specimens such as fine needle aspirates.40
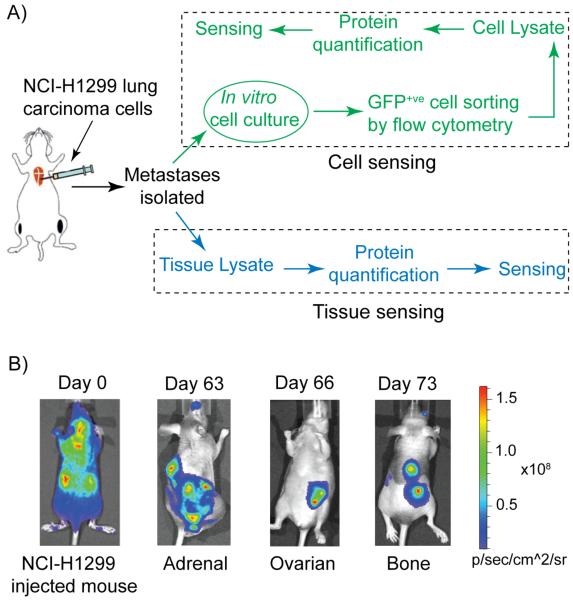
Xenograft models provide effective systems to isolate organ specific metastatic sublines and remain the models of choice for clinically relevant studies. For this study, we developed new metastatic sublines following arterial inoculation of parental human NCI-H1299 cells stably expressing the EGFP-luc2 reporter gene in athymic (nu/nu), beige (NIH-III) mice (Figure 2a). Metastasis development, monitored via bi-weekly bioluminescence imaging (using Xenogen/Caliper IVIS Lumina), revealed the development of bone and soft-tissue metastasis (adrenal and ovarian) (Figure 2b). Necropsy and histopathology provided further validation of the organ-specific sites of metastases (Figure S3 in the Supporting Information). Next, GFP-expressing cells from various metastatic tumors were expanded in vitro, purified by fluorescence-activated cell sorting (FACS), and subsequently reintroduced into the mice. When compared to the parental lines, cells isolated from metastatic lesions (adrenal, bone, and ovary) demonstrated a significantly enhanced metastatic capacity with a higher number of metastases as well as a variable degree of tissue tropism, and a reduction in overall survival.
Figure 2.
(a) Schematic presentation of generating site-specific metastatic cells, and tissues and their sensing using the selective array-based sensing approach. (b) Representative bioluminescence images (BLI) of mice after intracardiac injection of EGFP-luc2 expressing NCI-H1299 cells (Day 0) and at different days showing adrenal, ovarian, and bone metastases.
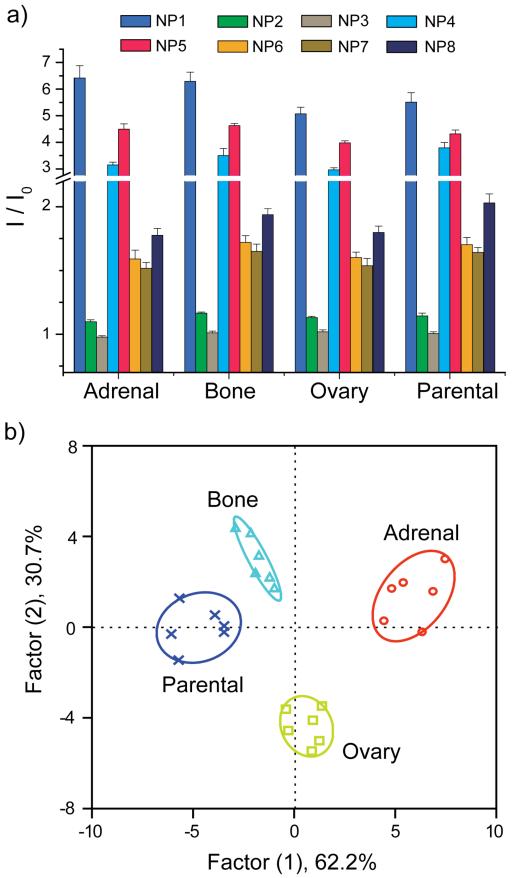
Our initial sensing studies were performed in vitro on cultured cells. Parental, bone-, ovary- and adrenal-derived NCI-H1299 cells were cultured then lysed. Next, 200 ng of total cell lysate protein from each sample was incubated with each of the NP-GFP supramolecular complexes. Changes in the fluorescence of GFP upon lysate incubation are shown in Figure 3a. Background fluorescence of the lysates was measured as well, with no detectable fluorescence signal. The fluorescence response patterns from the different lysates were found to be distinct, reproducible, and characteristic of each cell type. Linear discriminant analysis (LDA) was used as a clustering protocol to statistically analyze the fluorescence responses (see Supporting Information for the description of LDA).41,42 Analysis of the fluorescence pattern (8 NP-GFP conjugates × 4 cell lines × 6 replicates) resulted in three canonical factors (62.2, 30.7, and 7.1% of total variance), with the two most significant factors plotted in Figure 3b. Significantly, the different cell types clustered into four non-overlapping groups (using 95% confidence level ellipses). These results validate the ability of the sensor to differentiate between the parental NCI-H1299 and its metastasis-derived (adrenal, bone, and ovary) sublines, as well as between the organ-specific populations (adrenal vs. ovary, adrenal vs. bone, and ovary vs. bone) based on the composition of lysates.
Figure 3.
(a) Ratio of fluorescence intensities after (I) and before (I0) addition of the different metastatic cell lysates to the NP-GFP supramolecular complexes. The responses are averages of six replicate data and the error bars represent the standard deviations. (b) Canonical score plot for the fluorescence patterns as obtained from LDA against different parental and sublines.
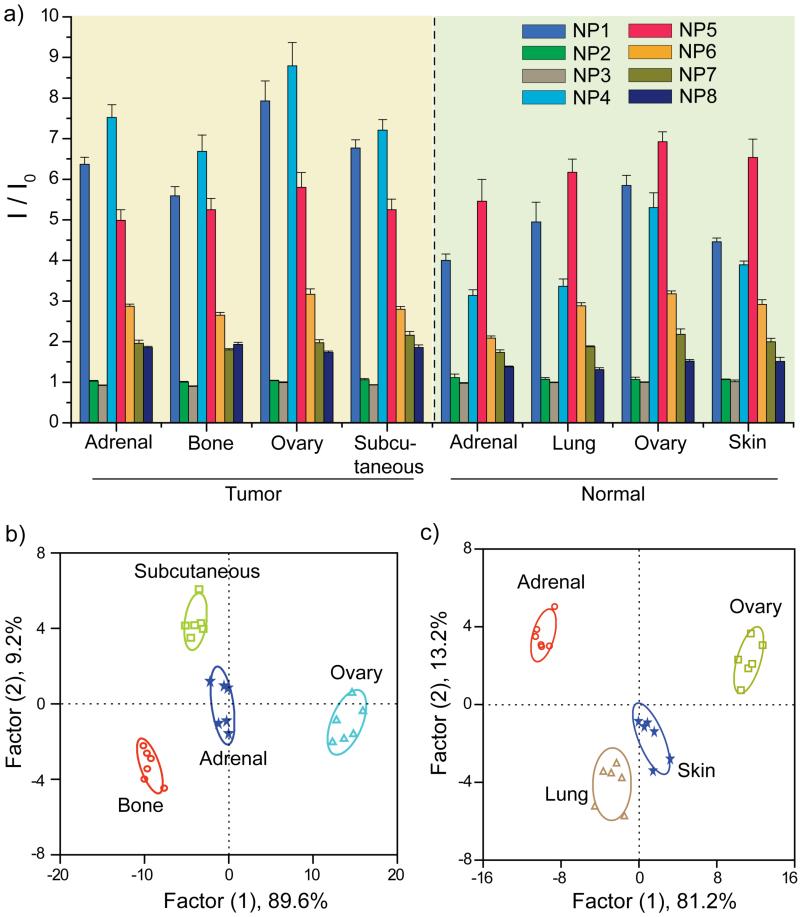
Building upon the discrimination of cultured cells using the selective sensor array, we next focused on tissue sensing as a clinically relevant sensing target. For these studies we collected tissues from three different NCI-H1299 metastatic tumors (bone, adrenal, and ovarian), as well as a tumor generated by subcutaneous injection of NCI-H1299 cells. Tumor tissue lysates were prepared, quantified using BCA assays, and sensing assays were performed as above. Figure 4a represents the ratio of the fluorescence intensities of the tissue lysates toward the NP-GFP complexes. The differential responses indicate that these complexes detect differences in protein ratios in diverse tumor tissues as the protein concentration of each lysates were kept constant. LDA classifies the tissues into four distinct clusters through three canonical factors (containing 89.6, 9.2, and 1.2% of the variation), with 100% identification accuracy among these tissues (Figure 4b).
Figure 4.
(a) Ratio of fluorescence intensities after (I) and before (I0) addition of the tumor and normal tissue lysates to the NP-GFP supramolecular complexes. The responses are averages of six replicate data and the error bars represent the standard deviations. (b) Canonical score plot of the fluorescence patterns as obtained from LDA against the four tumor lysates. (c) The LDA score plot derived from the fluorescence changes for the four healthy tissues.
To validate the detection efficiency of our selective array-based sensing strategy, we performed tests to identify unknown samples from random tissue lysates chosen from the training set (see Supporting Information). We observed 94% accuracy of 32 unknown samples (30 out of 32) of tumor tissues (Table S9 in the Supporting Information). Taken together, these studies demonstrate that rapid and efficient discrimination between metastatic tissues in preclinical models can be achieved based on their lysate compositions.
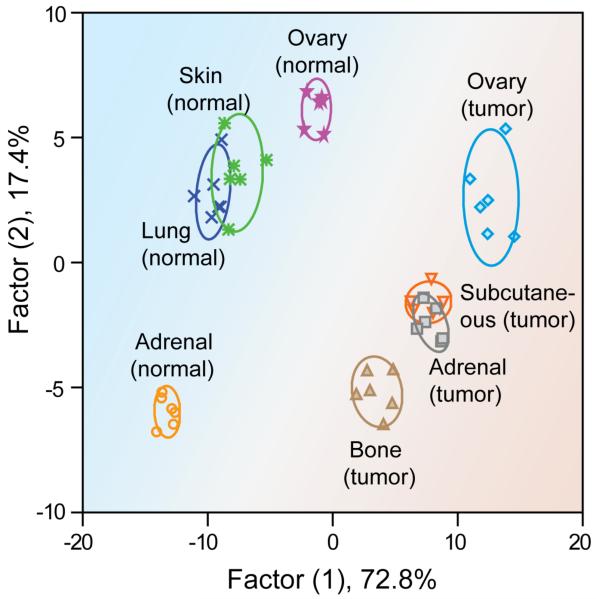
The ability to differentiate tumors from normal tissues in biopsies represents a key requirement for diagnostic applications. The ability of our sensors to discriminate between healthy and metastasized cancerous tissues was determined using lysates isolated from four normal tissues (adrenal, ovary, skin, and lung) and four NCI-H1299 metastatic deposits (adrenal, ovary, subcutaneous, and bone). Also, we compared healthy tissue from two different normal mice to probe subject variability (the same was done for tumor tissues as well). Sensing assays were performed as above using 200 ng of proteins. A distinct and reproducible fluorescence pattern was observed for the four normal tissues (Figure 4a). The canonical score plots obtained from LDA of the fluorescence responses (Figure 4c) showed that complete differentiation was achieved amongst the normal tissues. Next, the fluorescence response data from the tumor and healthy tissue lysates were combined and analyzed by LDA. Significantly, the normal and malignant tissues clustered into two completely separate regions (Figure 5), indicating a dramatic difference between the fingerprint patterns. Complete differentiation was also seen between each of the different tumors. While overlap is observed in the two-dimensional plot (Figure 5), 100% classification accuracy of all the eight analytes was obtained using additional dimensions provided by LDA (Figure S4 in the Supporting Information). Overall, application of array-based sensing to lysates provides complete discrimination between healthy and tumor tissues, as well as differentiating between tumor types, providing pre-clinical validation of this approach for cancer detection. Therefore, the gold nanoparticle based sensor array provides new opportunities for cancer diagnosis in the clinic by creating a global signature pattern of different cancer types and comparing new cases with the database.
Figure 5.
A 2-Dimensional LDA score plot derived from combining the fluorescence response patterns of tumor and healthy tissues (Fig. 4), with 95% confidence ellipses. The color shading is drawn to show the distinct regions between healthy and tumor tissue.
It should be noted that the present sensor array based on selective interactions presents a versatile tool that is potentially applicable to other tumor types. However, distinguishing various tumor types might entail a general set of NP receptors with higher recognition ability. Isolating the effect of the NP structure from the present study can provide us the NP surface chemistry important for discrimination between wide ranges of tumors. Despite the contributions from each NP toward the final differentiation accuracy in the above results, it can be observed from the raw fluorescence patterns that NP1, NP4, and NP5 produce good differentiations, consistent with other cell sensing studies30,43 (the individual contribution from each NP is shown through the Jackknifed classification, Supporting Information). Hence, hydrophilic and aromatic recognition play vital roles in the differential interactions with different cells and tissues. These NP structures provide a starting point for designing NP platform with greater selectivity toward different bioanalytes, a systematic structure-activity correlation study that is underway.
CONCLUSION
The array-based sensing using the NP-GFP complexes described here provides an unbiased strategy to distinguish between normal and metastatic cells and tissues. This sensing approach provides a complementary strategy to traditional biomarker-based methods for diagnosis or prognostication.44 In the present sensing, the discrimination relies upon the phenotypic differences within the overall proteomic signatures of the respective cells and tissues. Using the lysates for these sensors offers distinct advantages compared to whole cell sensing, such as increased homogeneity of the test samples leading to reduced error in identification, increased reproducibility, and higher sensitivity. Furthermore, the sensor is efficient in discriminating between samples with as little as 200 ng of cell- or tissue-lysed proteins, minimizing biopsy size. In addition to the high sensitivity, the simple sensor could differentiate effectively between low (parental) and high (bone, adrenal, and ovary) metastases, as well as between site-specific cells. Notably, this proteomic-based approach is the first successful application of selective array-based systems to normal and spontaneously developed metastatic tissues, providing a simple but generic approach to phenotypically distinguish disease states. Overall, this array-based sensing strategy presents the prospect of unbiased phenotype screening of tissue states arising from genetic variations and differentiation state, a strategy we are currently pursuing.
METHODS
Materials and ethical statement
All the reagents/materials required for nanoparticle synthesis and GFP expression were purchased from Fischer Scientific, except for gold salt that was from Strem Chemicals Inc. Human NCI-H1299 cells were purchased from ATCC. 5-6 week old athymic (nu/nu), beige (NIH-III) female mice were purchased from Charles River Laboratories (St-Constant, QC). Mice were housed in viral antibody-free conditions in the University of Calgary Animal Resources Center. All experiments were conducted in compliance with Canadian Council of Animal Care guidelines and with ethical approval from the University of Calgary Animal Care Committee.
Nanoparticle synthesis and GFP expression
Nanoparticles26,30,43 and GFP45 were synthesized following previous reports. The syntheses of the nanoparticles are detailed in the Supporting Information.
Establishing in vivo NSCLC metastases and isolation of the metastatic sublines
First, NCI-H1299 cells were stably transfected with a CMV-based vector expressing the EGFP-Luc2 fusion protein. To generate metastases, 6 week old female NIH-III mice were anaesthetized by intraperitoneal (i.p.) injection of ketamine (100 mg/kg) and xylazine (6 mg/kg). Then EGFP-Luc2 expressing NCI-H1299 cells suspended in 100 μl of sterile PBS were injected into the left ventricle of the mice. Following luciferin administration, imaging was performed immediately after each injection to verify that intracardiac injections resulted in systemic distribution of the bioluminescent cells. Development of metastases was monitored by bi-weekly bioluminescence imaging of anesthetized mice. At necropsy, organs were harvested for ex vivo evaluation of bioluminescence to confirm the anatomical distribution of the metastasis. In addition, adrenal, ovary, and bone metastasis was confirmed by hematoxylin and eosin (H&E) staining. Metastatic lesions (adrenal, ovary, and bone) were extracted and cultured in vitro for 2-3 weeks. To confirm enhanced metastasis and organ specificity of these sublines, FACS sorted GFP+ve cells were expanded in culture for 1-2 weeks and then re-administered to NIH-III mice by intracardiac injection.
Bioluminescence imaging (BLI)
BLI was performed using a sensitive cooled charge coupled device (CCD) camera mounted above a light-tight specimen box (Xenogen IVIS Lumina system, Caliper Life Sciences). For imaging, each mouse was injected intraperitoneally with D-luciferin (150 mg/kg dissolved in PBS), anesthetized with isofluorane, and then placed onto a warmed stage inside the specimen box. Anesthesia was maintained with 1.5-2% isofluorane. Exposure times ranged from 20 sec to 2 min depending on the photon emission rates from each metastatic site. Results were analyzed by using Living Image 3.2 software (Caliper Life Sciences). For ex vivo imaging, tissues were first excised, placed into 24-well tissue culture plates containing 300 μg/ml D-luciferin in PBS, and exposed for 30-120 sec in the IVIS Lumina instrument.
Histopathology
To confirm the presence of metastatic cells in soft and skeletal tissues, selected tissues were excised from the mice at necropsy and were preserved in 10% formalin solution immediately after ex vivo imaging. Tissues were processed and paraffin embedded before sectioning (5 μm) and staining with H&E. After fixation in 4% paraformaldehyde, bone metastases were decalcified in 14% EDTA prior to processing and sectioning.
Cell culture
The NCI-H1299-EGFP-luc2 (parental), bone, adrenal, and ovary subline cells were grown in RPMI-1640 media supplemented with 10% FBS and 1% antibiotics in 100 mm plates. The cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2. Cells were regularly passaged by trypsinization with 0.25% trypsin-EDTA, 1X solution (Invitrogen) in PBS (pH 7.2).
Preparation of the lysates
The metastatic tumors and normal organs were isolated from NIH-III mice. Tissue homogenates (10% w/v) were prepared in lysis buffer [0.15 mmol/L NaCl, 5 mmol/L EDTA, 1% Triton-X 100, 10 mmol/L Tris-HCl (pH 7.4), plus half of a tablet of complete protease inhibitor cocktail in 50 mL buffer (Roche Diagnostics GmbH)]. Next, whole tissue lysates were spun down at 4°C for 15 min at 14,000 rpm and the supernatants were used for protein quantification and sensing experiments. Similarly, for cell lysate preparation, confluent 100 mm plates were first washed with cold Ca2+/Mg2+-free PBS followed by treatment with 500μL of lysis buffer (with protease inhibitor) for 10 min at 4°C. Then, cells were scraped using a cell scraper and lysate was collected in an eppendorf tube. To isolate the pure cellular proteins, cell lysates were spun down at 4°C for 15 min at 14,000 rpm. The supernatants containing the cellular proteins were used for sensing experiments.
Fluorescence titrations
In the fluorescent titration experiment between nanoparticles and GFP, the change of fluorescence intensity at 510 nm was measured with an excitation wavelength of 475 nm at various concentrations of NPs from 0 to 100 nM on a Molecular Devices SpectraMax M3 microplate reader (at 25 °C). Decay of fluorescence intensity arising from 100 nM GFP was observed with increasing NP concentration. Nonlinear least-squares curve fitting analysis was carried out to estimate the binding constant (Kb) and association stoichiometry (n) using a 1:1 binding model.45
Sensing studies
NP-GFP conjugates were generated by mixing appropriate stoichiometries of nanoparticles and GFP (100 nM) in 5 mM sodium phosphate buffer (pH 7.4). Then, 200 μL of each NP-GFP complex solution was loaded into a 96-well microplate and initial fluorescence intensities of the quenched complexes were measured at 510 nm. Then, 200 ng different of cell/tissue lysates were incubated with these complexes to determine the changes in fluorescence of the NP-GFP complexes using Molecular Devices SpectraMax M3 microplate reader (at 25 °C).
LDA analysis
The raw data matrix was processed by classical linear discriminant analysis (LDA) using SYSTAT software (version 11.0, SystatSoftware, Richmond, CA, USA). In LDA, all variables were used in the model (complete mode) and the tolerance was set as 0.001. The raw fluorescence response patterns were transformed to canonical patterns where the ratio of between-class variance to the within-class variance was maximized according to the pre-assigned grouping. To identify the unknown samples, the fluorescence response patterns of the new cases were first converted to canonical scores using the discriminant functions established on the training cases. Then, the Mahalanobis distance,47,48 the distance of a case from the centroid of a training group in the multidimensional discriminant space, was calculated for the new cases. The new case was assigned to the group with the shortest Mahalanobis distance from the case.
Supplementary Material
ACKNOWLEDGMENT
V.R. acknowledges support from the NIH (GM077173) and F.R.J. from the Canadian Cancer Society Research Institute. F.R.J. was the recipient of a Canada Research Chairs award.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Syntheses, description of linear discriminant analysis, fluorescence titrations, histopathology of the metastases, three-dimensional LDA plot for normal and tumor tissues, fluorescence response data, Jackknifed analyses of the fluorescence responses, and identification of unknown samples. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
S.R., A.B., A.K.S., and V.M.R. conceived and designed the experiments. S.R., A.K.S., A.B., S.G.E., O.R.M., R.M., and B.Y. performed all the experiments. S.R., F.R.J., A.K.S., and V.M.R. analyzed the data. S.R., and V.M.R. co-wrote the manuscript, A.K.S., and F.R.J. revised it, and all of the authors commented on the manuscript.
REFERENCES
- 1.Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang PG, Cassivi SD, Schild SE, Adjei AA. Non-Small Cell Lung Cancer: Epidemiology, Risk factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ihde DC, Minna JD. Non-Small-Cell Lung-Cancer .1. Biology, Diagnosis, and Staging. Curr. Prob. Cancer. 1991;15:61–104. doi: 10.1016/0147-0272(91)90014-2. [DOI] [PubMed] [Google Scholar]
- 5.Srinivas PR, Kramer BS, Srivastava S. Trends in Biomarker Research for Cancer Detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 6.Tyers M, Mann M. From Genomics to Proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 7.Roesli C, Borgia B, Schhemann C, Gunthert M, Wunderli-Allenspach H, Giavazzi R, Neri D. Comparative Analysis of the Membrane Proteome of Closely Related Metastatic and Nonmetastatic Tumor Cells. Cancer Res. 2009;69:5406–5414. doi: 10.1158/0008-5472.CAN-08-0999. [DOI] [PubMed] [Google Scholar]
- 8.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. In Vivo Cancer Targeting and Imaging with Semiconductor Quantum Dots. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 9.Hanash S. Disease Proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 10.O’Dwyer D, Ralton LD, O’Shea A, Murray GI. The Proteomics of Colorectal Cancer: Identification of a Protein Signature Associated with Prognosis. PloS one. 2011;6:e27718. doi: 10.1371/journal.pone.0027718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Mann M. Mass Spectrometry-Based Proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Carbayo M. Antibody Arrays: Technical Considerations and Clinical Applications in Cancer. Clin. Chem. 2006;52:1651–1659. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 13.Borrebaeck CAK. Antibody Microarray-Based Oncoproteomics. Expert Opin. Biol. Ther. 2006;6:833–838. doi: 10.1517/14712598.6.8.833. [DOI] [PubMed] [Google Scholar]
- 14.Simpson RJ, Dorow DS. Cancer Proteomics: From Signaling Networks to Tumor Markers. Trends Biotechnol. 2001;19:S40–S48. doi: 10.1016/S0167-7799(01)01801-7. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Multi-Parameter Phenotypic Profiling: Using Cellular Effects to Characterize Small-Molecule Compounds. Nat. Rev. Drug Discov. 2009;8:567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 16.Malek A, Catapano CV, Czubayko F, Aigner A. A Sensitive Polymerase Chain Reaction-Based Method for Detection and Quantification of Metastasis in Human Xenograft Mouse Models. Clin. Exp. Metastasis. 2010;27:261–271. doi: 10.1007/s10585-010-9324-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang J-W, Peng S-Y, Li J-T, Wang Y, Zhang Z-P, Cheng Y, Cheng D-Q, Weng W-H, Wu X-S, Fei X-Z, Quan, et al. Identification of Metastasis-Associated Proteins Involved in Gallbladder Carcinoma Metastasis by Proteomic Analysis and Functional Exploration of Chloride Intracellular Channel 1. Cancer Lett. 2009;281:71–81. doi: 10.1016/j.canlet.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Hedegaard M, Krafft C, Ditzel HJ, Johansen LE, Hassing S, Popp J. Discriminating Isogenic Cancer Cells and Identifying Altered Unsaturated Fatty Acid Content as Associated with Metastasis Status, Using K-Means Clustering and Partial Least Squares-Discriminant Analysis of Raman Maps. Anal. Chem. 2010;82:2797–2802. doi: 10.1021/ac902717d. [DOI] [PubMed] [Google Scholar]
- 19.Turner APF, Magan N. Electronic Noses and Disease Diagnostics. Nat. Rev. Microbiol. 2004;2:161–166. doi: 10.1038/nrmicro823. [DOI] [PubMed] [Google Scholar]
- 20.Miranda OR, Creran B, Rotello VM. Array-Based Sensing with Nanoparticles: ‘Chemical Noses’ for Sensing Biomolecules and Cell Surfaces. Curr. Opin. Chem. Biol. 2010;14:728–736. doi: 10.1016/j.cbpa.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright AT, Anslyn EV. Differential Receptor Arrays and Assays for Solution-Based Molecular Recognition. Chem. Soc. Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 22.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J, Subramanian A, Ross KN, et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 23.Lavigne JJ, Anslyn EV. Sensing a Paradigm Shift in the Field of Molecular Recognition: From Selective to Differential Receptors. Angew. Chem. Int. Ed. 2001;40:3119–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Greene NT, Shimizu KD. Colorimetric Molecularly Imprinted Polymer Sensor Array Using Dye Displacement. J. Am. Chem. Soc. 2005;127:5695–5700. doi: 10.1021/ja0468022. [DOI] [PubMed] [Google Scholar]
- 25.Zhou HC, Baldini L, Hong J, Wilson AJ, Hamilton AD. Pattern Recognition of Proteins Based on an Array of Functionalized Porphyrins. J. Am. Chem. Soc. 2006;128:2421–2425. doi: 10.1021/ja056833c. [DOI] [PubMed] [Google Scholar]
- 26.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UH, Rotello VM. Sensing of Proteins in Human Serum Using Conjugates of Nanoparticles and Green Fluorescent Protein. Nat. Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright AT, Griffin MJ, Zhong ZL, McCleskey SC, Anslyn EV, McDevitt JT. Differential Receptors Create Patterns that Distinguish Various Proteins. Angew. Chem. Int. Ed. 2005;44:6375–6378. doi: 10.1002/anie.200501137. [DOI] [PubMed] [Google Scholar]
- 28.Phillips RL, Miranda OR, You C-C, Rotello VM, Bunz UHF. Rapid and Efficient Identification of Bacteria Using Gold-Nanoparticle-Poly(para-phenyleneethynylene) Constructs. Angew. Chem. Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 29.Duarte A, Chworos A, Flagan SF, Hanrahan G, Bazan GC. Identification of Bacteria by Conjugated Oligoelectrolyte/Single-Stranded DNA Electrostatic Complexes. J. Am. Chem. Soc. 2010;132:12562–12564. doi: 10.1021/ja105747b. [DOI] [PubMed] [Google Scholar]
- 30.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Detection and Differentiation of Normal, Cancerous, and Metastatic Cells Using Nanoparticle-Polymer Sensor Arrays. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, Huang X. Magnetic Glyco-Nanoparticles: A Tool to Detect, Differentiate, and Unlock the Glyco-Codes of Cancer via Magnetic Resonance Imaging. J. Am. Chem. Soc. 2010;132:4490–4499. doi: 10.1021/ja100455c. [DOI] [PubMed] [Google Scholar]
- 32.Scott MD, Dutta R, Haldar MK, Guo B, Friesner DL, Mallik S. Differentiation of Prostate Cancer Cells Using Flexible Fluorescent Polymers. Anal. Chem. 2012;84:17–20. doi: 10.1021/ac202301k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durr E, Yu JY, Krasinska KM, Carver LA, Yates JR, Testa JE, Oh P, Schnitzer JE. Direct Proteomic Mapping of the Lung Microvascular Endothelial Cell Surface In Vivo and in Cell Culture. Nat. Biotechnol. 2004;22:985–992. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
- 34.Seike M, Kondo T, Fujii K, Yamada T, Gemma A, Kudoh S, Hirohashi S. Proteomic Signature of Human Cancer Cells. Proteomics. 2004;4:2776–2788. doi: 10.1002/pmic.200300795. [DOI] [PubMed] [Google Scholar]
- 35.Singer SJ, Nicolson GL. Fluid Mosaic Model of Structure of Cell-Membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 36.De M, Ghosh PS, Rotello VM. Applications of Nanoparticles in Biology. Adv. Mater. 2008;20:4225–4241. [Google Scholar]
- 37.Rakow NA, Suslick KS. A Colorimetric Sensor Array for Odour Visualization. Nature. 2000;406:710–713. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 38.Reddy MM, Wilson R, Wilson J, Connell S, Gocke A, Hynan L, German D, Kodadek T. Identification of Candidate IgG Biomarkers for Alzheimer’s Disease via Combinatorial Library Screening. Cell. 2011;144:132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, et al. An Unbiased Cell Morphology-Based Screen for New, Biologically Active Small Molecules. Plos Biol. 2005;3:764–776. doi: 10.1371/journal.pbio.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H, Yoon TJ, Figueiredo JL, Swirski FK, Weissleder R. Rapid Detection and Profiling of Cancer Cells in Fine-Needle Aspirates. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurs PC, Bakken GA, McClelland HE. Computational Methods for the Analysis of Chemical Sensor Array Data from Volatile Analytes. Chem. Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 42.Palacios MA, Wang Z, Montes VA, Zyryanov GV, Anzenbacher P. Rational Design of a Minimal Size Sensor Array for Metal Ion Detection. J. Am. Chem. Soc. 2008;130:10307–10314. doi: 10.1021/ja802377k. [DOI] [PubMed] [Google Scholar]
- 43.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Cell Surface-Based Differentiation of Cell Types and Cancer States Using a Gold Nanoparticle-GFP Based Sensing Array. Chem. Sci. 2010;1:134–138. [Google Scholar]
- 44.Ludwig JA, Weinstein JN. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 45.De M, Rana S, Rotello VM. Nickel-Ion-Mediated Control of the Stoichiometry of His-Tagged Protein/Nanoparticle Interactions. Macromol. Biosci. 2009;9:174–178. doi: 10.1002/mabi.200800289. [DOI] [PubMed] [Google Scholar]
- 46.You CC, De M, Han G, Rotello VM. Tunable Inhibition and Denaturation of Alpha-Chymotrypsin with Amino Acid-Functionalized Gold Nanoparticles. J. Am. Chem. Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 47.Mahalanobis PC. On the Generalized Distance in Statistics. Proc. Natl. Inst. Sci. India. 1936;2:49–55. [Google Scholar]
- 48.Gnanadesikan R, Kettenring JR. Robust Estimates, Residuals, and Outlier Detection with Multiresponse Data. Biometrics. 1972;28:81–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.