Abstract
Mono- and multispecies microbial populations alter the chemistry of their surrounding environments during colony development thereby influencing multicellular behavior and interspecies interactions of neighboring microbes. Here we present a methodology that enables the creation of three-dimensional (3D) models of a microbial chemotype that can be correlated to the colony phenotype through multimodal imaging analysis. These models are generated by performing matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) imaging mass spectrometry (IMS) on serial cross-sections of microbial colonies grown on 8 mm deep agar, registering data sets of each serial section in MATLAB to create a model, and then superimposing the model with a photograph of the colonies themselves. As proof-of-principle, 3D models were used to visualize metabolic exchange during microbial interactions between Bacillus subtilis and Streptomyces coelicolor, as well as, Candida albicans and Pseudomonas aeruginosa. The resulting models were able to capture the depth profile of secreted metabolites within the agar medium and revealed properties of certain mass signals that were previously not observable using two-dimensional MALDI-TOF IMS. Most significantly, the 3D models were capable of mapping previously unobserved chemical distributions within the array of sub-surface hyphae of C. albicans and how this chemistry is altered by the presence of P. aeruginosa, an opportunistic pathogen known to alter virulence of C. albicans. It was determined that the presence of C. albicans triggered increased rhamnolipid production by P. aeruginosa, which in turn was capable of inhibiting embedded hyphal growth produced beneath the C. albicans colony at ambient temperature.
Keywords: MALDI, imaging mass spectrometry, microbial interactions
Introduction
In nature, most microbes exist as part of a larger biological community consisting of many microbial neighbors as well as host organisms. With each microbial component of these communities typically capable of generating metabolic exchange factors affecting the local environment, the chemo- and phenotypic development of neighboring organisms is constantly being influenced. First observed by Alexander Fleming after an inadvertent fungal contamination of Staphylococcus aureus in a Petri dish that resulted in inhibition of the pathogen by penicillin, it is now common knowledge that microbes can utilize their arsenal of unique metabolic exchange factors to affect the composition of their surrounding environment (Ng and Bassler, 2009; Davies, 2010; Romero et al., 2011; Phelan et al., 2012). Such metabolic exchange factors enable the creation of unique biological niches and affect microbial multicellular behavior, community behavior and colony development (Phelan et al., 2012). Therefore, it is important to study microbial interactions not only to gain greater insight into their role in a larger microbial consortium, but to also observe how production of metabolic exchange factors triggered by competing neighboring organisms direct these microbial interactions.
Agar-based culturing techniques have been used to discover pharmacotherapies, develop biofuels and are used for disease diagnostics. Despite the central role of microbial culturing in academia, biotechnology and clinical microbiology, non-microscopy-based analytical approaches for monitoring the production of metabolic exchange factors directly on solid growth media have seen little advancement since Julius Petri developed the Petri dish (Zhang, 2004). Recently, we developed a two-dimensional (2D) agar-based matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) imaging mass spectrometry (IMS) technique for detection of microbial metabolites directly from colonies grown on solid media (Yang et al., 2009). Although MALDI-TOF IMS has traditionally been heavily utilized in histopathology of tissue sections (Chughtai and Heeren, 2010; Schwamborn and Caprioli, 2010; Seeley et al., 2011), our modification of MALDI-TOF IMS allows for chemical characterization of individual and interacting microbes (Yang et al., 2009; Liu et al., 2010 ). This technique has enabled a 2D top–down view of the chemical make-up from numerous microbial systems in a spatial manner allowing chemical characterization of observed phenotypes.
Although 2D MALDI-TOF IMS of microbes is capable of revealing tens to thousands of discreet chemical signals in a spatial manner from the sample surface, the limited overhead perspective of the analysis obscures information regarding the depth profile of secreted metabolites throughout the agar medium. Such information could lead to better characterization of a microbial system because the ability of secreted compounds to penetrate and diffuse through the agar medium will determine the capacity in which these compounds are capable of influencing neighboring bacteria. Within the field of biology, three-dimensional (3D) IMS has been used to determine molecular distributions within mammalian tissue and has been correlated with other 3D imaging data such as magnetic resonance imaging to provide a more complete picture of the chemical environment within the organ (Crecelius et al., 2005; Sinha et al., 2008; Seeley et al., 2012). Herein, we describe an adaptation of 3D MALDI-TOF IMS for capturing the multiplexed molecular nature of developing microbial colonies and the chemical interplay of microbial interactions. Using a Bacillus subtilis–Streptomyces coelicolor interaction as a proof-of-principle experiment, we describe a workflow adapted for routine 3D imaging of secreted metabolites from microbial colonies grown on agar media.
To further illustrate the utility of this method, we describe 3D MALDI-TOF IMS of Candida albicans, an opportunistic fungal pathogen that produces an extensive array of vegetative hyphae that burrow deep into the agar. Many microbes produce extensive arrays of sub-surface vegetative hyphae for a wide variety of purposes (Flardh and Buttner, 2009; Brand, 2012). As the local chemical environment within this sub-surface structure may influence its behavior, such as chemotaxis (Chet and Mitchell, 1976; Wadhams and Armitage, 2004) or filament differentiation (Truckses et al., 2004), elucidating the localized chemical output within the hyphae may provide valuable insight into its biology. In agar-based phenotypic experiments, these vegetative hyphae burrow deep into the agar (Burns et al., 1998; Sudbery, 2011) and have been shown to have a role in the well-investigated microbial interaction of C. albicans with Pseudomonas aeruginosa (Hogan and Kolter, 2002; Hogan et al., 2004; Mcalester et al., 2008). As the limited overhead perspective of traditional 2D MALDI-TOF IMS would not be able to fully observe molecular distributions within this subsurface structure, we used 3D modeling to determine if unique molecular distributions were in fact present within the vegetative hyphae and if compounds secreted by P. aeruginosa could inhibit their proliferation.
Materials and Methods
Culturing of bacteria and fungi
Starter cultures for B. subtilis PY79, P. aeruginosa PAO1 and C. albicans ySN250 were prepared by inoculating 3 ml of LB liquid media (Fisher Scientific, Pittsburgh, PA, USA) from a 20% glycerol cell stock and incubating at 28 °C until an OD600 of 0.5 was obtained (12–20 h of incubation). S. coelicolor A3(2) was inoculated directly from 20% glycerol frozen spore stocks. All samples to be analyzed using 3D MALDI-TOF IMS were grown on 8 mm thick nutrient limited ISP-2 agar prepared by combining 15 g agar (Sigma-Aldrich, St Louis, MO, USA), 1 g yeast extract (Sigma-Aldrich), 1 g dextrose (Fisher Scientific) and 1.5 g malt extract (Sigma-Aldrich) in 1 liter of Milli-Q grade water followed by sterilization via autoclave. Although growing the microbes on deep agar allowed for better molecular images of secreted metabolites within the agar, the media needed to be optimized in order to maintain the phenotype observed under normal growth conditions. As the amount of agar media used when culturing 3D samples is five times greater than what is typically used for 2D samples (Yang et al., 2009), we found that using 1/5th the amount of nutrients in the media allowed for a phenotype and colony size consistent with what we have previously reported (Yang et al., 2009, Watrous et al, 2012).
For B. subtilis PY79 versus S. coelicolor A3(2) interactions, three 1 μl aliquots of S. coelicolor A3(2) spore stock were inoculated with 5 mm spacing between each spot in a linear fashion. This inoculum was allowed to grow at 30 °C for 48 h after which a 0.5 μl aliquot of the B. subtilis PY79 starter culture was placed 2 mm from the terminal spot of S. coelicolor A3(2) and allowed to grow at 30 °C for an additional 24 h before MALDI IMS analysis. Individual control colonies were prepared by inoculating 1 μl of the S. coelicolor A3(2) spore stock or 0.5 μl of the B. subtilis PY79 starter culture on ISP-2 deep agar media and allowing to grow at 30 °C for 72 and 24 h, respectively.
For C. albicans ySN250 versus P. aeruginosa PAO1 interactions, two 0.5 μl aliquots of the C. albicans starter culture were inoculated roughly 10 mm apart and allowed to grow for 48 h at 30 °C after which two 0.5 μl aliquots of the P. aeruginosa starter culture were inoculated in line with the two C. albicans colonies with one aliquot 2 mm from one of the C. albicans colonies and the second aliquot roughly 10 mm from the first P. aeruginosa aliquot. After an additional 4 days of incubation at 30 °C, samples were prepared for MALDI IMS experiments. Single control colonies for C. albicans and P. aeruginosa were prepared similarly where single 0.5 μl inoculates were grown separately for 6 and 4 days, respectively.
Sample preparation protocol for 2D MALDI-TOF IMS
A single colony of C. albicans ySN250 was prepared by inoculating 0.5 μl of starter culture onto 1.5 mm deep ISP2 nutrient agar, which was prepared by combining 15 g agar (Sigma-Aldrich), 5 g yeast extract (Sigma-Aldrich), 5 g dextrose (Fisher Scientific) and 7.5 g malt extract (Sigma-Aldrich) in 1 liter of Milli-Q grade water followed by sterilization via autoclave. The reason the sample for 2D MALDI-TOF IMS was grown on 1.5 mm deep agar as opposed to the 8 mm deep agar used for 3D MALDI-TOF IMS analysis is due to signal intensity being optimal at this agar depth (this is also why serial cross-sections from samples prepared for 3D analysis were cut at 1 mm thickness). As the agar becomes deeper there is a steep decline in overall signal intensity and eventually the sample becomes too tall to fit into the instrument, which has a <1 mm vertical clearance. The sample was allowed to grow for 6 days at 30 °C at which time the agar was excised and transferred directly to a MALDI target plate followed by immediate deposition of Universal MALDI matrix (Sigma-Aldrich) using a 53 μm test sieve (Hogentogler & Co, Columbia, MD, USA). Universal MALDI matrix is a 1:1 mixture of 2,5-dihydroxybenoic acid and α-cyano-4-hydroxy-cinnamic acid matrices. The sample was dried for 5 h at 37 °C after which excess matrix was removed using compressed air and immediately imaged in linear positive mode from m/z 50–5000 using a Bruker Autoflex Speed MALDI mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany) at 400 × 400 μm spatial resolution with 200 shots per raster location using Random Walk shot pattern. Data were collected and analyzed using flexControl (version 3.0; Bruker Daltonik GmbH) and flexImaging (version 2.0; Bruker Daltonik GmbH) software. Calibration was done before the imaging run using Bruker Pepmix4 calibration standard (Bruker Daltonik GmbH).
Sample preparation protocol for 3D MALDI-TOF IMS
A 15 mm × 35 mm × 8 mm section of agar was excised from the Petri plate and placed colony-side down on a homemade slicing instrument (Figure 1). The excised agar was cut width-wise at room temperature using microtome blades into 1.0 mm sections with each section placed on its side on a stainless steel MALDI target plate. The MALDI targets used allowed for a maximum of six sections per plate. To ensure that cellular material from the colony was not distributed throughout the surface of each slice during sectioning, each sample was inverted before it was sectioned so that the samples were cut from the bottom of the agar down through to the surface of the agar. Also, after each slice, the cutting blade was washed sequentially in methanol, ethanol and water with the blade being wiped clean after each solvent with a fresh KimWipe to ensure no carry over (Supplementary Figure S1). Once the desired sample area was sectioned (typically resulting in a 15 mm × 15 mm piece of the excised sample being left over, which was done purposefully to help stabilize the sample during sectioning), Universal MALDI matrix (Sigma-Aldrich) was immediately deposited using a 53 μm test sieve (Hogentogler & Co). Samples were dried for 3 h at 37 °C; however, microbes producing high amounts of viscous metabolites, such as C. albicans, required up to 5 h to dry. The drying process reduces sample thickness of each slice from 1.0 mm to about 100 μm (Yang et al., 2012). After drying, excess matrix was removed using compressed air and immediately imaged in quick succession using the same instrument parameters as previously stated for the 2D MALDI IMS experiments. The mass spectrometry data were gathered for each serial cross-section resulting in a total imaging area of about 2000–3000 mm2.
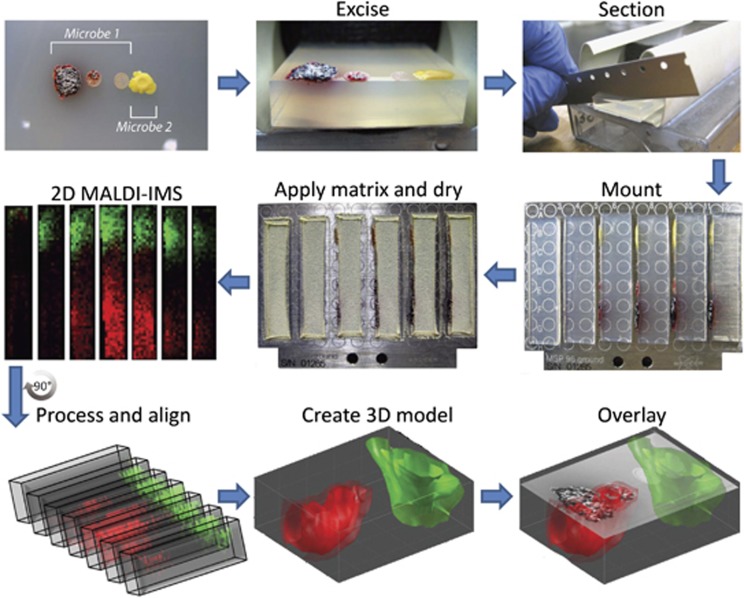
Figure 1.
General workflow for 3D MALDI-TOF IMS of microbial colonies on agar. A 15 mm × 35 mm × 8 mm area of the interacting microbes were excised and 1 mm cross-sections from the sample were cut and placed on its side on the MALDI target. Matrix is then applied to the MALDI target using a 53 μm sieve followed by drying at 37 °C for 3–5 h (depending on the system). Each MALDI target plate is then subjected to 2D MALDI-TOF IMS after which the data for each slice is loaded into MATLAB software where individual slices are virtually aligned, and finally visualized as a 3D model. In this paper, we represent a 3D mass spectrometry model as three isosurfaces (that is, layers) corresponding to different levels of normalized signal intensity (for example, 90%, 75% and 50% normalized intensity with the 50% isosurface being the most transparent). This model can then be overlaid with the optical image of the sample to correlate chemistry with observed phenotypes. The system used for this example, S. coelicolor A3(2) and Staphylococcus aureus MSSA, was chosen because of its intense color allowing for better optical images to be taken.
Generation of 3D models from MALDI-TOF IMS data
MALDI IMS spectra were converted into mzXML format using CompassXport software (Bruker Daltonics, Bremen, Germany) and loaded into MATLAB software (The Mathworks Inc., Natick, MA, USA). After normalizing to the total ion count followed by baseline correction, a 3D volume data set for a given m/z-value was created by co-registering all slices that involved aligning the slice edges as each slice was approximately the same size and shape. For each spectrum, its slice location was used as its z-coordinate, producing for each m/z-value a volume data set, which was further processed by reducing intensities of hot spots corresponding to abnormally intense spectra typically observed during MALDI IMS, volume denoising and contrast enhancement applied for better visualization. The resulting 3D volume data set corresponding to an m/z-value was visualized using 50%, 75% and 90% semi-transparent isosurfaces indicating 50%, 75% and 90% relative abundance of the molecular compound within the imaging area, respectively, with the 50% isosurface having highest transparency and the 90% isosurface appearing solid. Each isosurface was smoothed for better visualization. This simplified way of visualization was found to be well suited for microbial data where many mass signals are typically present as gradients of concentration because of their secreted nature. See Supplementary Methods for additional information and details.
Preparation for tandem mass spectrometry analysis
Tandem mass spectrometry experiments were performed on a Thermo LTQ-FT mass spectrometer (Thermo Electron Corp., Bremen, Germany) equipped with Nanomate and nanoDESI electrospray sources (Watrous et al., 2012). Microbial colonies were grown identically to samples that were analyzed by MALDI-TOF IMS and subjected to liquid surface extraction followed by direct infusion. A solvent of methanol:0.1% formic acid (70:30, v/v) was used to perform the extraction. Electrospray parameters included a spray voltage of 2 kV, heated inlet temperature of 200 °C, nebulizing gas pressure of 40 p.s.i. (for Nanomate), collision-induced dissociation energy of 25 V and collision time of 500 ms. Tandem mass spectra were annotated by hand and compared with standard MS2 spectra.
Purification and structural elucidation of rhamnolipids
Results
3D MALDI-IMS of B. subtilis PY79 and S. coelicolor A3(2)
To demonstrate that 3D MALDI-TOF IMS of microbial interactions can be used to obtain new chemical information, we set out to study the interaction between soil bacteria B. subtilis PY79 and S. coelicolor A3(2). Previously, we described the metabolic exchange between B. subtilis PY79, a lab domesticated strain of B. subtilis, and S. coelicolor A3(2) by MALDI-TOF IMS, imprint desorption electrospray ionization (DESI) IMS, and nanoDESI direct profiling (Yang et al., 2009; Watrous et al., 2010, 2012). Together, these complimentary techniques were able to characterize portions of the chemical output of both bacteria, which was essential in generating hypotheses as to their role in the dynamics of the interaction. Although chemical information from the sample surface is incredibly useful, micro-organisms are capable of producing unique chemical environments beneath the colony that are different from the conditions on the surface. In addition, growth and analysis of colonies grown on 8 mm deep agar allows one to observe differences in the ability of specific compounds to diffuse through the agar medium and therefore influence neighboring populations.
The resulting 3D models of the MALDI-TOF IMS data (Figure 2) were consistent with chemotypes previously observed from 2D MALDI-TOF IMS and imprint DESI IMS as well as with previously observed phenotypes with B. subtilis PY79 accelerating metabolic output and areal hyphae production in neighboring colonies of S. coelicolor (Yang et al., 2009; Watrous et al., 2010). Within S. coelicolor, increased production of actinorhodin, a purple pigmented polyketide, and SapB, a lantibiotic peptide necessary for aerial hyphae formation, could be seen in the S. coelicolor colony closest to the B. subtilis colony (Bystrykh et al., 1996; Kodani et al., 2004; Yang et al., 2009; Watrous et al., 2010). In addition, increased production of the cyclic lipopeptide calcium-dependent antibiotic (CDA) in S. coelicolor was also seen at the interface between the two colonies (Hojati et al., 2002). As the PY79 mutant of B. subtilis has had most of its polyketide synthase and non-ribosomal peptide synthetase machinery rendered inoperable (Zeigler et al, 2008), the only major product observed in both the control colony and the interaction was subtilosin, a macrocyclic bacteriocin (Kawulka et al., 2004).
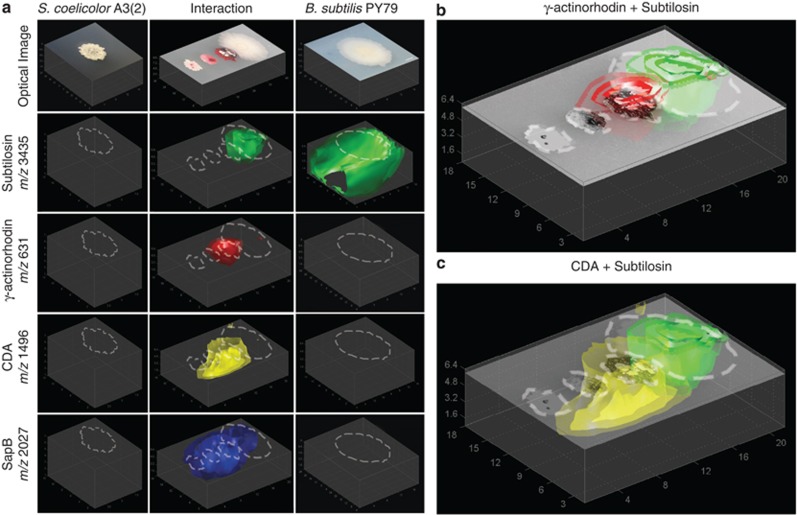
Figure 2.
3D models of metabolic exchange observed in the interaction of S. coelicolor with B. subtilis (negative ion mode). (a) The 3D models of S. coelicolor A3(2) alone (left column), S. coelicolor A3(2) interacting with B. subtilis PY79 (middle column) and B. subtilis PY79 alone (right column). Ion intensities in the control colonies were normalized to the signal from the interacting colonies. (b) Overlay of the 3D MALDI models of γ-actinorhodin from S. coelicolor (red) with subtilosin from B. subtilis (green) with the optical image of the interacting colonies. (c) Overlay of the 3D MALDI models of CDA from S. coelicolor (yellow) with subtilosin from B. subtilis (green) with the optical image of the interacting colonies.
Although these results have been previously observed, modeling of the mass signals in 3D allowed for additional information to be obtained (See Supplementary Figure S2 for all additional observed signals). Although previous 2D IMS studies showed the distributions of the red pigment prodiginine as well as actinorhodin to be localized to the S. coelicolor colony, their localizations by 3D modeling show that while actinorhodin is heavily secreted into the agar (Figures 2a and b), prodiginine was not (Supplementary Figure S3), which is consistent with previous studies stating that actinorhodin is secreted into the environment while prodiginine remains associated with the membrane (Vaidyanathan et al., 2008). Similarly, although 2D MALDI-TOF IMS showed the distributions of the secreted peptides CDA and SapB to be similar, 3D MALDI-TOF analysis revealed their diffusion patterns within the agar medium to be strikingly different owing to their specific chemical properties. In addition, 3D MALDI-TOF analysis showed signals for γ-actinorhodin, CDA and SapB can all be seen present beneath the interacting edge of the B. subtilis colony (especially CDA and SapB), which would be impossible to observe using 2D MALDI-TOF IMS as the MALDI laser is not capable of penetrating the bacterial colony to reach the agar beneath. These results illustrate how 3D modeling of MALDI-TOF IMS data collected from bacteria grown on 8 mm deep agar are not only able to reproduce previous observations from 2D IMS experiments, but also grants additional information on the chemical environment beneath the microbial colonies.
3D MALDI-IMS of C. albicans ySN250 and P. aeruginosa PAO1
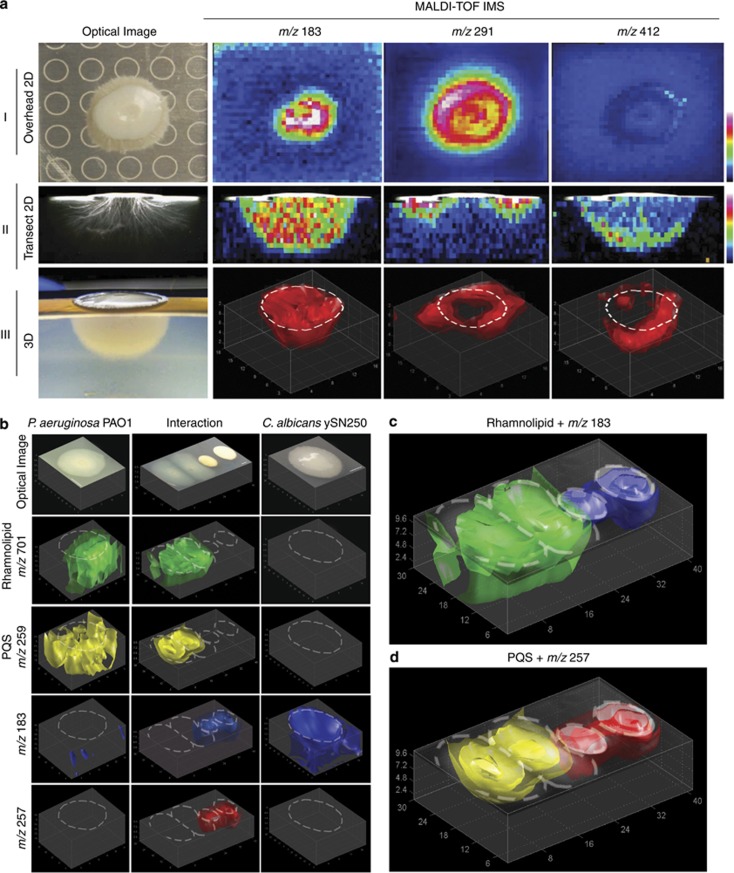
To investigate if specific chemical localizations within the embedded hyphae of C. albicans could be observed, we compared MALDI-TOF IMS data collected from a single C. albicans ySN250 colony using a traditional overhead perspective MALDI-TOF IMS (Figure 3a and I) with a 2D MALDI-TOF image of a single 1.0 mm cross-sectional slice through the center of a similar C. albicans colony (Figure 3a and II). Visually, colonies of C. albicans grown on 1.5 and 8 mm deep agar media appeared very similar with the colony consisting of a raised solid white circular center (roughly 8 mm across) and an off-white outer ring caused by horizontally growing embedded hyphae. Although the structure of the embedded hyphae was slightly obscured in the 1.5 mm sample because of the hyphae extending to the bottom of the Petri plate, the sample grown on deep agar showed a semi-spherical growth pattern for the hyphae (Figures 3a and II and III). Comparison of the two data sets shows that while many of the signals detected in the overhead 2D MALDI-TOF image are also present in the image of the cross-sectional slice, their distributions were drastically different (Figures 3a and I and II). For instance, the mass signals at m/z 183 and 291 showed similar signal distributions in the overhead image while the cross-sectional image showed that m/z 183 exhibits an even distribution across the embedded hyphae while m/z 291 is present only directly beneath the outer edge of the colony. Other signals were only observed in the cross-sectional image because of their localization being exclusively beneath the agar surface, such as m/z 412 whose signal was only observed at the far edges of the hyphae (Figure 3a and II). When 3D modeling of the MALDI-TOF IMS data were performed (Figure 3a and III), the distributions observed in the cross-sectional slice (Figure 3a and II) were explained with 3D models of m/z 183 showing an even distribution throughout the hyphae, m/z 291 forming a ring shaped distribution directly beneath the outer edge of the colony and m/z 412 being present only in the far reaches of the hyphae.
Figure 3.
(a) Comparison of 2D and 3D MALDI-TOF IMS of individual colonies of C. albicans (positive ion mode). Ion intensity for a-I and a-II is visualized using gradient color map where blue indicates low signal intensity and red indicates high signal intensity. Sample for a-I was grown on 1.5 mm thick ISP2 nutrient agar while a-II, a-III and b–d were grown on 8 mm thick nutrient limited ISP2 nutrient agar. (a-I) 2D MALDI-TOF overhead image of a single colony of C. albicans. Note that the rings in the optical image are from the MALDI target plate. (a-II) 2D MALDI-TOF image of a single cross-sectional slice of a C. albicans colony. Optical image of slice was taken using a transmitting light microscope (a-III) 3D MALDI-TOF image of a single colony of C. albicans. Note that optical image is taken from a semi-overhead perspective of the full excised sample similar to Figures 1. (b–d) 3D MALDI-TOF IMS of interacting colonies of C. albicans ySN250 and P. aeruginosa PAO1. (b) The 3D models of P. aeruginosa PAO1 alone (left column), P. aeruginosa PAO1 interacting with C. albicans ySN250 (middle column) and C. albicans ySN250 alone (right column). Ion intensities in the control colonies were normalized to the signal from the interacting colonies. (c) Overlay of the 3D MALDI images of the rhamnolipid at m/z 701 from P. aeruginosa (green) with m/z 183 from C. albicans (blue) with the optical image of the interacting colonies. (d) Overlay of the 3D MALDI images of pseudomonas quorum signal (PQS) from P. aeruginosa (yellow) with m/z 257 from C. albicans (red) with the optical image of the interacting colonies.
As the chemical distribution of specific compounds within the vegetative hyphae of C. albicans may have a role in its interactions with other microbes, we set out to study the interaction between C. albicans and P. aeruginosa, an opportunistic pathogen commonly co-isolated with C. albicans from lung sputum of immunocompromised individuals (Pierce, 2005) including cystic fibrosis patients (Burns et al., 1998; Davies, 2002). Interactions between P. aeruginosa PAO1 and C. albicans ySN250 were analyzed similarly to the interaction between B. subtilis PY79 and S. coelicolor A(3)2 as described earlier in this work. The visual phenotype of the interaction was consistent with published data where embedded hyphae formation in C. albicans was inhibited at the colony interface by P. aeruginosa as well as the C. albicans colony producing a yellowish-brown pigment (Gibson et al., 2009).
The resulting data showed that while many molecules remained unaffected by the presence of the competing microbe (See Supplementary Figure S4 for all additional observed signals), other signals, such as m/z 183, were inhibited in the region of interaction (Figures 3b and c). Additional signals, such as m/z 257 from C. albicans, were clearly more abundant when challenged with a competing bacterium (Figure 3b). Surprisingly, the pseudomonas quorum signal at m/z 259 from P. aeruginosa, which controls production of many metabolites within P. aeruginosa (Venturi, 2005), did not show increased production in the area of interaction with C. albicans (Figures 3b and d and Supplementary Video 1). Several signals, however, did show increased intensity in the region of interaction between the two colonies and could be associated with a family of compounds secreted by P. aeruginosa. The most prominent of these signals were observed at m/z 673, 699 and 701 (Figures 3b–d, Figure 4 and Supplementary Video 2) with additional signals also observed (Supplementary Figure S4).
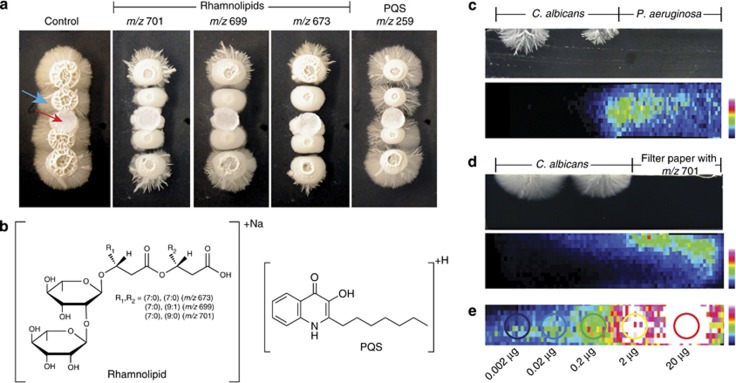
Figure 4.
The effect of Pseudomonas metabolites on C. albicans vegetative hyphae. (a) Bioassay of purified rhamnolipids (70 μg for m/z 701 and 699; 68 μg for m/z 673) and pseudomonas quorum signal (PQS) (26 μg) on individual colonies of C. albicans grown on 1.5 mm thick ISP2 nutrient agar. The red arrow points to the filter paper disc while the blue arrow points to the vegetative hyphae. (b) Structure of the rhamnolipids detected and PQS tested in the bioassay in (a). (c–e) To determine the concentration of the rhamnolipid (m/z 701) present in the interaction, a replicate interaction between P. aeruginosa and C. albicans was prepared where a single cross-sectional slice was imaged using 2D MALDI-TOF imaging alongside a blank section of agar spotted with known concentrations of m/z 701. All three data sets in c–e were collected on one MALDI plate in a single run and thus the instrument parameters were identical and the relative intensities can be directly compared. (c) A cross-sectional overview of the concentration of the rhamnolipid at m/z 701 when P. aeruginosa is spotted adjacent to C. albicans. (d) A cross-sectional overview of the m/z 701 ion distribution when 200 μg of purified material was deposited on a filter disc and placed next to C. albicans, which was determined by IMS to diffuse to similar concentrations in the agar as per the interaction of the P. aeruginosa in the 3D assay (8 mm agar plates) as determined in (c). (e) Estimation of the local concentration of rhamnolipid in the interactions. It was determined that the highest local rhamnolipid concentration (m/z 701) within the interacting Pseudomonas colony was roughly equivalent to the 0.2 μg spot when taken in account the diameter of the spot after diffusion (∼4 mm) this represents an estimated 16 μg ml−1 concentration (Figures 3b and c).
This increased production and location hinted that these signals may possibly have a role in the alteration of the C. albicans hyphae. Therefore, these molecules were isolated and structurally characterized by high-resolution Fourier transform ion cyclotron resonance (FT-ICR) tandem mass spectrometry (MS/MS) and nuclear magnetic resonance (NMR) (Supplementary Figures S5–S10 and Supplementary Table 1). This analysis confirmed that these signals belong to the rhamnolipid class of metabolites (Figure 4b; Soberon-Chavez et al., 2005). To determine the approximate concentration of rhamnolipids present in the agar during the interaction with C. albicans, an additional three samples were prepared and analyzed simultaneously on a single MALDI target plate: a 1 mm thick cross-sectional slice taken from the center of an interaction identical to Figure 3b (Figure 4c), a 1 mm thick cross-sectional slice taken from the center of an interaction between C. albicans and a filter disc containing 200 μg of purified rhamnolipid at m/z 701 (Figure 4d), and lastly a series of known amounts of rhamnolipid (m/z 701) deposited directly on a 1 mm thick slice of blank agar (Figure 4e). Previously, we have used a similar approach to estimate the concentration of surfactin produced by B. subtilis while others have estimated concentrations of antibiotics from ants (Gonzales et al., 2011; Schoenian et al., 2011). Using this approach, we estimated that the amount of the m/z 701 rhamnolipid, at its highest concentration in the interaction region, is approximately 16 μg ml−1. As expected, no effect was observed when pseudomonas quorum signal was tested in a similar manner (Figure 4a). This confirmed that the rhamnolipid class of molecules is capable of inhibiting growth of the vegetative hyphae of C. albicans at the same amounts found in the agar of the interaction (Figures 3b and 4). We obtained similar results for all three purified rhamnolipids (m/z 673, 699, 701) that were tested (Figure 4a).
Discussion
Multicellular behavior of microbes and interspecies interactions on solid media is mainly dictated by metabolic exchange factors and nutrient availability in the growth media (Phelan et al., 2012). MALDI-TOF IMS of serial sections followed by 3D rendering allows for visualization of metabolic exchange factors produced by microbes within the agar in 3D, which dictate colony and community behavior. As biology is inherently 3D, mapping the molecular distributions of all metabolic exchange factors in 3D is likely to better reveal a link between chemotypes and phenotypes as well as provide a more complete image of the true chemical makeup of the biological system (McMahon et al., 2006). Although performing 3D IMS analysis on tissue samples can prove difficult and very time intensive because of the expertise and instrument time required to precisely prepare and analyze each tissue section (Andersson et al., 2008; Nemes et al., 2009; Eberlin et al., 2010), the method presented here has proven to be fast and reproducible allowing for 3D analysis of a single sample (from sectioning to 3D modeling) to be completed within a single day. With matrix application using the test sieve being very reproducible, virtual alignment of each cross-section simplified because of each slice having the same size and shape, and with mass signals spanning several sequential cross-sections (because of the secreted nature of most microbial metabolites) providing a good metric of data quality, we have found this method to be a reliable way to gain further insight into the unique chemical environments beneath the agar surface. Finally, although the larger agar volume does lead to more diffuse signal, imaging the agar in a cross-sectional manner as opposed to the colony itself results in lower pixel-to-pixel variability because of reduced variation in surface morphology and more consistent matrix crystallization. Direct imaging of the intact colonies can yield imaging artifacts, such as lack of signal from areas of the colonies covered in aerial hyphae because of lack of matrix adherence (Yang et al., 2012).
As a proof-of-principle experiment, we showed that 3D MALDI-TOF IMS was capable of expanding on existing IMS technologies by allowing for an additional dimension of biological information to be visualized even when analyzing a very well studied system (Straight et al., 2006). Analysis of the interaction between B. subtilis PY79 and S. coelicolor A3(2) using 3D MALDI-TOF IMS not only confirmed previous observations (Yang et al., 2009; Watrous et al., 2010, 2012), but was also able to show differences in metabolite distributions that were not observable using 2D IMS (Figure 2). We were able to confirm that while actinorhodin is secreted into the surrounding environment, prodiginine remains associated with the membrane (Figures 2a and band Supplementary Figure S3) as well as show that while the distributions of SapB and CDA appear similar when analyzed by 2D MALDI-TOF IMS, their ability to diffuse through the agar medium is clearly distinguishable when looking at their corresponding 3D models (Figures 2a and c). One surprising result discovered by using 3D modeling of the MALDI-TOF data were the presence of most of the secreted S. coelicolor ions beneath the B. subtilis PY79 colony (Figures 2b and c). This was not previously observed using 2D MALDI-TOF IMS as the MALDI laser cannot penetrate the bacterial colony.
The ability to observe the diffusion of metabolites within the local agar environment, as opposed to just on the surface, becomes incredibly important when studying microbial systems (especially pathogens) that develop sub-surface structures. One example is a native member of the human mucosal biofilm community, C. albicans. This opportunistic fungal pathogen is responsible for superficial and systemic infections in immunocompromised patients (Pierce, 2005). Although the human immune system is partially responsible for controlling C. albicans infections, microbes commonly co-isolated from patients suffering from such infections have been shown to produce metabolites capable of modulating C. albicans virulence (Hogan and Kolter, 2002; Morales and Hogan, 2010). As one such organism, P. aeruginosa uses a variety of physical and chemical methods to limit the growth of C. albicans (Gibson et al., 2009; Bandara et al., 2010). It attaches exclusively to the hyphal germ tubes of C. albicans and secretes several virulence factors, including 3-oxo-C12 homoserine lactone and pyocyanin, to inhibit growth of C. albicans (Hogan et al., 2004). In response to this attack, C. albicans reverts back to its yeast state and therefore eliminates the ability of P. aeruginosa to adhere to the C. albicans colony.
As the interaction between C. albicans and P. aeruginosa occurs almost exclusively beneath the surface of the media within the vegetative hyphae of the C. albicans colony, we showed that 3D MALDI-TOF IMS was capable of capturing unique chemical distributions within this sub-surface structure as well as to pinpoint molecules that had a putative role in inhibiting hyphae formation in C. albicans (Figures 3 and 4). Although many of these compounds are currently unknown, knowledge of their spatial distributions helps provide insight into their possible functions with ions, such as m/z 412 possibly being involved in hyphae elongation because of its localization to the outer edge of the hyphae. When analyzing the interaction between C. albicans and P. aeruginosa, we showed that the presence of P. aeruginosa was able to modulate certain chemical signals within the C. albicans hyphae (Figures 3b–d). By using the 3D models to pinpoint signals intensified in P. aeruginosa at the colony interface (Supplementary Video 2), we were able to rapidly identify that a major contributor to this chemical modulation were a family of rhamnolipids (Figures 3b and cand Figure 4). Rhamnolipids have been reported to have antifungal activity against zoosporic plant pathogens and have a role in allowing P. aeruginosa to inhibit growth of certain bacteria (Stanghellini and Miller, 1997; Soberon-Chavez et al., 2005). By isolating and purifying these compounds, we were able to confirm the ability of rhamnolipids to inhibit hyphae formation in C. albicans at relatively low concentrations (Figures 4c–e).
Collectively, we have demonstrated that the expansion of 2D MALDI-TOF IMS into 3D for microbial analysis provides valuable additional information regarding the spatial distribution of metabolic exchange factors of microbes cultured in Petri dishes. IMS offers the unique vantage point that enables spatially resolved biochemical analysis and correlates the chemistry in a spatial manner with the corresponding phenotypes. In turn, 3D MALDI-TOF IMS enables the formulation of novel hypotheses regarding the biological implications of certain mass signatures based on their distribution within the 3D model from the biological sample as we demonstrated with the roles of rhamnolipids in the interaction between C. albicans and P. aeruginosa. The increased production of the rhamnolipids and its localization in the region where the vegetative hyphae were decreased was revealed by 3D MALDI-TOF IMS enabling us to demonstrate that the rhamnolipids contributed to this phenotype. This information can be used to better understand the fundamental dynamics of microbial molecular encounters by allowing us to determine the origin, identity and assign a purpose of each signal.
Acknowledgments
This work was supported by the Keck Foundation and National Institute of General Medical Sciences Grant NIH GM086283, GM094802 (PCD), NIH Molecular Biophysics Training Grant NIH GM08326 (JDW), and European Union 7th Framework Programme grant 305259. We thank Suzanne Noble, University of California San Francisco, for kindly providing us with C. albicans ySN250.
Author contributions
JDW, TA and PCD designed the research plan; JDW, VVP and C-CH performed the experiments; TA performed 3D modeling of the MALDI-TOF IMS data; C-CH, WJM and BMD performed NMR analysis; JDW, VVP, C-CH, WJM and PCD analyzed MS data; JDW, VVP, C-CH, TA and PCD wrote the paper.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Meth. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- Bandara HMHN, Yau JYY, Watt RM, Jin LJ, Samaranayake LP. Pseudomonas aeruginosa inhibits in-vitro Candida biofilm development. BMC Microbiol. 2010;10:125. doi: 10.1186/1471-2180-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol. 2012;2012:517529. doi: 10.1155/2012/517529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Ermerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, et al. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- Bystrykh LV, Fernandez-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L. Production of actinorhodin-related ‘blue pigments' by Streptomyces coelicolor A3(2) J Bacteriol. 1996;178:2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chet I, Mitchell R. Ecological aspects of microbial chemotactic behavior. Ann Rev Microbiol. 1976;30:221–239. doi: 10.1146/annurev.mi.30.100176.001253. [DOI] [PubMed] [Google Scholar]
- Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J Am Soc Mass Spectrom. 2005;16:1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Davies J. How to discover new antibiotics: harvesting the parvome. Curr Opin Chem Biol. 2010;15:5–10. doi: 10.1016/j.cbpa.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Davies JC. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev. 2002;3:128–134. doi: 10.1016/s1526-0550(02)00003-3. [DOI] [PubMed] [Google Scholar]
- Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-dimensional visualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew Chem. 2010;49:873–876. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flardh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. App Environ Microbiol. 2009;75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DJ, Haste NM, Hollands A, Fleming TC, Hamby M, Pogliano K, et al. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology. 2011;157:2485–2492. doi: 10.1099/mic.0.048736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vika A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Fett F, et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol. 2002;9:1175–1187. doi: 10.1016/s1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- Kawulka KE, Sprules T, Diaper CM, Whittal RM, McKay RT, Mercier P, et al. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to alpha-carbon cross-links: formation and reduction of alpha-thio-alpha-amino acid derivatives. Biochemistry. 2004;43:3385–3395. doi: 10.1021/bi0359527. [DOI] [PubMed] [Google Scholar]
- Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene rams in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2004;101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-T, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci USA. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcalester G, O'Gara F, Morrissey JP. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol. 2008;57:563–569. doi: 10.1099/jmm.0.47705-0. [DOI] [PubMed] [Google Scholar]
- McMahon G, Glassner BJ, Lechene CP. Quantitative imaging of cells with multi-isotope imaging mass spectrometry (MIMS)-Nanoautography with stable isotope tracers. Appl Surf Sci. 2006;252:6895–6906. [Google Scholar]
- Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, Barton AA, Vertes A. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81:6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan VV, Liu W-T, Pogliano K, Dorrestein PC. Microbial metabolic exchange—the chemotype-phenotype link. Nat Chem Biol. 2012;8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce GE. Pseudomonas aeruginosa, Candida albicans, and device-related nosocomial infections: implications, trends, and potential approaches for control. J Ind Microbiol Biotechnol. 2005;32:309–218. doi: 10.1007/s10295-005-0225-2. [DOI] [PubMed] [Google Scholar]
- Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenian I, Spiteller M, Ghaste M, Wirth R, Herz H, Spiteller D. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci USA. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry-looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- Seeley EH, Caprioli RM. 3D imaging mass spectrometry: a new frontier. Anal Chem. 2012;84:2105–2110. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley EH, Schwamborn K, Caprioli RM. Imaging of intact tissue sections: moving beyond the microscope. J Biol Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, et al. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Met. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberon-Chavez G, Lepine F, Deziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- Stanghellini ME, Miller RM. Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Dis. 1997;81:4–12. doi: 10.1094/PDIS.1997.81.1.4. [DOI] [PubMed] [Google Scholar]
- Straight PD, Willey JM, Kolter R. Interaction between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Truckses DM, Garrenton LS, Thorner J. Jekyll and Hyde in the microbial world. Science. 2004;306:1509–1511. doi: 10.1126/science.1104677. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan S, Fletcher JS, Goodacre R, Lockyer NP, Micklefield J, Vickerman JC. Subsurface biomolecular imaging of Streptomyces coelicolor using secondary ion mass spectrometry. Anal Chem. 2008;80:1942–1951. doi: 10.1021/ac701921e. [DOI] [PubMed] [Google Scholar]
- Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2005;20:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cel Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Watrous J, Hendricks N, Meehan M, Dorrestein PC. Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal Chem. 2010;82:1598–1600. doi: 10.1021/ac9027388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC, et al. A primer to agar-based microbial imaging mass spectrometry. J Bacteriol. 2012;194:6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-L, Xu Y, Kersten RD, Liu WT, Meehan MJ, Moore BS, et al. Connecting chemotypes and phenotypes of cultured marine microbial assemblages using imaging mass spectrometry. Angew Chem. 2011;50:5839–5842. doi: 10.1002/anie.201101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-L, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. The origins of 168, W23 and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Beyond the Petri dish. Nat Biotechnol. 2004;22:151–152. doi: 10.1038/nbt0204-151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.