Abstract
Although microorganisms largely drive many ecosystem processes, the relationship between microbial composition and their functioning remains unclear. To tease apart the effects of composition and the environment directly, microbial composition must be manipulated and maintained, ideally in a natural ecosystem. In this study, we aimed to test whether variability in microbial composition affects functional processes in a field setting, by reciprocally transplanting riverbed sediments between low- and high-salinity locations along the Nonesuch River (Maine, USA). We placed the sediments into microbial ‘cages' to prevent the migration of microorganisms, while allowing the sediments to experience the abiotic conditions of the surroundings. We performed two experiments, short- (1 week) and long-term (7 weeks) reciprocal transplants, after which we assayed a variety of functional processes in the cages. In both experiments, we examined the composition of bacteria generally (targeting the 16S rDNA gene) and sulfate-reducing bacteria (SRB) specifically (targeting the dsrAB gene) using terminal restriction fragment length polymorphism (T-RFLP). In the short-term experiment, sediment processes (CO2 production, CH4 flux, nitrification and enzyme activities) depended on both the sediment's origin (reflecting differences in microbial composition between salt and freshwater sediments) and the surrounding environment. In the long-term experiment, general bacterial composition (but not SRB composition) shifted in response to their new environment, and this composition was significantly correlated with sediment functioning. Further, sediment origin had a diminished effect, relative to the short-term experiment, on sediment processes. Overall, this study provides direct evidence that microbial composition directly affects functional processes in these sediments.
Keywords: ecosystem functioning, common garden, reciprocal transplant, estuary
Introduction
Microbial composition—the identity and relative abundance of microbial taxa—varies over space and time (Fuhrman et al., 2006; Martiny et al., 2006; Gilbert et al., 2012; Hanson et al., 2012) and in response to abiotic perturbations (Allison and Martiny, 2008). However, the question remains where and when such variation affects the functioning of ecosystems (Schimel, 1995; Balser and Firestone, 2005; Strickland et al., 2009). To test the effect of microbial composition on process rates, composition must be manipulated and maintained, ideally in a natural ecosystem. In this way, one can directly tease apart the effects of composition, the environment and their interactions (Reed and Martiny, 2007). In larger organisms, composition can often be controlled by seeding or caging (for example, Power, 1990; Tilman et al., 1996), but such precise manipulations of microorganisms in a field environment are usually untenable.
Instead, microbial ecologists have three main options for explicitly testing the effect of microbial composition on ecosystem functioning, each with their own benefits and limitations. The first strategy is to use laboratory microcosms, then measure one or more aggregate processes of the assemblage. For instance, Bell et al. (2005) constructed communities from cultivatable bacteria in sterile microcosms and found that bacterial richness and composition affected respiration rates. Similarly, direct manipulation of mychorrhizal fungal diversity altered plant biomass and soil nutrients in a greenhouse experiment (van der Heijden et al., 1998). To increase the community complexity, other studies have also inoculated microcosms with diverse, intact microbial communities (for example, Langenheder et al., 2005). For instance, Strickland et al. (2009) found that respiration varied greatly among leaf litter microcosms inoculated with three different soil communities. In contrast, Langenheder et al. (2006) and Dimitriu et al. (2010) found weak or no effects of the microbial inoculum on functioning in aquatic and peat microcosms. Still other studies have diluted a single intact community to alter taxon richness and composition to test for effects of composition on microcosm functioning. In general, such dilution experiments have found weak or no effects of the inoculum on functioning in a variety of systems (Griffiths et al., 2001; Franklin and Mills, 2006, Wertz et al., 2006; Dimitriu et al., 2010). Of course, a downside to all microcosm experiments is that they are carried out in the laboratory or greenhouse, which may or may not be a good proxy of the natural environment.
A second approach is to remove or inhibit a target group of microorganisms using chemicals in the field. For instance, studies using a fungicide (benomyl) to suppress arbuscular mychorrizal fungi demonstrate that fungi can have large effects on plant communities (Hartnett and Wilson, 1999; Daleo et al., 2007). Similarly, nitrapyrin applications that inhibit ammonia-oxidizers suggest that these microorganisms influence not only soil nitrogen cycling, but also plant biomass and litter decomposition (Austin et al., 2007). Although these studies can be carried out in a natural environment, a caveat to these approaches is that they may have unintended consequences for the abiotic environment and other organisms. Indeed, what is toxic to some microorganisms, may stimulate the growth of another (for example, Dantas et al., 2008).
Finally, the influence of microbial composition can be tested by using in situ transplants of whole microbial communities (for example, Bottomley et al., 2004; Waldrop and Firestone, 2006). The upside of this approach is that it considers a complex community in a natural setting. For instance, Balser and Firestone (2005) reciprocally transplanted soil cores between a forest and grassland site. Changes in microbial composition were associated with changes in soil functioning and in particular, nitrogen cycling. However, two limitations with such experiments are that the soil matrix differs along with the microbial community and that the transplanted cores may not fully equilibrate with the surrounding environment. Further, such studies do not usually prevent migration of microorganisms after transplantation (but see Gasol et al., 2002), such that the effect of composition on functioning becomes more ambiguous over time; it is then unclear whether changes in microbial composition over time are because of microbial migration or growth within the soil cores.
To build on these previous studies, we aimed to demonstrate an approach that can explicitly test whether variability in microbial composition affects functional processes in a field setting, while controlling for both the abiotic environment and microbial migration. To do this, we reciprocally transplanted riverbed sediments between two different abiotic environments: an oligohaline (salinity ∼2) and an euhaline (15–30) location. (For simplicity, we refer to these locations as the ‘freshwater' and ‘saltwater' sites.) We placed the transplanted sediments into microbial ‘cages' (Gasol et al., 2002; Reed and Martiny, 2007) to prevent the movement of microorganisms, while allowing the sediments to experience the abiotic conditions (such as temperature, salinity and nutrients) of the surroundings. We focused on estuarine sediments for three reasons. First, the aquatic environment allows the use of cages made of dialysis tubing (Figure 1). Second, salinity is known to be a key abiotic factor determining microbial composition (Lozupone and Knight, 2005), particularly along estuarine gradients (for example, Freitag et al., 2006; Mohamed and Martiny, 2010); therefore, we predicted that changes in salinity would quickly select for distinct microbial communities. Third, by sampling two locations from the same river, differences in the substrate matrix between locations could be minimized.
Figure 1.
Dialysis tubing sediment cages with PVC collars. United States quarter dollar (24.26-mm diameter) shown for scale.
We report the results from two experiments at these locations. In the first experiment—hereafter, the ‘short-term' experiment—we reciprocally transplanted sediment cores in cages from the freshwater and saltwater environments. After 1 week, we removed the cages from the riverbed and immediately assayed a variety of functional processes. This time frame was chosen to allow time for the sediments to equilibrate with the environment, but not enough time for a discernable change in microbial composition because of altered growth rates. In the second, ‘long-term' experiment, we allowed the sediments to remain in the riverbed for 7 weeks before measuring the functional parameters. In both experiments, we examined the composition of bacteria generally (targeting the 16S rDNA gene) and sulfate-reducing bacteria (SRB) specifically (targeting the dsrAB gene) using terminal restriction fragment length polymorphism (T-RFLP). SRB are ubiquitous in freshwater and marine sediments, and their composition appears to vary along estuarine gradients (for example, Hawkins and Purdy, 2007).
If microbial composition directly affects the functional processes of these sediments, we would expect to find support for three hypotheses: (1) In the short-term experiment, sediment processes would depend on both the sediment's origin (reflecting differences in microbial composition between salt and freshwater sediments), as well as the surrounding environment. (2) In the long-term experiment, microbial composition in the transplanted sediments would shift in response to their new environment. (3) As a result of this compositional shift, the sediment's origin would have a diminished effect (relative to the short-term experiment), and the surrounding environment a greater effect, on sediment processes.
Materials and Methods
The experimental sites were located 2.5 km apart along the riverbed of the Nonesuch River in Maine, USA. The saltwater, euhaline (salinity >25) site bordered the Scarborough Marsh. Plant composition in this marsh is typical of northern New England salt marshes and dominated by Spartina patens and S. alterniflora (Ewanchuck and Bertness, 2004). The ‘freshwater,' oligohaline (salinity <5) site upstream bordered a diverse marsh, similar to other oligohaline marshes in the area (Crain, 2007).
Microbial cages and transplant experiments
To limit the migration of microorganisms while allowing for solutes to permeate, we constructed cages around the 6-cm diameter by 6-cm deep sediment cores out of dialysis tubing. We used large diameter tubing (6.4 cm) with molecular weight cutoff of 8 kD (Spectrum Laboratories, Inc, CA, USA), which should exclude the movement of microbial eukaryotes, bacteria, archaea and most viruses.
To test the potential for bacterial migration, we performed an initial experiment in the lab. Dialysis ‘cages' were filled with sterile saline solution (0.87% NaCl) and placed in 1-l beakers filled with saline and inoculated with Xanthomonas sp. (average 2.7 × 107 CFU ml−1). After 5 days, only 247 CFU ml−1 (±87) were detected inside the cages, while an average of 1.1 × 108 CFU ml−1 (±2.3 × 106) were detected in external media. Thus, only about 1 per million cells penetrated the cages over 5 days.
For the field experiments, dialysis tubing pieces (32-cm long) were rinsed thoroughly at least 48 hours before use, incubated in sterile water at 4 °C for 24 h and then rinsed again in sterile DI water before use in the field. After sliding the sediment core into the dialysis tubing, each end was folded over a polyvinyl chloride (PVC) cylinder placed just inside the tubing, fitting an o-ring over the folded end onto the internal cylinder, and slipping an outer PVC cylinder over the o-ring.
For both experiments, 32 sediment cores were taken from exposed riverbed sediment at low tide and placed into microbe cages. The cages were placed in river water from their native site until all cages were transplanted back into the riverbed of the appropriate site. The sediments were placed in the holes made by the original cores in their original vertical orientation. For the short-term experiment, the cores were incubated for 7 days during mid-June.
Within 2 weeks of completing the short-term experiment (the beginning of July), we initiated the long-term experiment. Before placing the cores into the cages, a subset of sediment was sampled and frozen for initial sediment analyses and DNA extraction. These cores were incubated for a total of 61 days. To prevent degradation of the dialysis tubing during this longer incubation period, the tubing was replaced (while preventing contamination of the sediments) every 7–10 days. During this experiment, the integrity of many cages at the freshwater marsh site was compromised, possibly because of runoff from nearby development. This treatment was therefore eliminated from the analyses.
Gas sampling
At the end of both experiments, we removed the cores from the riverbed and opened the dialysis bag to reveal the top of the sediment core. The entire cage was placed into a 6-cm long 5-cm diameter schedule 40 PVC, which acted as collars for vented static chambers. The enclosed cores were placed in two gallon buckets containing ∼500 ml of river water. PVC caps fitted with rubber septa were placed over the enclosed cages. Total headspace volume was 150 ml.
At the same time in both locations, headspace gas was sampled every 20 min for a period of 60 min. Gas samples were stored in 2-ml gas sampling vials. While in the field, a set of control vials were filled with 100 ppm of CH4 or 1000 ppm of CO2 to test for leakages, but none were detected. The gas samples were analyzed by gas chromatography (Shimadzu GC2014 equipped with a thermal conductivity detector to measure CO2 and a flame ionization detector to measure CH4, and fitted with a 1-ml sample loop; Shimadzu Corporation, Kyoto, Japan). We calculated the gas production rates as the slope of the linear regression of gas concentration over time.
Sediment biogeochemistry
Salinity, sediment moisture, inorganic N and potential gross nitrification were analyzed for each replicate after both the short- and long-term experiments. Total C and N and sediment texture were measured during the short-term experiment of the initial samples. After the short-term experiment, pH and SO4 concentrations were also measured.
Salinity was measured by refractometer on pore water obtained after centrifuging 1.5-ml sample of sediment taken from the center of the core. Sediment moisture was measured by determining weight loss after drying 10 g of soil for 24 h (or until constant weight) at 105 °C. pH was measured using a 1:1 DI water sediment mixture and pH meter. Sediment texture was measured on four random samples of native freshwater sediments and saltwater sediments taken at the beginning of the long-term experiment (Elliott et al., 1999).
Inorganic N was measured by extracting 10 g of wet sediment with 75 ml of 2 ℳ KCl. Concentrations of NH4 and NO3 were measured using a microplate technique (Weatherburn, 1967; Doane and Horwath, 2003). Extractable SO4 was extracted from 10 g of wet sediment using 40 ml of 0.016 ℳ KH2PO4, and analyzed with a Hach pocket colorimeter (Hach, Loveland, CA, USA). Half the samples were also analyzed by IC (Maine Soil Testing Laboratory, University of Maine) with similar results. Total C and N of sediments were measured via Carla Erba combustion (ANR Analytical Labs, University of California, Davis).
Potential gross nitrification was estimated by the pool dilution technique (Hart et al., 1994). For each core, two equivalents of ∼15 g of dry sediment were placed into 50-ml specimen cups. To remove any NH4+ limitation, each sample was amended with 3 ml of 100 mℳ NH4Cl (∼30 mℳ NH4+ final concentration). Samples then received 5 ml of 0.1 mℳ K15NO3 solution (50 μg of K15NO3), which was slowly injected into the sediment. The initial samples were immediately extracted with 75 ml of 2 ℳ KCl and capped tightly, while the final samples were capped and incubated at 25 °C for 24 h before being extracted with 2 ℳ KCl.
Enzyme activities
A 10 g sub-sample of sediment (stored at −20 °C until analyzed) was used for enzyme activity analyses. Activities of three ecto-enzymes, leucine aminopeptidase, phosphatase and sulfatase were measured in both acidic (pH 5) and basic (pH 8) buffers. Leucine aminopeptidase, phosphatase and aryl sulfatase acitivities were measured with methylumbelliferyl-linked substrates (Saiya-Cork et al., 2002).
Microbial community analysis
For community analysis by T-RFLP, sediment cores 1 cm in diameter and 1-cm deep were taken from the top of each sediment cage in the field, placed on ice for transport and stored at −80 °C until analyzed. DNA was extracted from 0.5 g of sediment. To minimize shearing and PCR inhibition, we used a protocol combining the PowerSoil DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, USA) and the FastDNA Spin kit for soil (Qbiogene Inc., Montreal, CA, USA). Briefly, samples were placed in MO BIO Power Clean bead tubes and homogenized via the MPBio FastPrep for 15 s at setting 4. Homogenized samples were transferred to FastDNA Spin Kit tubes, and the manufacture's instructions were followed. Genomic DNA was screened for shearing and quantified. For all samples, four 25-μl PCR reactions were combined. The reaction mixtures contained 2 μl of genomic DNA (∼20 ng of DNA), 23-μl Pre-Mix D (Epicentre Biotechnologies, Madison, WI, USA), 2.5 U of Platinum Taq polymerase (Invitrogen Corporation, Carlsbad, CA, USA), and 12.5 pmoles of each primer. The 5′ end of the forward primers of each primer pair was labeled with 6-FAM. The dsrAB fragment (1.9 kb) was amplified using the forward primer dsr1fF and reverse primer dsr4R (Wagner et al., 1998). We used the following conditions for PCR amplification of dsrAB: initial denaturing step at 94 °C for 6 min, 32 cycles of 94 °C at 90 s, 58 °C annealing for 45 s, 72 °C extension for 90 s and final extension at 72 °C for 10 min. The 16S rRNA gene fragment was amplified using the forward primer 8F (Lane, 1991) and reverse primer 1492R (positions1492–1509) (GYTACCTTGTTACGACTT) (modified from Lane, 1991). The PCR conditions were a modified touchdown protocol: initial denaturing step at 94 °C for 3 min, 23 cycles of 94 °C for 45 s, 58 °C for 20 s 72 °C for 90 s, 10 additional cycles with the annealing temperature decreasing by 0.2 °C each cycle and 5 final cycles with the annealing temperature at 56 °C. The dsrAB PCR products were purified using the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA, USA) and 16S PCR products were gel extracted using the Gel DNA Recovery kit (Zymo Research Corp., Orange, CA, USA). PCR products were quantified using a NanoDrop spectrophotometer (NanoDrop Products, Wilmington, DE, USA).
For T-RFLP analysis, 200 ng of products were digested with 20 U of enzyme for 4 h, according to the manufacturer's protocol. The dsrAB products were digested with NdeII (Promega Corporation, Madison, WI, USA) and 16S rDNA products were digested with MspI (New England BioLabs, Ipswich, MA, USA). After digestion, samples were de-salinated using a QIAquick Nucleotide Removal kit (Qiagen). Digested products were submitted to Laragen, Inc. (Los Angeles, CA, USA) for electrophoresis. Peak areas were quantified using the GeneMarker software (SoftGenetics, LLC, State College, PA, USA), and peak areas were standardized and ‘true peaks' identified using the methods of Abdo and coworkers (Abdo et al., 2006).
To confirm the repeatability of the fragment patterns, 7% of the samples were processed in duplicate with independent PCR reactions and enzyme digestion. The T-RFLP patterns were highly correlated between replicates; the average R2 between band lengths of replicated samples was 0.96±0.06 s.d.
Statistical analyses
For the short-term experiment, we used a two-way ANOVA (JMP software, v. 5.1, SAS, Cary, NC, USA) to test for the effects of sediment origin (a proxy for microbial composition), the incubating environment and their interaction on sediment characteristics and functional processes. For the long-term experiment where we lost one of the treatments, we used a one-way ANOVA to test for differences between the treatments and a Tukey's multiple means comparison for post-hoc comparison of means. We used t-tests to test whether sediment characteristics differed by origin in the saltwater environment after the 2-month incubation. To correct for non-normality and heteroscedacity, in the short-term experiment NH4+concentrations was log transformed, methane production rates were rank transformed and gross nitrification data was arcsin-square root transformed; in addition, appropriate outliers were removed (<5%). In the long-term experiment, basic leucine aminopeptidase activity was also log transformed, and one outlier was removed.
To test whether community composition differed among cores types, we calculated Bray–Curtis similarities between the T-RFLP samples and tested for differences using an analysis of similarity (ANOSIM) (Primer 6 software, PRIMER-E Ltd, Ivybridge, UK) (Clarke and Warwick, 2001). To examine overall functional properties, we calculated a standardized, Euclidean distance of all nine functional parameters between each pairwise combination of the sediment cores. We then used ANOSIM to test whether sediment origin or environment affected this overall functional metric. Finally, to compare sediment composition and functioning directly, we used RELATE (Primer 6) to test for a correlation between compositional and functional similarity among sediment cages.
Results
A main goal of our experimental design was to minimize differences in the sediment matrix contained within the cages, while maximizing differences in the abiotic environment that could permeate the cages. Sediment texture was not significantly different between the sites; average percent sand, clay and silt across all samples were 28.4%, 13.4% and 58.1%, respectively. The carbon to nitrogen ratio (C:N) was also similar in the sediments at both locations. Total C and N differed slightly, however, with the freshwater sediments containing a higher percentage of both C and N than the saltwater sediments (Table 1).
Table 1. Characteristics of the sediment cores from freshwater and saltwater environments. Freshwater and saltwater values are means with 1 s.e. in parentheses.
| Freshwater | Saltwater | t (d.f.) | P | |
|---|---|---|---|---|
| % Sand | 27.7 (2.0) | 29.1 (1.8) | −0.047 (8) | 0.65 |
| % Silt | 60.2 (3.2) | 56.3 (3.0) | 0.86 (10) | 0.40 |
| % Clay | 12.1 (1.9) | 14.6 (1.8) | −1.01 (10) | 0.33 |
| C:N | 13.2 (0.4) | 13 (0.3) | 0.45 (527) | 0.65 |
| % C | 4.3 (0.1) | 2.3 (0.1) | 12.00 (27) | <0.0001 |
| % N | 0.4 (0.04) | 0.2 (0.009) | 5.6 (16) | <0.0001 |
A t-test assuming unequal variances was used to test for differences between the two locations.
Short-term experiment
Abiotic conditions
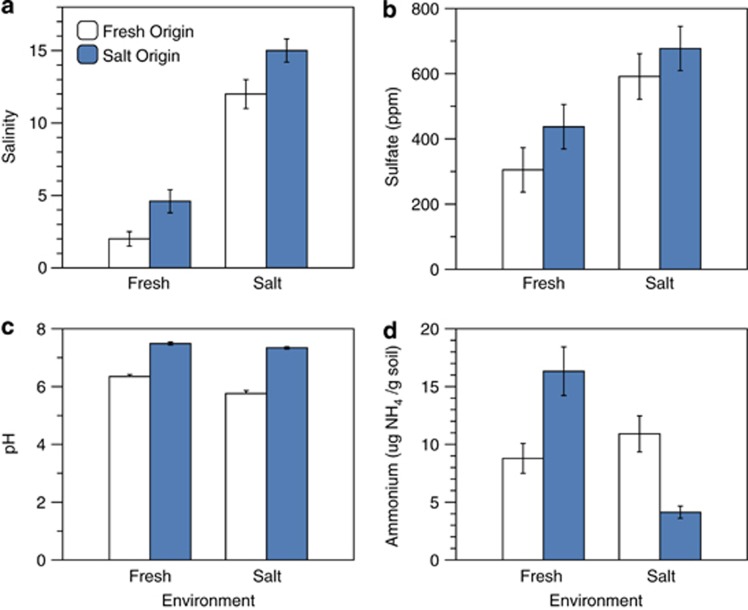
After 1 week, abiotic conditions within the transplanted sediments changed dramatically. Salinity in the saltwater sediments that were transplanted to the freshwater environment dropped to from 15 to 4.6, but remained significantly >2, the salinity of the native freshwater sediments (Figure 2a; Table 2). Sulfate concentrations equilibrated even more quickly, as they depended only on the surrounding environment and not on the sediments' origin (Figure 2b; Table 2).
Figure 2.
Means (±1 s.e.) of (a) salinity, (b) sulfate, (c) pH and (d) ammonium at the end of the short-term experiment.
Table 2. Effects of sediment origin and environment on sediment characteristic at the end of the short-term experiment.
| Origin | Environment | Origin x environment | |
|---|---|---|---|
| Salinity | F1,60=191, P<0.0001 | F1,60=2278, P<0.0001 | F1,60=1.44, P=0.2 |
| Sulfate | F1,59=2.5, P=0.1 | F1,59=15, P<0.0003 | F1,59=0.12, P=0.7 |
| pH | F1,60=428, P<0.0001 | F1,60=31, P<0.0001 | F1,60=11.0, P=0.001 |
| Ammonium | F1,56=0.20, P=0.7 | F1,56=11, P<0.002 | F1,56f=P<0.0001 |
F statistics, degrees of freedom, and significance values are reported for the two-way ANOVA tests.
In contrast to salinity and sulfate, pH was relatively static over 1 week and was primarily determined by the origin of the sediment. However, pH decreased slightly, but significantly, when freshwater sediments were transplanted to saltwater (Tukey post-hoc test: P<0.05), the opposite direction to that of the native saltwater sediments (Figure 2c; Table 2). Ammonium concentrations did not differ by sediment origin directly, but were affected by a significant origin by environment interaction (Figure 2d; Table 2). These results might indicate that the abiotic environment in the cages had not yet come to equilibrium with the new environment. Alternatively, the interactive effect of origin and environment on ammonium and the unexpected shift in pH in the saltwater environment could mean that abiotic conditions are also influenced by the outcome of sediment functioning, including the influence of microbial composition on these functions.
Bacterial composition
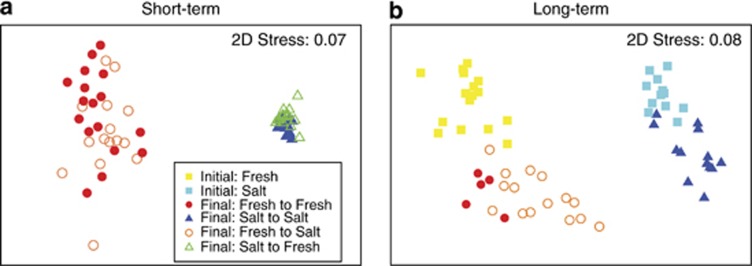
As expected, bacterial composition differed greatly between the saltwater and freshwater sediment cages and changed little after 1 week in the opposite environment. General bacterial composition, as determined by 16S rDNA T-RFLP profiling, differed greatly by sediment origin (two-way ANOSIM: R=0.995, P=0.001). In contrast, the surrounding environment (hereafter, environment) had a much weaker influence on bacterial composition. Bacterial composition in sediments from the same origin differed only slightly by environment (R=0.052, P=0.044; Figure 3a). The composition of SRB, as determined by T-RFLP of the dsrAB gene, showed a similar pattern as general bacterial composition. Composition differed significantly by the sediment origin (ANOSIM; R=0.518 and P=0.001), but not by the environment (R=−0.031 and P=0.85; Supplementary Figure S1a).
Figure 3.
NMDS (nonmetric multidimensional scaling) plots representing the Bray–Curtis similarity of general bacterial communities (on the basis of 16S rDNA T-RFLP profiles) in the sediments (a) at the end of the short-term experiment and (b) at the beginning (initial) and end (final) of the long-term experiment. The x-axis in both panels largely differentiates the communities by sediment origin. The y-axis in panel (b) differentiates the communities by the time of sampling (initial versus final samples).
Functional assays
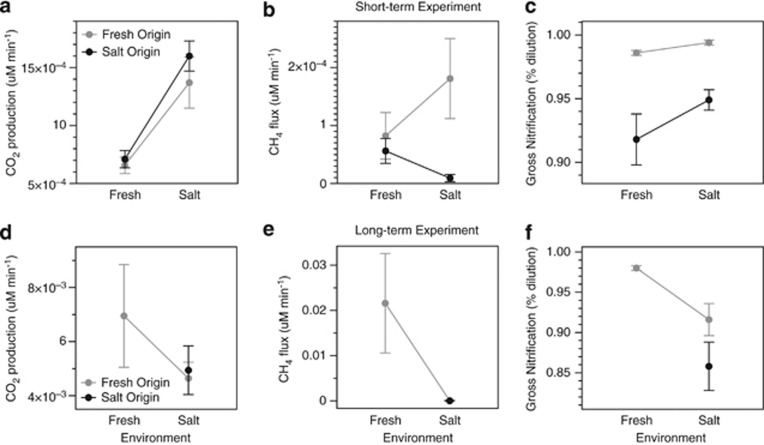
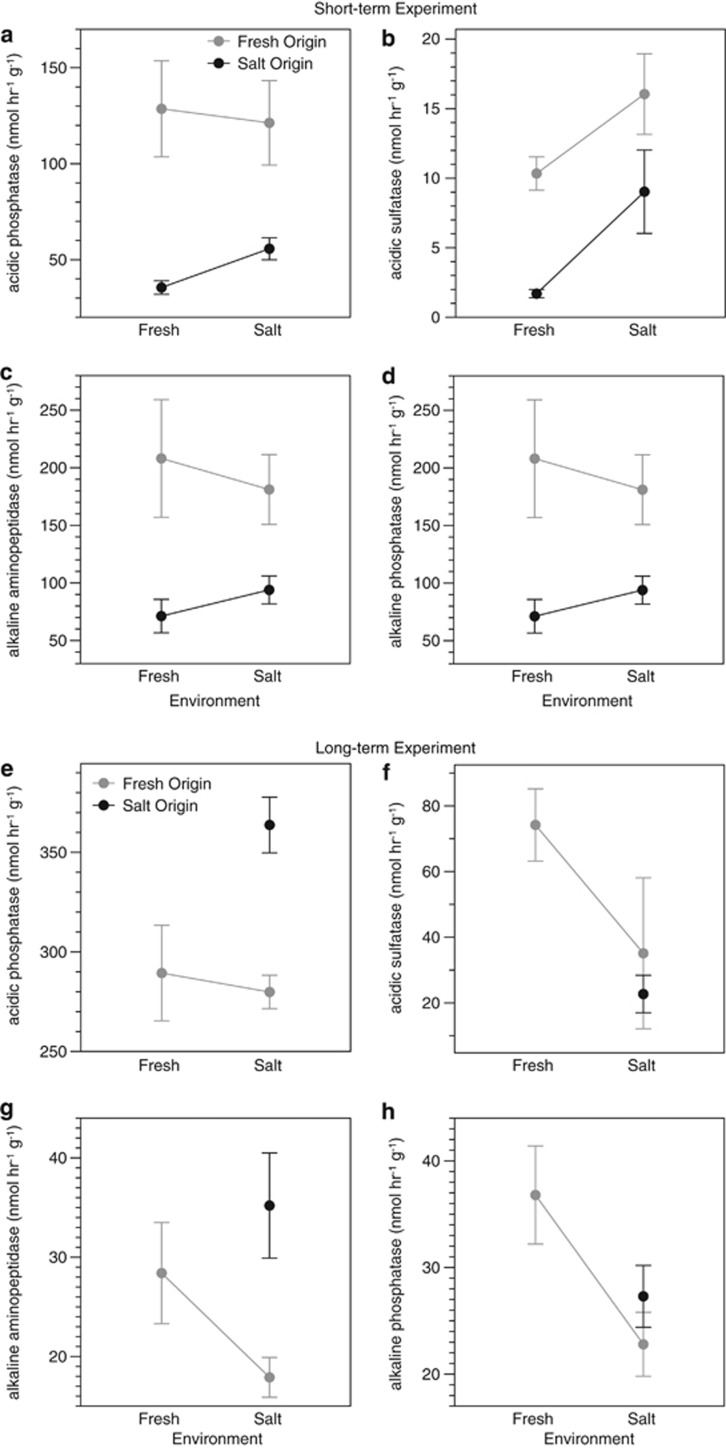
To test for the effect of these compositional differences on sediment functioning, we measured nine functional parameters on the sediments in each cage after the week-long experiment. A variety of the parameters depended on the origin of the sediment, even when controlling for the environment. Sediment origin had a significant main effect on nitrification rates and four of the six enzyme activities (Table 3; Figure 4c; Figure 5a and d). Further, sediment origin and environment interacted to affect methane production. The saltwater sediment produced significantly less methane in the saltwater than freshwater environment (Figure 4b), as expected from prior studies of methanogenesis in saltmarsh sediments (for example, Senior et al., 1982). Overall, sediment origin significantly affected six of the nine functional parameters, either directly or through an interactive effect with the environment. Given that bacterial composition differed greatly between the freshwater and saltwater sediments, these results are consistent with the hypothesis that sediment functioning is determined in large part by bacterial composition (Hypothesis 1).
Table 3. Effect of sediment origin and environment on sediment processes for the short- and long-term experiments.
|
Short-term |
Long-term |
||||
|---|---|---|---|---|---|
| Origin | Environment | Origin x Environment | Origin | Environment | |
| CO2 production | F1,60=2 | F1,60=17*** | F1,60=0.9 | F1,32=0.07 | F1,32=2 |
| CH4 flux | F1,58=2 | F1,58=0.1 | F1,58=4* | F1,31=2 | F1,31=28*** |
| Nitrification | F1,48=81*** | F1,48=9** | F1,48=0.09 | F1,29=4 | F1,29=6* |
| Acidic | |||||
| Aminopeptidase | F1,59=0.7 | F1,59=2 | F1,59=2 | F1,32=0.7 | F1,32=0.002 |
| Phosphatase | F1,60=36*** | F1,60=2 | F1,60=3 | F1,30=25*** | F1,30=0.03 |
| Sulfatase | F1,60=35*** | F1,60=8** | F1,60=2 | F1,32=2 | F1,32=14** |
| Alkaline | |||||
| Aminopeptidase | F1,54=36*** | F1,54=6** | F1,54=0.1 | F1,30=18** | F1,30=4* |
| Phosphatase | F1,60=11** | F1,60=0.2 | F1,60=1 | F1,32=1 | F1,32=6* |
| Sulfatase | F1,56=0.02 | F1,56=0.3 | F1,56=0.5 | F1,32=0.02 | F1,32=1.5 |
The F statistic and degrees of freedom are reported for the two-way ANOVA tests. Statistical significance is indicated by asterisks; *P⩽0.05, **P⩽0.005, and ***P⩽0.0001.
Figure 4.
Means (±1 s.e.) of net respiration, net methane production, and gross nitrification by sediment origin and incubating environment for the (a–c) short- and (d–f) long-term experiments. The gray lines connect the sediments originating from the freshwater environment, and the black lines connect the sediments originating from the saltwater environment.
Figure 5.
Means (±1 s.e.) of the activities of four enzymes by sediment origin and incubating environment for the (a–d) short- and (e–h) long-term experiments. Acidic aminopeptidase and alkaline sulfatase were not affected by sediment origin in either experiment and so are not shown. For ease of viewing, the gray lines connect the sediments originating from the freshwater environment, and the black lines connect those originating from the saltwater environment.
Not surprisingly, the environment also had a strong, direct influence on sediment functioning (Table 3). In particular, carbon dioxide (CO2) production was higher in saltwater than in freshwater (Figure 4a) and was not affected by sediment origin, either directly or through an interactive effect with the environment. The environment also directly affected nitrification rates and two of the enzyme activities (acidic sulfatase and alkaline aminopeptidase; Figure 4b and 4c).
To examine overall sediment functioning, we also calculated a standardized, Euclidean distance of all nine functional parameters between each pairwise combination of the sediment cores. This analysis upholds the results of the individual functional parameters that both origin and environment affect on sediment functioning (two-way ANOSIM; Origin: R=0.198, P=0.001; Env: R=0.224, P=0.001)(Supplementary Figure S2a).
Long-term experiment
In the second experiment, we assayed initial bacterial composition (just as the sediments were placed into the cages) and bacterial composition and sediment functioning after 2 months (61 days). Because of an unknown disturbance at the freshwater site, most of the saltwater sediment cores transplanted to this location were destroyed. However, we were still able to test our hypotheses with the sediment cores transplanted from the freshwater to the saltwater environment.
Abiotic conditions
After 2 months, salinity of the sediment cores equilibrated with the surrounding environment and did not depend on the core origin. Salinity of the sediments was 27 in saltwater environment, regardless of core origin (tdf=24=−0.56; P=0.58; Supplementary Figure S3a). In contrast, salinity in the cores from the freshwater sites was only 2.7. As in the short-term experiment, ammonium concentrations did not respond to the surrounding environment. Ammonium differed between the sediments of different origins incubating in the same saltwater site (tdf=22=−3.5, P=0.002) and was similar in the freshwater sediments regardless of the incubating site (tdf=6=0.37, P=0.72; Supplementary Figure S3b).
Bacterial composition
Bacterial composition changed over the course of the 2-month experiment. Composition in the sediment cores incubated in their native environment differed significantly from their initial composition (one-way ANOSIM: R=0.233; P=0.001; Figure 3b). These differences in initial and final bacterial composition may reflect an effect of the cages on sediment composition over time, temporal changes in the surrounding environment or a combination of both.
Incubating freshwater sediment cores in a saltwater environment also shifted bacterial composition significantly after 2 months, consistent with Hypothesis 2. At the end of the experiment, the environment had a much stronger affect on bacterial composition than it did in the short-term experiment (R=0.474, P=0.001 versus R=0.052, P=0.044), although sediment origin continued to have a significant effect on bacterial composition (Origin: R=0.988, P=0.001). Furthermore, bacterial composition in the transplanted freshwater sediments changed in the manner expected; they became more similar to the composition of the native saltwater cores. Although sediment origin continues to have a strong effect on bacterial composition, the transplanted freshwater sediments (open orange circles) are more similar to the native saltwater communities (blue triangles) than are the native freshwater sediments (solid red circles) (Figure 3b).
In contrast, we did not detect a significant change in the composition of the SRB. SRB composition differed by sediment origin, but not by the incubating environment (Origin: R=0.788, P=0.001; Env.: R=0.068, P=0.339; Supplementary Figure S1b). Thus, the freshwater sulfate reducing community appears resistant to change even after 2 months of incubation in the saltwater environment.
Functional assays
In support of our final hypothesis (Hypothesis 3), sediment origin had less of an influence on the functional assays in the long-term experiment than in the short-term experiment. Sediment origin directly affected only two of the nine functional assays (acidic phosphatase and alkaline aminopeptidase), compared with five in the short-term experiment (Table 3; Figures 4 and 5). Also as hypothesized, the surrounding environment had less of an influence in the long-term experiment; environment had a direct effect on five of the nine assays, compared with two in the short-term experiment (Table 3; Figures 4 and 5). Because of the loss of the sediment cores transplanted from the saltwater to freshwater environment, we were not able to test for the interactive effects of sediment origin and the environment on the functional assays.
Both sediment origin and environment influenced overall functioning in the sediment cages (Origin: R=0.201, P=0.002; Env.: R=0.442, P=0.016). Similar to the shift observed in bacterial composition, the functional profile of the sediment cores transplanted from freshwater to saltwater was intermediate to those native to either environment (Supplementary Figure S2b), such that the transplanted freshwater sediments were on average more similar to those of the native saltwater sediments than of the native freshwater sediments. Further, overall function was correlated to bacterial composition. Within the saltwater environment, pairwise functional distance and 16S compositional similarity were correlated, suggesting that more similar bacterial communities led to more similar sediment functional processes (RELATE test; Spearman's ρ=0.285, P=0.001).
Discussion
The results of the two transplant experiments support all three of our hypotheses, indicating that microbial composition affects functioning in these estuarine sediments. Several lines of evidence are particularly revealing. First, we observed significant effects of sediment origin in both the short- and long-term experiments. By itself, the origin effect in the short-term experiment is not definitive evidence that the two communities are functionally different. Salinity changed quickly, but did not completely equilibrate in the transplanted sediments after 1 week; thus, the origin effect might have reflected abiotic differences. In addition, the short-term origin effect might be ambiguous, if the transplanted microorganisms did not have enough time to acclimate to the new conditions. If this was the case, then with more time, the communities might have converged functionally despite their compositional differences (Comte and del Giorgio, 2011). However, the results of the long-term experiment suggest that these possibilities cannot entirely explain the origin effect in the short-term experiment. Even after 6 more weeks for the environment to permeate the cages and the microorganisms to acclimate, an origin effect persisted, providing additional evidence that the remaining differences in microbial composition were responsible for the differences in sediment functioning.
Second, as the transplanted sediment composition shifted in the long-term experiments, we observed a significant correlation between overall bacterial composition and sediment functioning. Such a correlation also supports the idea that bacterial composition has a direct influence on sediment processes (Strickland et al., 2009). At the same time, we did not observe a shift in SRB composition. This result was unexpected, given that sulfate reduction is generally greater in salt marshes than in tidal freshwater habitats (Odum, 1988). At least three reasons may account for this result. First, SRB may be more plastic to salinity changes than expected, such that SRB taxa can function equally well in both these locations. Second, SRB abundance (which we did not assay) may have increased in the saltier environment, even though the relative abundance among taxa (their composition) did not change. Finally, the genetic assay used (the particular primers and/or the fingerprinting method) may have missed finer-scale changes in SRB composition (for example, Santillano et al., 2010).
Third, although salinity in the transplanted cages equilibrated with the surrounding environment after 7 weeks, ammonium concentration did not. These counter results indicate that although the surrounding waters had permeated through the cages, sediment processes internal to the cages directly influenced ammonium concentrations. Microorganisms drive most processes in the nitrogen cycle, and previous studies provide evidence that their particular composition influences these processes rates (Cavigelli and Robertson, 2000, Avrahami et al., 2002; Balser and Firestone, 2005; Bernhard et al., 2007a). A related result is that sediment origin (either directly or interactively with environment) affected nitrification and methane flux, but not CO2 production. This outcome is consistent with Schimel's (1995) the hypothesis that microbial composition is more likely to influence ‘narrow' rather than ‘broad' processes. Narrow processes are defined as those carried out by relatively phylogenetically restricted groups of microorganisms. Taxa within these narrow groups may respond similarly to a new environment and therefore as their abundance changes, the processes these organisms carry out will likely change as well. In contrast, broad processes like respiration are performed by many taxa among a variety of phylogenetic groups and thus, even dramatic shifts in microbial composition may maintain similar respiration rates. For example, Balser and Firestone (2005) observed that microbial composition was correlated with soil nitrification rates but not soil respiration. Similarly, Dimitriu et al. (2010) found that the microbial community had a stronger effect on the activity of phenol-oxidase than that of more widely produced enzymes.
Of course, the main challenge of field transplant experiments is to prevent migration and control for all environmental parameters that might affect functional processes; only then can an origin effect be attributed entirely to differences in microbial composition. The cages probably prevented most migration of bacterial cells, but not all, as revealed by initial lab tests. Given the high cell densities in estuarine sediments, it seems unlikely that a few immigrants would grow to dominance in the cages after only 7 weeks, but we cannot be sure. Further, while we purposely compared communities along the same river to try to control for substrate, we were not entirely successful; most metrics were similar, but at least at the beginning of the experiment, carbon and nitrogen content differed slightly in sediments from the two locations. In fact, even if we did not detect any differences in the sediments, we could not exclude the possibility that an unmeasured abiotic variable was in part responsible for the effect of sediment origin. Thus, while every effort should be made to minimize this limitation, whole-community transplant studies will always require a caveat about potentially uncontrolled, abiotic variables.
In sum, this work supports previous correlative studies (for example, Bertilsson et al., 2007; Bernhard et al., 2007b; Comte and del Giorgio, 2010) and lab experiments (for example, Bell et al., 2005; Strickland et al., 2009), suggesting that microbial composition influences some ecosystem processes. To our knowledge, however, this study is only one of a few that directly demonstrates the effect of microbial composition on functioning in the field while controlling for microbial migration (Allison et al., in press; Gasol et al., 2002). Such experiments would also be useful to investigate further details of the relationship between microbial composition and ecosystem functioning. Indeed, using other approaches, many studies have already started to address when, where and why microbial composition is related to ecosystem functioning. For instance, a key to predicting the response of ecosystems to environmental change is to disentangle the importance of shifts in microbial composition versus the physiological plasticity of that community (Schimel et al., 2007; Singh et al., 2010; Comte and del Giorgio, 2011). Generally, ecosystem models assume that process rates—even when directly carried out by microorganisms—are determined by environmental conditions (Schimel and Gulledge, 1998), implicitly assuming that microbial communities are so physiological plastic that any differences in their composition are unimportant. Future transplant experiments might also consider more detailed temporal dynamics of microbial composition and functioning over time (for example, Berga et al., 2012). Such a design could test whether microbial composition influences the resilience of ecosystem processes to environmental change under field conditions.
Acknowledgments
We thank Caryl Ann Becerra, Teresa Conneely, Vicente Gomez-Alvarez, Marie Hennelly, Melissa Lage, Jay Lennon, Faye Lemieux, Adam Martiny, Kristin Matulich, Devon Mohamed, Klaus Nüsslein, Nicole Shapiro and Claudia Weihe for discussions, paper comments, and field and lab assistance. This research was supported by the National Science Foundation (MCB-0701494) and the Gordon and Betty Moore Foundation.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abdo Z, Schüette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- Allison SD, Lu Y, Weihe C, Goulden ML, Martiny AC, Treseder KK, et al. in pressMicrobial abundance and composition influence litter decomposition response to environmental change Ecology ( http://dx.doi.org/10.1890/12-1243.1 ). [DOI] [PubMed]
- Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin AT, Sala OE, Jackson RB. Inhibition of nitrification alters carbon turnover in the patagonian steppe. Ecosystems. 2007;9:1257–1265. [Google Scholar]
- Avrahami S, Conrad R, Braker G. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl Environ Microbiol. 2002;68:5685–5692. doi: 10.1128/AEM.68.11.5685-5692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balser TC, Firestone MK. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry. 2005;73:395–415. [Google Scholar]
- Bell T, Newman J, Silverman B, Turner S, Lilley A. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- Berga M, Székely AJ, Langenheder S. Effects of disturbance intensity and frequency on bacterial community composition and function. PLoS One. 2012;7:e36959. doi: 10.1371/journal.pone.0036959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard A, Tucker J, Giblin A, Stahl D. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol. 2007a;9:1439–1447. doi: 10.1111/j.1462-2920.2007.01260.x. [DOI] [PubMed] [Google Scholar]
- Bernhard AE, Tucker J, Giblin AE, Stahl DA. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol. 2007b;9:1439–1447. doi: 10.1111/j.1462-2920.2007.01260.x. [DOI] [PubMed] [Google Scholar]
- Bertilsson S, Eiler A, Nordqvist A, Jørgensen NOG. Links between bacterial production, amino-acid utilization and community composition in productive lakes. ISME J. 2007;1:532–544. doi: 10.1038/ismej.2007.64. [DOI] [PubMed] [Google Scholar]
- Bottomley P, Taylor A, Boyle S, Mcmahon S, Rich J, Cromack K, et al. Responses of nitrification and ammonia-oxidizing bacteria to reciprocal transfers of soil between adjacent coniferous forest and meadow vegetation in the Cascade Mountains of Oregon. Microb Ecol. 2004;48:500–508. doi: 10.1007/s00248-004-0215-3. [DOI] [PubMed] [Google Scholar]
- Cavigelli MA, Robertson GP. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology. 2000;81:1402–1414. [Google Scholar]
- Clarke KR, Warwick RM.2001Change in Marine Communities: An Approach to Statistical Analysis and Interpretation2nd edn.PRIMER-E: Plymouth, UK [Google Scholar]
- Comte J, del Giorgio PA. Linking the patterns of change in composition and function in bacterioplankton successions along environmental gradients. Ecology. 2010;91:1466–1476. doi: 10.1890/09-0848.1. [DOI] [PubMed] [Google Scholar]
- Comte J, del Giorgio PA. Composition influences the pathway but not the outcome of the metabolic response of bacterioplankton to resource shifts. PLoS One. 2011;6:e25266. doi: 10.1371/journal.pone.0025266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain CM. Shifting nutrient limitation and eutrophication effects in marsh vegetation across estuarine salinity gradients. Estuaries Coasts. 2007;30:26–34. [Google Scholar]
- Daleo P, Fanjul E, Casariego AM, Silliman BR, Bertness MD, Iribarne O. Ecosystem engineers activate mycorrhizal mutualism in salt marshes. Ecol Lett. 2007;10:902–908. doi: 10.1111/j.1461-0248.2007.01082.x. [DOI] [PubMed] [Google Scholar]
- Dantas G, Sommer MOA, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- Dimitriu PA, Lee D, Grayston SJ. An evaluation of the functional significance of peat microorganisms using a reciprocal transplant approach. Soil Biol Biochem. 2010;42:65–71. [Google Scholar]
- Doane TA, Horwath WR. Spectrophotometric determination of nitrate with a single reagent. Anal Lett. 2003;36:2713–2722. [Google Scholar]
- Elliott ET, Heil JW, Kelly EF, Monger HC.1999Soil structural and other physical propertiesIn: Robertson GP, Coleman DC, (eds).Standard Soil Methods for Long-Term Ecological Research Oxford University Press: New York, NY; 74–87. [Google Scholar]
- Ewanchuck PJ, Bertness MD. Structure and organization of a northern New England salt marsh plant community. J Ecol. 2004;92:72–85. [Google Scholar]
- Franklin RB, Mills AL. Structural and functional responses of a sewage microbial community to dilution-induced reductions in diversity. Microb Ecol. 2006;52:280–288. doi: 10.1007/s00248-006-9033-0. [DOI] [PubMed] [Google Scholar]
- Freitag TE, Chang L, Prosser JI. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol. 2006;2006:684–696. doi: 10.1111/j.1462-2920.2005.00947.x. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. P Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol JM, Comerma M, Garcia JC, Armengol J, Casamayor EO, Kojecka P, et al. A transplant experiment to identify the factors controlling bacterial abundance, activity, production, and community composition in a eutrophic canyon-shaped reservoir. Limnol Oceanogr. 2002;47:62–77. [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths B, Ritz K, Wheatley R, Kuan H, Boag B, Christensen S, et al. An examination of the biodiversity-ecosystem function relationship in arable soil microbial communities. Soil Biol Biochem. 2001;33:1713–1722. [Google Scholar]
- Hanson CA, Horner-Devine MC, Fuhrman JA, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Hart SC, Stark JM, Davidson EA, Firestone MK.1994Nitrogen mineralization, immobilization, and nitrificationIn: Weaver RW, Angle JS, Bottomley PS, (eds).Methods of Soil Analysis. Part 2. Microbiology and Biochemical Properties Soil Science Society of America: Madison: Wisconsin; 985–1018. [Google Scholar]
- Hartnett D, Wilson G. Mycorrhizae influence plant community structure and diversity in tallgrass prairie. Ecology. 1999;80:1187–1195. [Google Scholar]
- Hawkins RJ, Purdy KJ. Genotypic distribution of an indigenous model microorganism along an estuarine gradient. FEMS Microbiol Ecol. 2007;62:187–194. doi: 10.1111/j.1574-6941.2007.00376.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M, (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons: Chichester, England; 115–147. [Google Scholar]
- Langenheder S, Lindstrom E, Tranvik L. Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol. 2006;72:212–220. doi: 10.1128/AEM.72.1.212-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Lindstrom ES, Tranvik LJ. Weak coupling between community composition and functioning of aquatic bacteria. Limnol Oceanogr. 2005;50:957–967. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Mohamed DJ, Martiny JBH. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2010;5:379–388. doi: 10.1038/ismej.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odum WE. Comparative ecology of tidal fresh-water and salt marshes. Annu Rev Ecol Syst. 1988;19:147–176. [Google Scholar]
- Power ME. Effects of fish in river food webs. Science. 1990;250:811–814. doi: 10.1126/science.250.4982.811. [DOI] [PubMed] [Google Scholar]
- Reed HE, Martiny JBH. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol. 2007;62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- Saiya-Cork K, Sinsabaugh R, Zak D. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem. 2002;34:1309–1315. [Google Scholar]
- Santillano D, Boetius A, Ramette A. Improved dsrA-based terminal restriction fragment length polymorphism analysis of sulfate-reducing bacteria. Appl Environ Microbiol. 2010;76:5308–5311. doi: 10.1128/AEM.03004-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimel J, Balser TC, Wallenstein M. Microbiol stress-response physiology and its implications for ecosystem function. Ecology. 2007;88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- Schimel J.1995Ecosystem consequences of microbial diversity and community structureIn: Chapin FS, Koerner C, (eds).Arctic and Alpine Biodiversity: Patterns, Causes, and Ecosystem Consequences Springer Verlag: New York; 239–254. [Google Scholar]
- Schimel JP, Gulledge J. Microbial community structure and global trace gases. Global Change Biol. 1998;4:745–758. [Google Scholar]
- Senior E, Lindström EB, Banat IM, Nedwell DB. Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the east coast of the United Kingdom. Appl Environ Microbiol. 1982;43:987–996. doi: 10.1128/aem.43.5.987-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- Tilman D, Wedin D, Knops J. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature. 1996;379:718–720. [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- Wagner M, Roger A, Flax J, Brusseau G, DA S. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop MP, Firestone MK. Response of microbial community composition and function to soil climate change. Microb Ecol. 2006;52:716–724. doi: 10.1007/s00248-006-9103-3. [DOI] [PubMed] [Google Scholar]
- Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Freitag T, et al. Maintenance of soil functioning following erosion of microbial diversity. Environ Microbiol. 2006;8:2162–2169. doi: 10.1111/j.1462-2920.2006.01098.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.