Abstract
The human gastrointestinal (GI) tract microbiota acts like a virtual organ and is suggested to be of great importance in human energy balance and weight control. This study included 40 monozygotic (MZ) twin pairs to investigate the influence of the human genotype on GI microbiota structure as well as microbial signatures for differences in body mass index (BMI). Phylogenetic microarraying based on 16S rRNA genes demonstrated that MZ twins have more similar microbiotas compared with unrelated subjects (P<0.001), which allowed the identification of 35 genus-like microbial groups that are more conserved between MZ twins. Half of the twin pairs were selected on discordance in terms of BMI, which revealed an inverse correlation between Clostridium cluster IV diversity and BMI. Furthermore, relatives of Eubacterium ventriosum and Roseburia intestinalis were positively correlated to BMI differences, and relatives of Oscillospira guillermondii were negatively correlated to BMI differences. Lower BMI was associated with a more abundant network of primary fiber degraders, while a network of butyrate producers was more prominent in subjects with higher BMI. Combined with higher butyrate and valerate contents in the fecal matter of higher BMI subjects, the difference in microbial networks suggests a shift in fermentation patterns at the end of the colon, which could affect human energy homeostasis.
Keywords: BMI signatures, discordant BMI, gastrointestinal tract, microbiota, monozygotic twins
Introduction
The microbial cells inside the human gastrointestinal (GI) tract are collectively called the GI microbiota and provide an extensive genetic function counterpart to the host genome (Savage, 1977; Albert et al., 1980; Adlercreutz et al., 1984; Ramotar et al., 1984; Cummings and Macfarlane, 1997; Cebra, 1999; Metges, 2000; Shanahan 2002; Begley et al., 2005; O'Hara et al., 2006; Wei and Brent, 2006; Yang et al., 2009). Previous studies have shown that GI microbiota is host-specific and GI tract region-specific (Zoetendal et al., 2004; Rajilic-Stojanovic et al., 2009; Jalanka-Tuovinen et al., 2011), aberrant in composition and stability in patients suffering from GI disorders such as Crohn's disease (Seksik et al., 2003), and associated to host energy homeostasis (Backhed et al., 2004; Ley et al., 2006; Turnbaugh et al., 2006; Backhed et al., 2007; Samuel et al., 2008). Analysis of global fecal microbiomes introduced the concept that human GI microbiota appeared to have three distinct structural biome-types called enterotypes (Arumugam et al., 2011). Although the enterotype distinction did not appear to be correlated to health status or host demography, recent 16S rRNA-based studies by Wu et al. (2011) and by Huse et al. (2012) suggest that the distinctive biomes among the human GI microbiota appear to be more like a continuum with gradients of the main enterotype driving taxa, which could be driven by long-term dietary habits.
Human energy homeostasis varies greatly between persons but monozygotic (MZ) twins show more resemblance in the variations of their energy balance (Bouchard et al., 1990). Furthermore, current literature putatively links a large number of human genes to variations in body mass index (BMI). Recruitment of MZ twins discordant for BMI in studies exploring human transcript profiles has revealed several potential obesity marker-genes (Naukkarinen et al., 2010) and potential (mitochondrial) pathways that are associated with major BMI increases (Pietilainen et al., 2008). However, the identified human genes account for a relatively small amount of the observed variance in energy homeostasis (Saris and Tarnopolsky, 2003; Heid et al., 2011). Recent findings suggest that the GI microbiota is important for the energy and metabolic homeostasis of its host. In mice, clear links have been observed between energy homeostasis and GI microbiota, for instance: the resistance to obesity development of Germ-free mice (Backhed et al., 2007), stimulation of weight gain by GI tract colonization (Samuel et al., 2008), interaction between GI microbiota and fatty acid storage mechanisms (Backhed et al., 2004; Backhed et al., 2007), variations between genetic obese (ob/ob) and lean mice in the relative abundances of the bacterial phyla Bacteroidetes, Firmicutes and Actinobacteria (Ley et al., 2005; Turnbaugh et al., 2006).
In contrast to the associations found in mice, studies on the relation between human energy homeostasis and GI microbiota have generated conflicting results. Analogous to results obtained in mice, Ley et al. (2006) detected fewer Bacteroidetes and more Firmicutes in obese subjects compared with lean controls (Ley et al., 2006). Moreover, this study also revealed that the relative abundances of Bacteroidetes increased while Firmicutes decreased when the subjects decreased their BMI by following either a fat restricted or carbohydrate restricted diet (Ley et al., 2006). Although Duncan et al. (2008) confirmed a significant decrease in Firmicutes when subjects followed a low-carbohydrate weight-loss diet (Duncan et al., 2008), several other human studies did not confirm these differences in Bacteroidetes to Firmicutes (B:F) ratio (Duncan et al., 2008; Zhang et al., 2009; Schwiertz et al., 2010). Schwiertz et al. (2010) even concluded that relative abundance of Firmicutes was reduced in obese subjects (Schwiertz et al., 2010), but they also reported that higher levels of SCFAs (short chain fatty acids) were present in fecal material of obese subjects compared with lean controls, possibly suggesting that the amount of SCFAs produced is a more prominent determinant of the BMI status than the phylogenic distributions of the microbiota (Schwiertz et al., 2010). The conflicting results in these studies may be explained by the heterogeneity among human subjects, with respect to their genotype and lifestyle as well as specificity of an individual's microbiota. Furthermore, these studies have compared subjects on the opposite extremes of the BMI scale (lean and obese), while the microbiota is exposed to fundamentally different ‘environmental' factors in both states that go beyond BMI alone, such as diet, host metabolic and hormonal factors (Manson et al., 1995; Stevens et al., 1998), and low-grade systemic inflammation (Visser et al., 1999).
Results of mice studies often lead to the hypothesis that the mammalian host-genotype, in particular factors for the immune system phenotype, has a huge impact on the GI microbiota characteristics (Benson et al., 2010). Genotype, however, is not determined so far for human subjects participating in microbiota studies. Consequently, these human studies do not take the host-genotype into account as a determinant of the phenotype, while the genotype is known to be heterogeneous among modern human populations.
Genotypic influences could be minimized by evaluating phenotypic variations in MZ twins allowing pair-wise comparisons within a fixed genotype. An early study of separately living MZ twins and their marital partners revealed that for co-twins the within-pair microbiota similarity is significantly higher compared with unrelated individuals, while the microbiota similarity for married couples is not significantly higher compared with unrelated individuals (Zoetendal et al., 2001). Similar observations were reported in later studies (Stewart et al., 2005; Turnbaugh et al., 2009). Furthermore, differences in GI microbiota could be related to disease phenotype by comparing MZ twin pairs concordant and discordant for inflammatory bowel diseases (Dicksved et al., 2008; Willing et al., 2010; Lepage et al., 2011). A study by Turnbaugh et al. (2009) on a cohort of obese and lean adult females, MZ and dizygotic twin pairs, demonstrated that the microbiota in obese pairs was reduced in bacterial diversity and contains an altered representation of bacterial genes and metabolic pathways compared with lean pairs (Turnbaugh et al., 2009). This study, however, only included twins with concordant phenotypes in terms of BMI, again making both host genetics and absolute BMI values confounding variables between the groups of subjects. To identify specifically microbiota signatures of BMI, we compared the microbiota composition in MZ twin pairs that are concordant and discordant in BMI. This enabled us to achieve our main objective, which was to define microbiota signatures that correlate directly with BMI differences independent of the host genotype and absolute BMI values.
Materials and methods
Subjects, sample size and sampling
This study was approved by the METC of Wageningen University. A selection of MZ twin pairs was contacted from the East Flanders Prospective Twin Study, which presently has over 7.000 twins. Subjects who used medication that may affect the GI microbiota, prebiotics or probiotics within 1 month before sampling were excluded. Subjects with pre-existing bowel diseases and subjects that were pregnant or breast feeding were excluded as well. Since there are no previous HITChip studies on MZ twins, the power calculation was based on intra-individual microbiota variation observed in healthy subjects (Jalanka-Tuovinen et al., 2011), which we expect to be close to that within paired MZ twin microbiotas. Based on the expectation that we will have at least a proportional difference of 0.18 (s.d. 0.04), we calculated that 19 pairs per group would suffice to address our objective (assuming α=0.05 and power 1−β=0.20). Therefore, our cohort consisted of 20 twin pairs of varying genders and ages (>18 years) that were previously recorded to have a BMI difference of more than 5, and 20 age- and gender-matched control MZ twin pairs with no significant difference in BMI. Subjects were able to understand the written study information and signed an informed consent. Fecal samplings, bodyweight and length measurements were collected from these volunteers (Supplementary Table S1). Furthermore, the volunteers filled in a questionnaire concerning changes in their dietary habits, medication and GI symptoms of the last 4 weeks prior to sampling (Supplementary Table S1). Fecal samples were collected at the volunteers home, frozen immediately, and transported on dry ice to the laboratory where they were kept at −80° until further analysis.
Twin pair classification into BMI phenotypes
No subjects were underweight (BMI<18.5). Only two twin pairs and one sibling of a third pair were obese (BMI>30). To define which twin pairs were discordant in terms of weight maintenance the recommendation by Stevens et al. (2006) was used (Stevens et al., 2006), which in a maximum BMI difference for within-pair concordance of 1.35 kg m−2 and a minimum BMI difference for within-pair discordance of 2.7 kg m−2 (for details see SI Materials and methods). This definition ensures that twins classified as discordant truly have different phenotypes. Discordant twins were subdivided into two groups based on their relative weight compared with their own sibling. This resulted in a ‘lower BMI' group for the leaner siblings and a ‘higher BMI' group for the heavier siblings. At the moments of sampling 18 pairs were discordant, 16 were concordant and 6 occupied the gray area between our definitions of concordance and discordance (indistinct twins).
DNA extraction, microarray hybridization and data extraction
DNA was extracted utilizing the repeated bead beating protocol (Salonen et al., 2010). Fecal microbial diversity and composition was studied in detail using the Human Intestinal Tract chip (HITChip) as described previously (Rajilic-Stojanovic et al., 2009) (for further details see SI Materials and methods). This phylogenetic microarray has been shown to be a powerful tool for deep GI tract microbiota composition analysis and has been benchmarked against several classical 16S rRNA gene-based methodologies, such as qPCR, fluorescence in situ hybridization and 454 pyrosequencing (Booijink et al., 2007; Claesson et al., 2009; Rajilic-Stojanovic et al., 2009; van den Bogert et al., 2011) as well as metagenomics (Arumugam et al., 2011). HITChip probes are assigned to three phylogenetic levels: level 1, defined as order-like 16S rRNA gene sequence groups; level 2, defined as genus-like 16S rRNA gene sequence groups (sequence similarity >90%); and level 3, phylotype-like 16S rRNA gene sequence groups (sequence similarity >98%) (Rajilic-Stojanovic et al., 2009).
Organic acid and SCFA concentration measurement
To determine metabolic profiles, fecal samples were diluted in deionized water to a 10% (w/v) concentration and subsequent high-performance liquid chromatography (HPLC) analysis was performed as described previously (Stams et al., 1993) to determine citrate, malate, succinate, lacate, fumarate, formate, acetate, propionate, iso-butrate, butyrate and valerate concentrations. The HPLC system was equipped with a Shodex S1821 column and temperature was set to 70 °C.
Data analysis and statistical methods
For the total microbiota normalized signal values of all unique HITChip probes were used to calculate Simpson's Diversity index for each sample and the Spearman's correlation coefficient between different samples. Unique probe signal values per level 1 and per level 2 group (with >20 probes) group were used to calculate diversity and similarities for the groups of each phylogenetic level separately. Spearman's correlation coefficients between random unrelated subjects within this cohort were compared with Spearman's correlation coefficients within twin pairs by a Student's t-test with unequal groups.
For each sample, relative abundances were calculated for the groups of each specificity level by summing all signal values of the probes targeting a group and dividing by the total of all probe signals for the corresponding sample.
All comparisons between the discordant twin groups were pair-wise and significance were assessed with dependent two-group Wilcoxon signed rank tests. For all statistical tests that were performed on multiple parameters, the obtained P-values were adjusted by a Bonferroni correction. All P-values noted in text are adjusted P-values with P<0.05 being regarded as significant.
Results
MZ twins have highly similar microbiotas
A host-genotype controlled setup for this study was realized by recruiting MZ twins. Twin pairs enrolled in this study were contacted from the East Flanders Prospective Twin Study (Flanders, Belgium) (Derom et al., 2006). A total of 40 MZ twin pairs volunteered to donate fecal material from which DNA was extracted for microbiota composition analysis with phylogenetic microarray the HITChip (Rajilic-Stojanovic et al., 2009). This twin cohort consisted of 11 male pairs (age 19–43 years, BMI 18.5–34.7 kg m−2) and 29 female pairs (age 20–43 years, BMI 20.2–34.7 kg m−2). This selection consisted of 20 twin pairs of varying genders and ages that were previously recorded to have a BMI difference of more than 5 units (Supplementary Table S1). In addition, 20 age- and gender-matched control twin pairs with no significant difference in BMI were selected from the twin cohort (Supplementary Table S1).
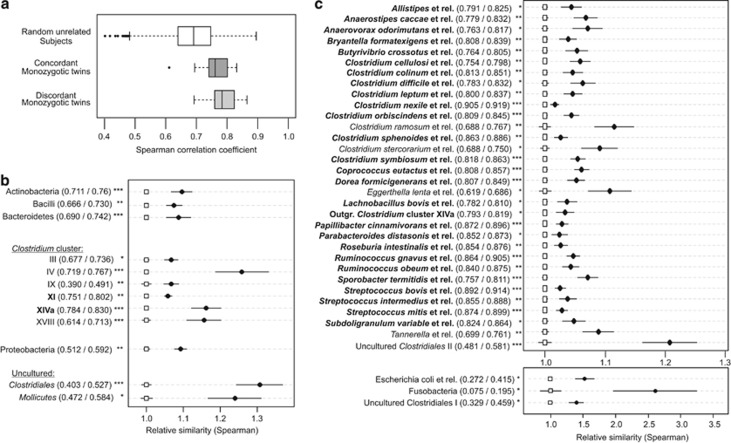
Similarity of the HITChip profiles between all subjects was calculated to determine the influence of the human genotype influence on the GI microbiota composition. Both, co-twins concordant (ΔBMI <1.35, n=16) and discordant (ΔBMI >2.7, n=18) for BMI showed a significantly higher similarity of their GI microbiota profile compared with random-paired subjects (P<0.001, Figure 1a). Similarity coefficients between co-twins did not significantly correlate to age or the length of time that the twins had been living separately, corroborating the previously reported impact of the host-genotypic factors on the microbiota composition (Zoetendal et al., 2001; Stewart et al., 2005; Turnbaugh et al., 2009).
Figure 1.
GI microbiota similarity in MZ twins. (a) Box-whisker plots of total microbiota profile similarity. Spearman's correlation coefficient was calculated for random unrelated subjects and between MZ twins. Average microbiota similarity between twins concordant for BMI and twins discordant for BMI are both significantly higher than between unrelated subjects (P=1 e−4, P=1 e−7, respectively). Dot-plots are shown of the mean order-like (b) similarity and the mean genus-like (c) similarity between random-paired subjects and MZ twins. Order-like and genus-like groups that were significantly different in similarity index are presented. The similarity indices of all order-like and genus-like groups are represented in Supplementary Figures S1 and S2, respectively. Mean within-pair Spearman's correlation coefficient values (depicted with black dots) and are relative to the mean Spearman's correlation coefficient values of random unrelated subjects within this cohort (depicted with open squares), for each phylogenetic group. Order-like and genus-like groups depicted in bold have within-pair similarities that are significantly higher than the total microbiota similarity. The plot-labels describe the actual mean Spearman's correlation coefficient value (unrelated subjects/MZ twins). Error bars represent 95% confidence intervals around the respective mean values. Asterisks indicate the level of significance of the corrected P-value: *P<0.05, **P<0.01, ***P<0.001.
To determine which microbial groups contributed the most to the high within-pair similarity, similarity indices between all subjects were calculated for all microbial subgroups. These subgroups can be defined at different phylogenetic levels as described previously (Rajilic-Stojanovic et al., 2009). Within-pair microbiota profile similarity of each phylogenetic subgroup was higher compared with random-paired subjects (Supplementary Figures S1 and S2), although this difference was not significant for every subgroup. The co-twins showed a significantly higher within-pair similarity compared with random-paired subjects for 12 order-like (level 1) and 35 genus-like (level 2) groups (Figures 1b and c). Moreover, for Clostridium clusters XI and XIVa and for 27 genus-like groups the within-pair similarities were even significantly higher than the total microbiota similarity (Figures 1b and c; Supplementary Table S2). A high similarity index within a bacterial group means that the probe signals occur in (nearly) the same ratios relatively to one another, implying that the presence and ratios of specific bacteria belonging to certain groups are highly conserved between the subjects. Hence, these structurally conserved bacterial groups within MZ twin pairs acknowledge the existence of a structural core in the human GI microbiota, which correlates with host genetics and shared (early life) environmental exposures and therefore can be considered as ‘imprinted structural cores'.
Next to an imprinted structural core the existence of a general core based on phylotype abundance was investigated. For HITChip data phylotype-like groups are defined as 16S rRNA gene groups with a sequence similarity of >98%. Above the array, background-specific signals of 96 phylotype-like were found in all subjects (100% prevalence). These phylotype-like groups, which comprise the general core in this twin cohort, are shown in Supplementary Table S3 and Supplementary Figure S3. In this twin cohort, the general core accounts on average for 34.7% (s.d. 12.8%) of the total microbiota. However, the abundance of this general core varies greatly between the subjects (from 10.6% for subject 10A to 81.5% for subject 26A), indicating enormous subject specificity at phylotype level in the GI tract.
Clostridium cluster IV is less diverse in higher BMI sibling group
To evaluate if microbial groups are associated to BMI and host genetic traits, microbiota composition was compared between twins that are concordant and discordant for BMI. At the moment of sampling, the BMI difference for 15 of the selected discordant twins was less than the 5 units recorded previously. Six twin pairs were between the ΔBMI thresholds used to determine concordance and discordance (1.35>ΔBMI >2.7) and were not taken into account. Each discordant twin pair was split up into two groups: the sibling with the lowest BMI of each pair was placed in the ‘lower BMI siblings' group and the sibling with the highest BMI of each pair was placed in the ‘higher BMI siblings' group. At the highest phylogenetic levels, no differences were observed between the two discordant twin pair groups. No consistent Bacteroides:Firmicutes (B:F) ratio differences were observed in pair-wise comparison of lower- and higher-BMI siblings (Supplementary Figure S4). Similarly, B:F ratios did not correlate with absolute BMI values. Moreover, these pair-wise comparisons did not reveal consistent differences between the total microbial diversity (inverse Simpson's index of diversity), which appeared to be hugely variable and between pairs the diversity differences ranged from 1.8 (twin pair 2; Supplementary Table S1) to 149.0 (twin pair 30; Supplementary Table S1). This variability indicates that the total microbiota diversity does not relate to the phenotypic differences in this twin cohort.
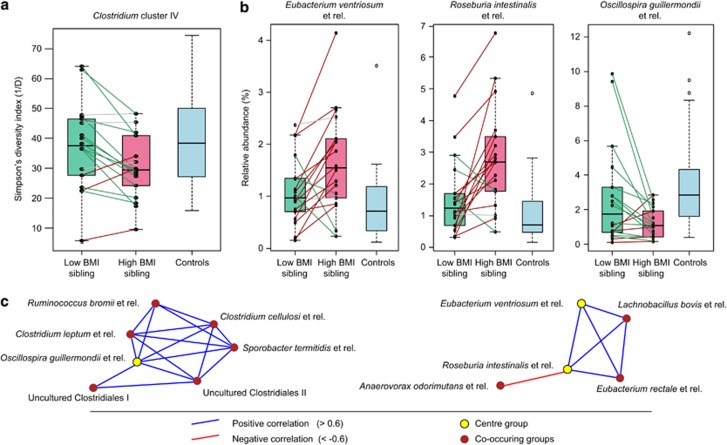
However, diversity at order-like (level 1) groups indicated that Clostridium cluster IV was significantly lower in diversity in the higher BMI siblings group (P=0.012), indicating that Clostridium cluster IV diversity decreases when BMI increases, independent of the absolute BMI value of the lower-BMI sibling (Figure 2a). Overall, the diversity of Clostridium cluster IV in the lower BMI siblings group is more comparable to the control group than the diversity in the higher BMI group.
Figure 2.
BMI phenotype influences on GI microbiota. (a) Box-whisker plots of inverse Simpson's index of diversity for Clostridium cluster IV in discordant and concordant twins. The results of each subject from the two discordant twins groups are visualized with black dots. Colored lines connect the dots of each twins pair. Gray lines indicate no change in diversity (or less than a factor of 0.1). A green line indicates that the diversity is higher in the lower BMI sibling, while a red line indicates a higher diversity in the higher BMI sibling. (b) Box-whisker plots of the relative abundances of the genus-like groups significantly differing in discordant twins: Eubacterium ventriosum et rel., Roseburia intestinalis et rel. and Oscillospira guillermondii et rel. The results of each subject from the two discordant twins groups are visualized with black dots. Colored lines connect the dots of each twins pair. Gray lines indicate no change in relative abundance (or less than a factor of 0.1). A green line indicates that the relative abundance is higher in the lower BMI sibling, while a red line indicates a higher relative abundance in the higher BMI sibling. (c) Co-occurrence networks in all subjects of the genus-like groups significantly differing in discordant twins. Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. (more abundant in higher BMI siblings) appear in a network of butyrate producers, and Oscillospira guillermondii et rel. (more abundant in lower BMI siblings) appears in a network of primary fiber degraders.
BMI phenotype correlates with signatures in genus-like microbial groups
To determine if quantitative differences in microbiota composition were consistently different between the higher BMI siblings and the lower BMI siblings groups given an identical genetic background, relative abundances of specific microbial groups were compared and contrasted. This revealed that the genus-like (level 2) groups Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. were significantly more abundant in the higher BMI siblings (P=0.014 and P=0.003, respectively; Figure 2b), while and Oscillospira guillermondii et rel. was significantly more abundant different in the lower BMI siblings (P=0.014; Figure 2b). These three genus-like (level 2) groups seem to co-occur in two distinct ecological networks with several other genus-like groups in all subjects when using a Spearman's correlation coefficient cutoff of >0.6 and <−0.6 (Figure 2c). Such ecological networks may visualize potential cooperation (mutualism or commensalism) and competition between the microbial groups. The first network, which is enriched in lower BMI sibling group, is centered around Oscillospira guillermondii et rel. and contains three other genus-like (level 2) groups which encompass isolates associated with degradation of plant material: Clostridium cellulosi et rel. Yanling et al. (1991), Ruminococcus bromii et rel. Klieve et al. (2007); Kovatcheva-Datchary et al. (2009) and Sporobacter termitidis et rel. Grech-Mora et al. (1996). Therefore, this network seems to be specialized in degradation of complex fibers, marking this network as a primary degrader network. Degradation of complex fibers can yield fermentation products, such as partially degraded oligosaccharides, acetate and lactate, which in turn can be used as substrates for those butyrate producers that act like scavengers (Pryde et al., 2002). The second network, which is enriched in the higher BMI sibling group, includes Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. which both contain known butyrate producing isolates capable of degrading fibers themselves (Barcenilla et al., 2000; Duncan et al., 2002). Furthermore, this network also includes Eubacterium rectale et rel. which is another group with known butyrate producers. Therefore, the second network seems to be a butyrate producing network.
SCFA profiles show within-pair differences in discordant twins
Since both discordant siblings were found to be enriched in different fermentation networks, metabolic profiling was performed to confirm if these differences were visible in the fermentation products of these sibling groups as well. Although several organic acids were not detected in this cohort (that is, citrate, lactate, fumarate and formate), acetate, propionate and butyrate were the most dominating metabolites in both groups. From all detected organic compounds only butyrate and valerate were present at significantly higher concentrations in higher BMI siblings compared with their lower BMI siblings (Table 1), which is in line with the predicted networks (Figure 2c).
Table 1. Fecal organic acid and short chain fatty acid concentration of twins discordant and concordant for BMI.
| Component | Lower BMI siblings (n=16) (mean±s.d.e) | Higher BMI siblings (n=16) (mean±s.d.e.) | P-value | Controls (n=32) (mean±s.d.e.) |
|---|---|---|---|---|
| Acetate | 56.44±6.38 | 74.41±9.04 | 0.252 | 56.97±6.49 |
| Butyrate | 13.76±1.59 | 25.12±3.51 | 0.013 | 16.40±1.90 |
| Iso-butyrate | 0.23±0.14 | 1.15±0.33 | 0.067 | 1.09±0.29 |
| Malate | 10.56±2.60 | 25.71±8.3 | 0.403 | 11.81±2.84 |
| Propionate | 16.82±1.76 | 25.25±4.04 | 0.193 | 15.86±1.42 |
| Succinate | 0.64±0.35 | 0.03±0.03 | 0.178 | 1.41±1.21 |
| Valerate | 1.47±0.30 | 3.43±0.62 | 0.006 | 2.00±0.30 |
Abbreviation: BMI, body mass index.
Citrate, lactate, fumarate and formate were not detected. Statistical testing of between the concentrations of lower BMI sibling and higher BMI sibling groups was performed with paired dependent two-group Wilcoxon signed rank tests and P-values are reported (significant P-values are in bold).
Discussion
The human GI microbiota and its relation to differences in BMI was investigated in a host-genotype controlled setup by analyzing fecal samples of MZ twins. The cohort was comprised of MZ twins discordant and concordant for BMI, allowing us to assess the influence of BMI differences independent of the absolute BMI value of the twins. With this MZ twin control study, we were also able to control for gender, age, birth weight and other prenatal and postnatal exposures shared by the co-twins. Furthermore, with 80 subjects containing 40 different human genotypes, this cohort allowed to assess more generic topics such as the human GI microbiota core.
Due to this variety of core definitions and molecular techniques employed in current literature, no consensus on a general human GI microbial core has emerged (Hamady and Knight, 2009; Tap et al., 2009; Turnbaugh et al., 2009; Qin et al., 2010; Claesson et al., 2011; Jalanka-Tuovinen et al., 2011; Salonen et al., 2012). In our cohort we could define a general core microbiota of 96 phylotype-like groups prevalent in all subjects that accounts for 34.7% of the total microbiota (s.d. 12.8%). However, in line with recent observations (Salonen et al., 2012), this general core is very dependent on detection threshold and furthermore highly subject specific, as even the most prominent phylotype-like groups comprised only 0.1–0.22% of the total microbiota in some subjects. Our assessment of a general phylogenetic GI microbiota core agrees with various previous studies; however, Turnbaugh et al. and Tap et al. did not find a common microbial core (Tap et al., 2009; Turnbaugh et al., 2009). Notably Turnbaugh et al. (2009) used a higher relative abundance threshold than we used here, that is, 0.5% (Turnbaugh et al., 2009). Although Tap et al. (2009) defined a phylogenetic core that accounted for 35.8% of the total sequences from their cohort (Tap et al., 2009), their core definition only accounts for 8.1% of the total signals in our twin cohort. Next to methodology and the actual core definitions, the high subject specificity of core phylotypes complicates assessment of the general GI microbiota core.
In contrast to a general genotype independent core based on phylotype abundance, a genotype-dependent structural core (at higher phylogenetic levels) was much more pronounced in our data set. This study demonstrates that the human genotype and possibly early life stimuli exhibit a strong influence on the GI microbiota structural composition and extends earlier findings on twins and their relatively high degree of inter-pair microbiota similarity (Van de Merwe et al., 1983; Zoetendal et al., 2001). Host metadata (Supplementary Table S1) revealed no factors that significantly influenced the microbiota profile similarity. Yet MZ twins have GI microbiota profiles that are more similar to each other than to random unrelated subjects, despite the fact that half of them is discordant in terms of BMI (Figure 1a). Within-pair microbiota similarity was not equally represented over the phylogenetic subgroups (Supplementary Figures S1 and S2). From different phyla, several subgroups displayed within-pair profiles which where conserved among MZ twin pairs (Figures 1b and c; Supplementary Table S2). Interestingly, most of the conserved phylogenetic subgroups are significantly higher compared with the total within-pair microbiota similarity. Several other genus-like groups were found to be very dissimilar compared with the total microbiota similarity, which include some facultative anaerobes that are known to be fastidious (Supplementary Figure S2). Although this could suggest that these groups likely respond quickly to changing conditions, this remains speculative. The conserved profiles do not necessarily mean that the corresponding groups are present at similar abundance levels, for instance, Roseburia intestinalis et rel. and Eubacterium ventriosum et rel. are less abundant in the lower BMI siblings compared with their higher BMI siblings (Figure 2b). Similarities of the conserved genus-like groups were independent of host phenotype (BMI related or otherwise). Therefore, the human microbiota appears to have an imprinted structural core. Some of the microbial groups of the imprinted structural core are more strongly genotype dependent, like Clostridium ramosum et rel., Escherichia coli et rel., Eggerthella lenta et rel., and genus-like groups of Fusobacteria and uncultured Clostridiales, as their within-pair similarity is at least 10% higher compared with random-paired subjects. From the imprinted structural core Clostridium nexile et rel., Ruminococcus gnaves et rel. and Streptococcus bovis et rel. have the highest within-pair similarity (>0.9 Spearman's correlation coefficient) but they are also highly similar in random-paired subjects. The existence of an imprinted structural core strengthens and extends earlier studies which link human genotype to GI microbiota composition (Van de Merwe et al., 1983; Zoetendal et al., 2001). Moreover, scientific findings have not yet established the exact potency of early life epigenetic imprinting on adult human beings. MZ co-twins have likely experienced the same stimuli and conditions during early life. Whether or not early life dietary influences are important, next to the host-genotype, for GI microbiota composition and the imprinted structural core remains an outstanding question. By design, this study circumvents early life influences by pair-wise analyses between co-twins with an identical genetic background.
Remarkably, approximately half of the genera that are structurally conserved within MZ twin pairs were previously reported to be driving genera for the classification of the three enterotypes (Arumugam et al., 2011). Although enterotypes are classifications based on microbial abundance levels (Arumugam et al., 2011) while the imprinted structural core members are not necessarily highly abundant in every subject, it is noteworthy that we observed a significant overlap between bacterial groups that are important for enterotype classification and the imprinted structural core. Finding these particular members to be part of the imprinted structural core adds to the debate on the classification of the GI microbiota structure, that is, whether the GI microbiota can truly be classified into distinct types (enterotypes) or if the GI microbiota should be regarded as a ‘state' (a part of a continuum). Our data indicate that the imprinted structural core harbors the capacity to form each enterotype (Supplementary Table S2), thereby favoring the previous observations that enterotypes can best be seen as distinct states, rather than distinct types. This finding deserves more attention in longitudinal dietary studies, especially since drivers from all three enterotypes are found to be structurally conserved per genotype and not just the drivers of one of the proposed enterotypes.
In previously reported studies, human genotype and phenotype could not be distinguished when reporting on the GI microbiota of lean and obese individuals. High expectations are raised in current literature about the role of GI microbiota on host energy homeostasis and weight control. This may not be justified given the fact that in this twin population with an identical genetic background discordance for BMI did not relate to spectacular differences in GI microbiota composition. It appears to be unlikely for genetically identical people to diverge in BMI as much as the subjects in cross-sectional studies on high and low BMI. Moreover, extreme differences in BMI and drastic weight-loss regimes are accompanied by many other potential confounding factors, such as (extreme) changes in diet, host physiology, host health status and change of physical activity and their consequences on the overall host metabolism. Our study design enabled the elimination of genotype influences, by pair-wise comparison of MZ twins with discordant BMIs independent of absolute BMI values. This allowed us to strongly corroborate the influence of the host-genotype on the GI microbiota structure, but this also unveiled specific microbiota differences associated to host phenotype.
In our twin cohort, no trend between BMI differences and total microbiota diversity or B:F ratio was detected, which agrees with the findings reported by Duncan et al. (2008). At lower phylogenetic levels, however, microbial differences related to differences in BMI phenotype were detected. A consistent pair-wise difference in diversity within the discordant twin pairs can be found for Clostridium cluster IV. Our results indicate that this group is associated with phenotypic changes in BMI, decreasing in diversity as BMI increases. Furthermore, from this Clostridium cluster we found the relatives of the long known, yet uncultivated Oscillospira guillermondii to be significantly higher in the lower BMI siblings. Interestingly, previous findings link low Clostridium cluster IV diversity to Crohn's disease (Manichanh et al., 2006). Therefore, it appears that Clostridium cluster IV diversity can be affected by systemic changes of the host phenotype. Moreover, depletion of several members of Clostridium cluster IV, in our case Oscillospira guillermondii et rel., is associated with phenotypic changes.
Members of the morphologically distinct genus Oscillospira are frequently seen in cattle and sheep rumen. Several Oscillospira species react to the diets of their host, increasing strongly when the hosts are feeding on fresh green fields (Mackie et al., 2003). It is likely that these organisms are adapted to degrade the fibers of young plants. Hence the plant fiber content in human diet might also influence the presence and abundance of Oscillospira species. Without further knowledge on the metabolism of Oscillospira guillermondii et rel. it is hard to elucidate the extent of their role in the host energy homeostasis. Possibly, the presence of Oscillospira species has an impact on the metabolism of nutritional fibers. In line with this hypothesis, is the finding of three other genus-like (level 2) groups that co-occur with Oscillospira guillermondii et rel. (Figure 2c) which have isolates associated with degradation of plant material: Clostridium cellulosi (Yanling et al., 1991), Ruminococcus bromii (Klieve et al., 2007; Kovatcheva-Datchary et al., 2009) and Sporobacter termitidis (Grech-Mora et al., 1996). It seems that the co-occurrence network of Oscillospira guillermondii et rel. is specialized in fermenting complex (plant) materials. Fermentation products from such a primary degrader network can be used by scavenging butyrate producing bacteria (Pryde et al., 2002).
Both the Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. groups were significantly higher in the higher BMI siblings compared with their corresponding lower BMI siblings. Cultured isolates belonging to these two taxonomic groups are known butyrate producers (Barcenilla et al., 2000; Duncan et al., 2002). However, other results from the HITChip data did not reveal additional differences in butyrate production (potential), as based on similar levels of other butyrate-producing organisms in the discordant twins. Analogously, the co-occurrence network of the Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. (Figure 2c) did not include other potential butyrate producing organisms that are detected by HITChip, such as: Butyrivibrio crossotus et rel., Coprococcus eutactus et rel., Eubacterium hallii et rel., Faecalibacterium prausnitzii et rel., Megasphaera elsdenii et rel. or Mitsuokella multiacida et rel. Moreover, Roseburia intestinalis et rel. was negatively correlated with Anaerovorax odorimutans et rel., which has a representative isolate capable of butyrate production as well (Matthies et al., 2000).
In contrast to the primary degrader network of Oscillospira guillermondii et rel., the network of Eubacterium ventriosum et rel. and Roseburia intestinalis et rel. is composed of butyrate producers that are likely able to degrade complex material on their own. We hypothesize that host BMI increase is accompanied by GI microbiota changes and therefore a metabolic shift in butyrate production structure inside the colon: from mainly scavenging fermentation products produced by primary degraders to produce butyrate to mainly fermenting fibers directly into butyrate. Such a metabolic shift is likely to alter the net energy production by the GI microbiota and subsequently affect host energy harvest. This hypothesis is strengthened by finding significantly more butyrate in the fecal content of the higher BMI siblings, which are enriched for the butyrate producing network, compared with their lower BMI co-twins. Moreover, more valerate is found in the higher BMI siblings as well. Valerate can be formed by fermentation of the amino-acid proline (Amos et al., 1971), which could indicate two (not mutually exclusive) possibilities: (1) no carbohydrates are left for a part of the microbial community, forcing this part to switch to amino-acid fermentation; (2) the microbial community is more efficient in utilizing different types of (polymer) nutrients. Hence, the valerate results are in line with the network predictions in both discordant sibling groups and add to our hypothesis on the net energy production by the GI microbiota. However, from our data it is not possible to determine causality. If primary degraders outcompete the butyrate producers that are capable of degrading fibers, then subjects with lower BMI may only have scavenging butyrate producers left due to a diet rich in complex fibers. This is in line with the observation that a higher intake of fiber leads to lower levels of BMI. On the other hand, subjects with lower BMI could have much less butyrate producers, scavenging or otherwise, to begin with and therefore these subjects do not possess a microbiota that can harvest all available energy from the diet. Furthermore, the higher BMI siblings could also have consumed more protein and therefore show the increase in valerate. Therefore, more information on dietary intake and fermentation products in the GI tract is needed to further elucidate the mechanism between GI microbiota and host energy harvest.
Overall, this study revealed the existence of an imprinted structural core, to which several enterotype drivers belong, in the human GI microbiota and that BMI-phenotypic signatures are observed that can be related to energy harvest potential. However, the results are not such that a clear weight regulatory effect can be explained. Nonetheless, we have shown that different microbial networks are associated to changes in BMI. A network of primary degraders was more prominent in subjects with lower BMI, while a network of butyrate fermenters was more prominent in subjects with higher BMI. Our data suggest that primary degraders, not capable of producing butyrate, could play a more important role in energy homeostasis than initially expected. Previous studies on obesity have mostly compared cross-sectional the microbiota of lean and obese people and reported contradicting results. Besides the differences in host-genotypes in these studies, the subject are at different ends of the BMI scale, that is, the lean and obese ‘states'. Here, we studied genetically identical human hosts and assessed BMI differences independent of the lean or obese characteristic of the individuals, enabling the detection of BMI-phenotype signatures in the GI microbiota. Known genetic background, or minimizing its influence in studies like this, will be pivotal in the deciphering of the effects of GI microbiota on mechanisms underlying phenotypic traits of the host, such as changes in BMI as reported here.
Acknowledgments
We thank Lut De Zeure for her excellent technical assistance and of course we thank the twins that participated. Since its origin, the East Flanders Prospective Survey has been partly supported by grants from the Fund of Scientific Research, Flanders (Belgium), and by the Association for Scientific Research in Multiple Births (Belgium). The funding sources had no role in the design and conduct of the study, in the collection, analysis and interpretation of the data, or in the preparation, review or approval of the manuscript. ST and EGZ received funding from the European Community's Seventh Framework Programme (FP7/2007–2013): MetaHIT, Grant agreement HEALTH-F4-2007-201052, coordinated by S Dusko Ehrlich (Institut National de la Recherche Agronomique, France).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adlercreutz H, Pulkkinen MO, Hämäläinen EK, Korpela JT. Studies on the role of intestinal bacteria in metabolism of synthetic and natural steroid hormones. J Steroid Biochem. 1984;20:217–229. doi: 10.1016/0022-4731(84)90208-5. [DOI] [PubMed] [Google Scholar]
- Albert MJ, Mathan VI, Baker SJ. Vitamin B12 synthesis by human small intestinal bacteria. Nature. 1980;283:781–782. doi: 10.1038/283781a0. [DOI] [PubMed] [Google Scholar]
- Amos HE, Little CO, Mitchell GE. Proline utilization during cellulose fermentation by rumen microorganisms. J Agric Food Chem. 1971;19:112–115. doi: 10.1021/jf60173a051. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CC, Zoetendal EG, Kleerebezem M, de Vos WM. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2007;2:285–295. doi: 10.2217/17460913.2.3.285. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Tremblay A, Després JP, Nadeau A, Lupien PJ, Thériault G, et al. The response to long-term overfeeding in identical twins. N Engl J Med. 1990;322:1477–1482. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108 (Suppl 1:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT. Colonic microflora: nutrition and health. Nutrition. 1997;13:476–478. doi: 10.1016/s0899-9007(97)00114-7. [DOI] [PubMed] [Google Scholar]
- Derom CA, Vlietinck RF, Thiery EW, Leroy FO, Fryns JP, Derom RM. The East Flanders Prospective Twin Survey (EFPTS) Twin Res Hum Genet. 2006;9:733–738. doi: 10.1375/183242706779462723. [DOI] [PubMed] [Google Scholar]
- Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, et al. Molecular analysis of the gut microbiota of identical twins with crohn's disease. ISME J. 2008;2:716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52 (Part 5:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Grech-Mora I, et al. Isolation and characterization of sporobacter termitidis gen. nov., sp. nov., from the digestive tract of the wood-feeding termite nasutitermes lujae. Int J Syst Bacteriol. 1996;46:512–518. [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. 2011;43:1164–1164. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Ye Y, Zhou Y, Fodor AA. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One. 2012;7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salonen A, Nikkilä J, Immonen O, Kekkonen R, Lahti L, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One. 2011;6:e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve AV, O'Leary MN, McMillen L, Ouwerkerk D. Ruminococcus bromii, identification and isolation as a dominant community member in the rumen of cattle fed a barley diet. J Appl Microbiol. 2007;103:2065–2073. doi: 10.1111/j.1365-2672.2007.03492.x. [DOI] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilić-Stojanović M, de Graaf AA, Smidt H, et al. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol. 2009;11:914–926. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Ley RE, khed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. 2003;69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson JE, Willett WC, Stampfer MJ, Colditz GA, Hunter DJ, Hankinson SE, et al. Body weight and mortality among women. N Engl J Med. 1995;333:677–685. doi: 10.1056/NEJM199509143331101. [DOI] [PubMed] [Google Scholar]
- Matthies C, Evers S, Ludwig W, Schink B. Anaerovorax odorimutans gen. nov., sp. nov., a putrescine-fermenting, strictly anaerobic bacterium. Int J Syst Evol Microbiol. 2000;50 (Part 4:1591–1594. doi: 10.1099/00207713-50-4-1591. [DOI] [PubMed] [Google Scholar]
- Metges CC. Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr. 2000;130:1857S–1864S. doi: 10.1093/jn/130.7.1857S. [DOI] [PubMed] [Google Scholar]
- Naukkarinen J, Surakka I, Pietiläinen KH, Rissanen A, Salomaa V, Ripatti S, et al. Use of genome-wide expression data to mine the ‘Gray Zone' of GWA studies leads to novel candidate obesity genes. PloS Genet. 2010;6:e1000976. doi: 10.1371/journal.pgen.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara AM, O'Regan P, Fanning A, O'Mahony C, Macsharry J, Lyons A, et al. Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology. 2006;118:202–215. doi: 10.1111/j.1365-2567.2006.02358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilainen KH, Naukkarinen J, Rissanen A, Saharinen J, Ellonen P, Keränen H, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. Plos Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar K, Conly JM, Chubb H, Louie TJ. Production of menaquinones by intestinal anaerobes. J Infect Dis. 1984;150:213–218. doi: 10.1093/infdis/150.2.213. [DOI] [PubMed] [Google Scholar]
- Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Salonen A, Salojärvi J, Lahti L, de Vos WM. The adult intestinal core microbiota is determined by analysis depth and health status. Clin Microbiol Infect. 2012;18 (Suppl 4:16–20. doi: 10.1111/j.1469-0691.2012.03855.x. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saris WH, Tarnopolsky MA. Controlling food intake and energy balance: which macronutrient should we select. Curr Opin Clin Nutr Metab Care. 2003;6:609–613. doi: 10.1097/00075197-200311000-00001. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F. The host-microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- Stams AJ, Van Dijk JB, Dijkema C, Plugge CM. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol. 1993;59:1114–1119. doi: 10.1128/aem.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Truesdale KP, McClain JE, Cai J. The definition of weight maintenance. Int J Obes. 2006;30:391–399. doi: 10.1038/sj.ijo.0803175. [DOI] [PubMed] [Google Scholar]
- Stevens J, Cai J, Pamuk ER, Williamson DF, Thun MJ, Wood JL. The effect of age on the association between body-mass index and mortality. N Engl J Med. 1998;338:1–7. doi: 10.1056/NEJM199801013380101. [DOI] [PubMed] [Google Scholar]
- Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Van de Merwe JP, Stegeman JH, Hazenberg MP. The resident faecal flora is determined by genetic characteristics of the host. Implications for Crohn's disease. Antonie Van Leeuwenhoek. 1983;49:119–124. doi: 10.1007/BF00393669. [DOI] [PubMed] [Google Scholar]
- van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–2080. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Bouter LM, McQuillan GM, Wener MH, Harris TB. ELevated c-reactive protein levels in overweight and obese adults. JAMA. 1999;282 (22:2131–2135. doi: 10.1001/jama.282.22.2131. [DOI] [PubMed] [Google Scholar]
- Wei C, Brent MR. Using ESTs to improve the accuracy of de novo gene prediction. BMC Bioinform. 2006;7:327. doi: 10.1186/1471-2105-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xie L, Li Y, Wei C. More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLoS One. 2009;4:e6074. doi: 10.1371/journal.pone.0006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanling H, Youfang D, Yanquan L. Two cellulolytic clostridium species: clostridium cellulosi sp. nov. and clostridium cellulofermentans sp. nov. Int J Syst Bacteriol. 1991;41:306–309. doi: 10.1099/00207713-41-2-306. [DOI] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, et al. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
- Zoetendal EG, Cheng B, Koike S, Mackie RI. Molecular microbial ecology of the gastrointestinal tract: from phylogeny to function. Curr Issues Intest Microbiol. 2004;5:31–47. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.