Abstract
The naturally transformable bacterium Streptococcus pneumoniae is able to take up extracellular DNA and incorporate it into its genome. Maintaining natural transformation within a species requires that the benefits of transformation outweigh its costs. Although much is known about the distribution of natural transformation among bacterial species, little is known about the degree to which transformation frequencies vary within species. Here we find that there is significant variation in transformation frequency between strains of Streptococcus pneumoniae isolated from asymptomatic carriage, and that this variation is not concordant with isolate genetic relatedness. Polymorphism in the signalling system regulating competence is also not causally related to differences in transformation frequency, although this polymorphism does influence the degree of genetic admixture experienced by bacterial strains. These data suggest that bacteria can evolve new transformation frequencies over short evolutionary timescales. This facility may permit cells to balance the potential costs and benefits of transformation by regulating transformation frequency in response to environmental conditions.
Keywords: Streptococcus pneumoniae, transformation, competence, fitness-associated recombination
Introduction
At variable times during growth, naturally transformable bacteria become transiently ‘competent' to take up environmental DNA, which is either stably incorporated into the bacterial chromosome or digested upon entry (Chen and Dubnau, 2004). Transformation is proposed to benefit bacteria in three possible ways: (1) it may provide nutrients in the form of nucleotides and nucleotide precursors (Redfield, 1993); (2) acquired DNA may be used as a substrate for genome repair (Bernstein et al., 1981); and (3) recombination between cells may assemble independent beneficial mutations into a single genetic background, thereby accelerating adaptation to novel or fluctuating environmental conditions (Vos, 2009; Didelot and Maiden, 2010). In addition to putative benefits of transformation, there are numerous possible costs. First, transformation can disrupt co-adapted gene complexes (Vos, 2009). Second, it may increase susceptibility to parasitic DNA or deleterious mutations from dead cells (Redfield, 2001). Third, DNA binding, uptake and incorporation may require a ‘handling' time that reduces rates of vegetative growth or leads to transient growth arrest (Haijema et al., 2001; Johnsen and Levin, 2010). Fourth, expression of the competence regulon may be energetically costly, and because in some species it involves a secreted peptide signal available as a potential ‘public good', the system is susceptible to cheats not producing the signal (Travisano and Velicer, 2004; West et al., 2006; Yang et al., 2010). Finally, in the focal species of this study, Streptococcus pneumoniae, there may be considerable costs associated with competence-induced cell lysis, wherein competent cells lyse non-competent members of the same population (Steinmoen et al., 2002). The maintenance of transformation within a bacterial isolate or species requires that the benefits of transformation outweigh its costs. Where this is not the case, bacterial strains would lose the ability to become transformed by fixing mutations in genes and regulatory regions required for competence. Alternatively, balancing the costs and benefits across space and time in bacteria may lead to changes in the per lineage rate of transformation.
S. pneumoniae is a Gram-positive human opportunistic pathogen that is a leading cause of global infant mortality and is responsible for up to 1 million deaths annually (O'Brien et al., 2009). Most often, the bacterium resides asymptomatically as part of the commensal flora of the human nasopharynx, where it can be detected in up to 60% of individuals, particularly in young children (Bogaert et al., 2004). S. pneumoniae is naturally transformable, and this species has been a model for understanding the mechanisms of transformation since the 1920s when transformation was first discovered by Griffith and then studied in detail by Avery and colleagues (Griffith, 1928; Avery et al., 1944). Cultures of pneumococci become transiently competent to take up DNA during a very brief interval during exponential growth (Tomasz, 1965). Competence is regulated by a two-component signalling system encoded by comC and comD, specifying the peptide signal CSP (competence stimulating peptide) and its cognate receptor, respectively. CSP is secreted into the extracellular environment where it binds to membrane-bound ComD. This causes ComD to phosphorylate its response regulator, ComE, inside the cell. At a certain threshold level of bound peptide, the signalling cascade is initiated and the cells enter a competent state (Claverys et al., 2007). Within S. pneumoniae there are two major comC and comD alleles (or pherotypes), type 1 and type 2, which are typically found at frequencies of 70% and 30% of isolates within sampled populations, respectively (Pozzi et al., 1996; Carrolo et al., 2009; Cornejo et al., 2010; Vestrheim et al., 2011). Although isolates from different pherotypes do not respond to non-self CSP, gene flow between pherotypes appears to be widespread, indicating that CSP type does not impose a barrier to recombination (Havarstein et al., 1997; Cornejo et al., 2010). Nevertheless, it remains unclear if there are differences in transformation rate as a function of pherotype.
Natural competence is phylogenetically patchy across bacteria, indicating that this ability has been independently gained and lost across species (Lorenz and Wackernagel, 1994). With notable exceptions (Sikorski et al., 2002; Fujise et al., 2004; Maughan and Redfield, 2009; Joloba et al., 2010), however, less is known about the variation in transformation rates across genotypes within bacterial species, the patterns by which transformation rates change and the consequences of these changes for bacterial populations. In addition, where studies of natural variation in transformation rates among pathogenic bacteria have been carried out, these have been done on clinical strains with a history of antibiotic exposure, and so may not be representative of these species overall (Didelot and Maiden, 2010). We overcome this limitation here, where we examine patterns of natural variation in transformation rates among a large collection of non-clinical commensal isolates of S. pneumoniae (the pneumococcus), and examine the scale and consequences of these changes using a population genetics framework. Finally, we determine the role of peptide signal polymorphism in modifying pneumococcal transformation rates. Briefly, we find that a large fraction of isolates are non-transformable, whereas there is significant variation in transformation rates among isolates that are. Moreover, we present evidence that these rates vary significantly among closely related lineages, and therefore appear to be rapidly evolving.
Materials and methods
Isolates and pherotyping
The 54 serotyped S. pneumoniae isolates included in this study were kindly provided by Professor Peter Hermans, and were originally obtained from healthy infants aged 3–36 months from the Amsterdam area in the Netherlands between January and March 1999 (Bogaert et al., 2001). Nine of the isolates were obtained from infants who had not attended day care centres, whereas the remaining 45 isolates were obtained from infants who had. Bacteria were stored at −80 °C in Complete Transformation Medium (CTM), pH 6.8 +20% glycerol. CTM contains 30 g l−1 tryptone soy broth (Lab M, Lancs, UK), 1 g l−1 yeast extract (Melford Laboratories Ltd, Suffolk, UK), 0.001 M CaCl2 (Fisher Scientific, Leicestershire, UK) and 0.2% BSA (Melford Laboratories Ltd, Suffolk, UK). CTM medium at pH 6.8 inhibits competence, whereas CTM medium at pH 7.8 allows cells to become competent (Tomasz and Mosser, 1966). All strains were preliminarily classified as pherotype 1, as determined by HinfI endonuclease restriction analysis of the comC gene (Cornejo et al., 2010), which discriminates between the comC-1 and comC-2 alleles. The sequences of the comC gene and the first 200 bp of the comD gene were determined by PCR amplification with primers comC–F (5′-AAAAAGTACACTTTGGGAGAAAAA-3′) (Cornejo et al., 2010) and comD–R (5′-ATCTCCTGAAGGAGTCATCG-3′) using Phusion DNA polymerase (New England Biolabs UK Ltd, Herts, UK), followed by Sanger sequencing at the University of Manchester Genomics facility. PCR conditions were: initial denaturing at 98 °C for 30 s; 35 cycles of 98 °C for 10 s, 51 °C for 30 s and 72 °C for 2 min; then a final extension of 72 °C for 5 min. Insertions in the competence pilus structural gene comYC were screened for using primers comYC–L (5′-TACGATTTGCCCCTCCATT-3′) and comYC–R (5′-GGTTTTTATCTTTGTGGCACTG-3′) (Croucher et al., 2011). These screens were repeated using primer comYC–L2 (5′-CCAAGAGACTTTCCAGCATG-3′) in place of comYC–L, as primer comYC–L was found to reside in a variable region of the comYC gene. Reaction conditions were: initial denaturing at 98 °C for 30 s; 35 cycles of 98 °C for 10 s, 56 °C for 30 s, and 72 °C for 1 min; then a final extension of 72 °C for 10 min. Neighbour-joining Jukes–Cantor protein trees for the ComC and ComD sequences were constructed using Geneious 5.5.2 (created by Biomatters, available from http://www.geneious.com/) from alignments utilising the Blosum62 cost matrix, a gap open penalty of 12 and a gap extension penalty of 3. Trees were visualised in FigTree v.1.2.3 (Andrew Rambaut, available from http://tree.bio.ed.ac.uk/software/figtree/).
Cell preparation
Bacteria were prepared for transformation by taking swabs from cultures stored at −80 °C and plating them on tryptic soy agar (Lab M, Lancs, UK) +3% defibrinated horse blood (Oxoid, Cambridge, UK) and incubating overnight at 37 °C+5% CO2 in a static incubator. Following growth, a swab of cells was taken and inoculated into Todd Hewit broth (Oxoid, Cambridge, UK), and the optical density of the culture adjusted to OD600<0.1. Cultures were grown to an OD600=0.3, equivalent to ∼3 × 108 cells, then frozen at −80 °C with the addition of 25% glycerol. Finally, frozen cell aliquots were diluted by a factor of 10 into CTM pH 6.8 without CaCl2 or BSA and cultures grown to an OD600=0.3, then frozen at −80 °C with the addition of 25% glycerol.
Transformation
Prepared aliquots of bacteria were taken from the freezer and diluted by a factor of 10 into CTM pH 6.8 without CaCl2 or BSA. Cultures were grown to an OD600=0.3, then diluted ten-fold into CTM pH 7.8. For transformation, 300 μl from each of these cultures was transferred to an eppendorf containing a saturating concentration of 1 μg ml−1 gDNA from S. pneumoniae strain R304 and 0.1 μg ml−1 synthetic competence stimulating peptide 1, CSP-1 (Sigma-Aldrich, Dorset, UK). The rpsL gene in strain R304, coding for the 30S ribosomal subunit S12, contains a point mutation that confers streptomycin resistance. This marker, known as str41, was used to quantify transformation frequency (Mortier-Barriere et al., 1998). The same R304 gDNA preparation was used in all experiments. Transformation was allowed to proceed at 30 °C for 30 min. Next, 1 μl of DNase I (New England Biolabs UK Ltd, Herts, UK) was added to each tube to digest any remaining extracellular DNA, and the tubes incubated for 1 h at 37 °C. Cells were plated onto tryptic soy agar +3% blood, and tryptic soy agar +3% blood +150 μg ml−1 streptomycin (Sigma-Aldrich) at appropriate dilutions to estimate total cell numbers and the number of transformants, respectively. As we are estimating transformation of the streptomycin marker, this assay quantifies locus-specific transformation frequencies rather than genome-wide frequencies. Plates were incubated for 48 h before colonies were counted. Transformation frequencies were estimated twice independently for each isolate, and are expressed as the mean number of transformants per colony forming unit. The detection limit for the assay is a transformation frequency of ∼1 in 108 cells. The variation in transformation frequency between isolates within serotypes was determined using generalised linear models in SPSS v.18 (SPSS Inc, Chicago, IL, USA), with transformation frequency as the dependent variable and strain nested within serotype as the factor. The model was run both with the inclusion of all transformation frequencies, and with zero values omitted.
Bacterial population structure
Multilocus sequence typing (MLST) of the isolates included in this study was performed as described previously (Enright and Spratt, 1998). Briefly, fragments from the seven housekeeping genes aroE, gdh, gki, recP, spi, xpt and ddl from each strain were amplified by PCR using Phusion DNA polymerase (New England Biolabs UK Ltd, Herts, UK) and sequenced. Primer sequences used for amplification and sequencing were modified as described by the Centres for Disease Control and Prevention (http://www.cdc.gov/ncidod/biotech/strep/alt-MLST-primers.htm). Allele numbers were assigned to sequences, and allelic profiles were assigned sequence types using the S. pneumoniae MLST database (http://spneumoniae.mlst.net/). All further analyses excluded the ddl locus as it has been shown to be linked to the penicillin-binding protein 2b gene (Enright and Spratt, 1999). The nucleotide sequences from the fragments of the remaining six genes were used to estimate population structure in the programme BAPS version 5.3 (Corander et al., 2008). The mixture analysis was performed under the clustering of linked molecular data option using the codon linkage model. For the admixture analysis, the upper bound on the number of populations, K, was set to 10, and admixture coefficients were estimated with 100 iterations. In order to remove the potential for allelic variation within populations to confound estimates of allelic exchange between populations, an estimate of within-population allelic variation was determined through sampling 200 reference individuals from each population and estimating admixture coefficients with 20 iterations. For comparative purposes, the degree of admixture of an isolate was calculated from the product of the membership coefficients for each population for that isolate, resulting in higher values for isolates with greater degrees of admixture and lower values for isolates with less admixture. The concatenated sequence of the six MLST genes were used to calculate genetic distances among isolates using the neighbour-joining method under the JukesCantor model of sequence evolution. The sum of branch lengths between each isolate and strain R6, the ancestral strain to strain R304, were regressed against the transformation frequency of each strain. Statistical tests were performed in SPSS v.18 (SPSS Inc, Chicago, IL, USA).
Susceptibility testing
Antibiotic susceptibilities of the isolates were tested by disk diffusion using the MASTRING-S system (MAST Group, Merseyside, UK). Antibiotic disk rings M11 and M43 were used containing chloramphenicol, erythromycin, fusidic acid, methicillin, novobiocin, penicillin G, streptomycin, tetracycline, clindamycin, gentamicin, trimethoprim and sulfamethoxazole. Tests were carried out according to the British Society for Antimicrobial Chemotherapy (BSAC) guidelines version 9.1 (Andrews, 2010).
Results
Transformation frequencies and population structure
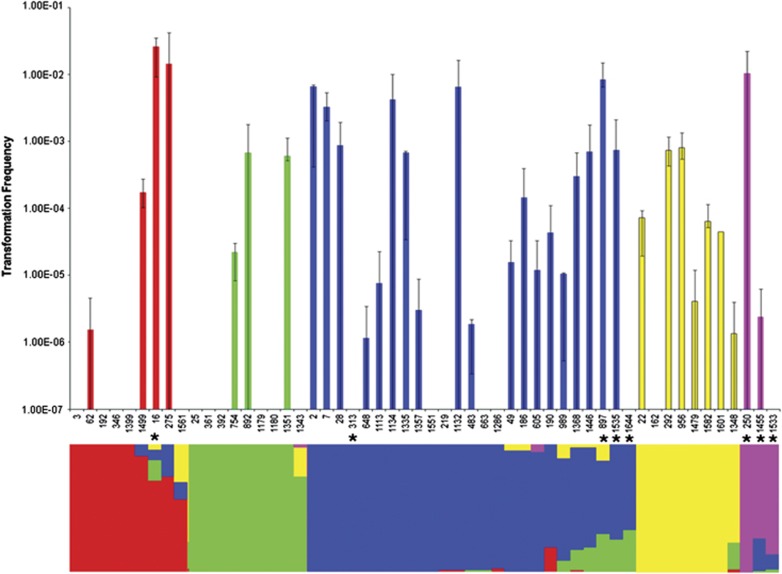
We observed significant variation in the frequency of transformation between isolates within serotypes, both including and excluding zero values from non-transformable isolates (generalised linear models; df=53, P<0.0001; df=34, P<0.0001) (Figure 1). Strikingly, no transformation was detectable for 18 strains (34%), whereas mean transformation frequencies for the other isolates (66%) varied over four orders of magnitude between 2.60 × 10−2 and 1.16 × 10−6 (Figure 1). To investigate whether inactivation of the competence pilus structural gene comYC due to prophage insertion could explain our inability to detect transformation in some isolates, as has been previously described (Croucher et al., 2011), we amplified this gene for all isolates. A fragment of the correct size for an uninterrupted comYC gene was seen in all isolates, so the absence of detectable transformation in some isolates is not due to this phenomenon. Antimicrobial resistance profiles were indistinguishable across all isolates.
Figure 1.
Top; isolate transformation frequencies. Error bars represent 95% CIs. Bottom; population structure as estimated using BAPS. The bar for each isolate is subdivided to indicate the relative genetic contribution that the isolate carries from each of the five populations. Red, population 1; green, population 2; blue, population 3; yellow, population 4; pink, population 5. Asterisks indicate the eight isolates with non-type 1 pherotypes.
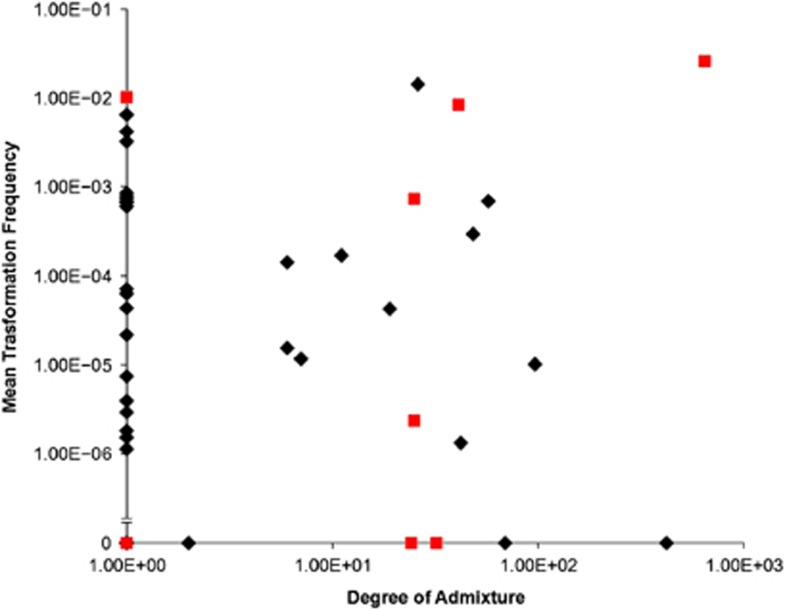
The 54 isolates belong to one of five different populations (Figure 1) comprised of 9, 9, 25, 8 and 3 isolates, respectively. The distribution of serotypes within four of the five populations was significantly different from that found in the whole data set following correction for multiple testing, with population 1 associated with serotype 14 (χ2 test P=5.5 × 10−5), population 2 associated with serotype 9V (χ2 test P=2.7 × 10−7), population 3 associated with serotype 23F (χ2 test P=1.4 × 10−111) and population 4 with serotype 6B (χ2 test P=4.1 × 10−5). The majority of isolates (57%) had no detectable admixture. There was no difference in the levels of admixture observed between each of the five populations (Kruskal–Wallis test, P=0.274), or between serotypes (Kruskal–Wallis test, P=0.502). Also there was no correlation between transformation frequency and the degree of admixture (Figure 2; Spearman rank-order correlation, P=0.353).
Figure 2.
Mean isolate transformation frequency plotted against the degree of admixture within each isolate. Black diamonds indicate isolates with comC-1 and comD-1, grey squares indicate isolates with comC and comD sequences other than type 1.
Isolates were mapped onto population structure to determine if different populations varied in transformation frequency (Figure 1). There were no differences in the distribution of transformation frequencies across the five populations detected (Kruskal–Wallis test, P=0.316). Previous studies conducting cross-species comparisons have found a log-linear relationship between isolate relatedness and transformation frequency (Majewski and Cohan, 1999; Majewski et al., 2000). This is due to higher levels of sequence homology between the transformed marker and closely related transformation recipients. To test if transformation frequencies were related to genetic distance in the pneumococcal isolates studied here, MLST data were used to calculate genetic distances between each isolate and S. pneumoniae strain R6 (the ancestral strain from which R304 is derived), and regressed against transformation frequency. There was no detectable relationship between isolate relatedness to strain R6 and mean transformation frequency (Supplementary Figure 1; linear regression, R2=0.044, P=0.13).
The role of pherotype
Sequencing of comC and the first 200 bp of comD confirmed that the majority of isolates carried the canonical comC-1 and comD-1 alleles, termed pherotype 1. However, eight isolates were found to carry different sequences at both the comC and comD loci, hereafter referred to as the non-pherotype 1 isolates. Of these eight isolates, seven carried a comC allele that differed from comC-1 by a single silent polymorphism, here termed comC-1.1 (Figure 3). The eighth isolate encoded a highly divergent ComC peptide, termed ComC-4; Figure 3 (Whatmore et al., 1999). These eight isolates also carried alleles differing from comD-1 over the first 133 amino acid positions. These non-comD-1 alleles were associated with particular populations, and were not shared across populations (Figure 3). A sequence termed ComD-1.2, differing from ComD-1 by five amino acids, was coded for in an isolate belonging to population 1; population 3 contained isolates coding for sequences termed ComD-1.4, and ComD-1.1, both differing from ComD-1 by five amino acids, and a silent variant of an allele described by Iannelli et al. (2005), here termed ComD-1.3; population 5 exclusively contained isolates coding for ComD-1.3, which varies at five amino acids from ComD-1. Isolate 313 carrying the comC-4 allele coded for a ComD sequence differing from ComD-1 by 21 amino acids. This protein was termed ComD-3.1, as it differs from ComD-3 (Whatmore et al., 1999) at a single amino acid site. There was no difference in transformation frequency between isolates with pherotype 1 or non-pherotype 1 alleles (Mann–Whitney U-test, P=0.384); this difference becomes marginally significant if strains lacking any detectable transformation are excluded from this analysis (Mann–Whitney U-test, P=0.059). However, we observed significantly more admixture in isolates carrying non-pherotype 1 alleles (Figure 2; Mann–Whitney U-test, P=0.009), which persists even if we exclude isolates for which no transformation was detected (Mann–Whitney U-test, P=0.029).
Figure 3.
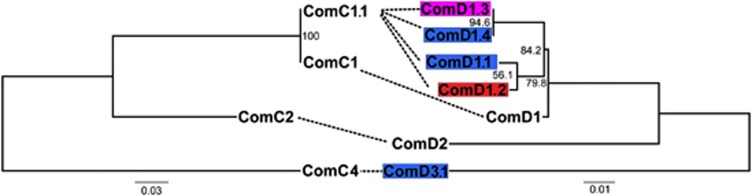
Protein trees for ComC (left) and ComD (right) sequences identified in isolates in this study. The sequences ComC-2 and ComD-2, not identified in this study, were also included. The ComC-1.1 sequence has been previously deposited in GenBank with the accession number CP000918. For ComD variants, the black shaded sequence is found in population 1, the mid-grey shaded sequences in population 3 and the light-grey shaded sequence in population 5. Canonical ComD-1 is found in all but population 5. Dashed lines connect ComC and ComD sequences that are found in the same strain. ComD-1.4 is identical to the comD sequence in the genome of strain AP200; ComD-1.1 is identical to that with GenBank accession number AJ240754; ComD-1.2 is identical to that with accession number AJ240779. ComD-3.1 refers to the ComD sequence differing from ComD-3 described by Whatmore et al (1999) (accession number AJ240793) at a single amino acid site.
Discussion
We find that the mean transformation frequency among transformable strains in this collection of carriage isolates of S. pneumoniae varied by up to four orders of magnitude, between 2.60 × 10−2 and 1.16 × 10−6, whereas transformation was not detectable in 34% of isolates. Two previous studies examining clinical pneumococcal isolates observed a similarly broad range of transformation frequencies (Hsieh et al., 2006; Joloba et al., 2010) and a moderate fraction of isolates (33%) where transformation could not be detected. Significant variation in transformation frequency between isolates is a feature also seen in other bacterial species including Actinobacillus actinomycetemcomitans (Fujise et al., 2004), Campylobacter jejuni (Wilson et al., 2003), H. influenzae (Maughan and Redfield, 2009) and Pseudomonas stutzeri (Sikorski et al., 2002). It therefore appears that the variation we observe is a general feature of naturally transformable bacterial species.
Differences or similarities in transformation frequency between strains could partially be explained by the degree of relatedness between these co-occurring isolates, whereby groups of closely related isolates share similar transformation frequencies that differ from those possessed by other groups. According to this explanation, the rate of transformation would evolve at a rate less than the time of divergence between phylogenetically disparate groups. In studies utilising collections of more distantly related isolates to those studies here (Majewski and Cohan, 1999; Majewski et al., 2000), transformation frequency declines with sequence divergence. However, this possibility is not supported from our analyses. When the 54 isolates from this study are clustered into groups of related genotypes using BAPS, we find that there is no correspondence between genotypic cluster and transformation rate. That is, there is as much variation observed within groups in transformation rate as across groups. In addition, we find no relationship between transformation frequency and the genetic distance of each strain to the donor isolate (Supplementary Information Figure 1). Likewise, although Sikorski et al. (2002) found that in some cases of closely related strains of P. stutzeri showed similar transformation frequencies, there were also instances where closely related isolates varied markedly in their transformation rate. These observations are consistent with the idea that transformation rate, like the mutation rate (Gutierrez et al., 2004), evolves readily among closely related bacterial isolates.
At present, it remains unclear what mechanisms underlie inter-strain differences in transformation rate. In S. pneumoniae, the competent state for transformation is induced when the local concentration of CSP reaches a certain threshold level. This, in turn, initiates the phosphorylation cascade that results in the transduction of over 100 genes, only some fraction of which are required for transformation (Peterson et al., 2004). Mutations in the signalling system inducing competence, or in genes crucial for DNA uptake and incorporation could reduce or eliminate transformation. Our results suggest three reasons why signalling is unlikely to underlie the differences we observe. First, because we induced competence using exogenous addition of CSP, our approach bypassed lesions that may exist in either peptide production or secretion. Second, sequencing determined that all comC alleles are intact. Third, differences in transformation rate may also arise due to mutations in the CSP receptor gene, comD. However, transformation rates in the eight strains in our sample carrying divergent receptors (Figure 3) did not differ from strains carrying the canonical ComD-1 receptor, nor were these isolates any more likely to lack transformation entirely. At the same time, these strains displayed higher rates of admixture, which may indicate that these receptors permit recombination with a broader range of inducing strains during co-colonisation. Mutations downstream of signalling, or potentially outside of the com regulon itself, are thus more likely causes for the differences we see. We observed no prophage insertion into comYC in any of the strains, but have not exhaustively examined other mutational causes and will hope to address these differences in the future. Another possibility is the differences between strains arising due to serotype. However, we find no difference in transformation frequencies between serotypes, and significant variation in frequencies within serotypes, demonstrating that differences in the capsular structure of the isolates is not responsible for the differences. Further detailed molecular and genetic studies are required to shed more light on the mechanisms underlying inter-strain differences in transformation frequency.
Whereas there is clear importance in understanding the mechanisms of inter-strain differences in transformation rate, it is equally important to begin understanding the evolutionary factors that underlie the maintenance of this variation (in analogy to identifying the evolutionary factors maintaining eukaryotic sex, see for example, Otto, 2009). Several advantages may accrue to bacteria from transformation, including direct and immediate benefits of nutritional gain, or gains derived from the import or recombination of beneficial alleles from different genetic backgrounds. Following Hanage et al. (2009) and other previous work describing the acquisition of antibiotic resistance through natural transformation (Dowson et al., 1989, 1990; Ferrandiz et al., 2005), we predicted that more transformable strains would exhibit greater levels of antibiotic resistance. However, we found that this was not the case (Supplementary Information Table 1). Indeed, we observed little variation in resistance at all among these isolates, which may be a consequence of the fact that these strains were isolated from carriage rather than disease. In addition to the benefits, there are several potential costs of competence, from the metabolic expense of transcribing the competence apparatus to the risk of importing deleterious alleles. One explanation of the variation we observe is that cells experience and balance these costs and benefits of transformation differently across time and space. By this explanation, transformation rate can increase or decrease rapidly by mutation as a function of the environment in which cells are found. S. pneumoniae is already known to modify the induction of transformation as a function of environmental cues, for example the presence of certain antibiotics (Prudhomme et al., 2006).
Such plasticity in transformation rate would be consistent with predictions of the Fitness Associated Recombination hypothesis (Redfield, 1988; Hadany and Beker, 2003), wherein recombination is more likely and advantageous in less fit individuals than in more fit individuals. Theoretical models have shown that populations adopting plastic recombination rates are more fit than the asexual ones, and are able to maintain fitness even when the DNA they take up carries deleterious mutations (Redfield, 1988). Furthermore, they are able to adapt more quickly to a changing (Hadany and Beker, 2003) or stable environment (Wylie et al., 2010). Empirical evidence for FAR has been presented for the fungus Aspergillus nidulans, where the production of more sexual fruiting bodies was associated with slower rates of fungal growth, with the effect replicated across three different growth environments (Schoustra et al., 2010). Data from the bacterium C. jejuni also point toward FAR, where transformation frequencies were found to be lower when bacteria were grown in more favourable conditions (Wilson et al., 2003). Our own preliminary data from long-term chemostat evolution experiments with S. pneumoniae is also consistent with this possibility, where we found that the benefits of transformation were conditional on environmental stress (Engelmoer and Rozen, unpublished data). Testing the FAR hypothesis in more detail requires a greater mechanistic understanding of the factors causing increases or decreases of the transformation rate. On the other hand, bacterial populations may adopt a bet-hedging strategy, where different members of the population transform at different frequencies in a manner that is blind to the surrounding environment. This would enable the population to maintain itself at its local fitness optimum, that under natural conditions is likely to be frequently changing as the bacteria interact with the host immune system, neighbouring flora, and pass through population bottlenecks during initial colonisation (Beaumont et al., 2009; Libby and Rainey, 2011). Alternatively, individual bacteria may sense their environment and modify their transformation frequency accordingly depending upon whether they are currently close to or far away from their local fitness optimum. Future work in this system will aim to clarify these possibilities.
To summarise, this study is the first to relate transformation frequencies in the human opportunistic pathogen S. pneumoniae with population genetic structure, and we find no association between the two. The absence of such an association combined with the knowledge that this bacterium is able to modulate transformation frequency in response to its environment provide support for the hypothesis that the transformation rate of naturally transformable bacteria can evolve rapidly in response to environmental conditions.
Acknowledgments
We would like to thank Peter Hermans for providing the bacterial strains used in this study, and Cath Jobbings for help measuring bacterial transformation rates. In addition, we acknowledge the helpful comments of three anonymous reviewers on an earlier version of this manuscript. We acknowledge the use of the pneumococcal MLST database that is located at Imperial College London and is funded by the Wellcome Trust. This work was funded by a BBSRC grant (BBF0020681) to DER.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andrews JM.2010. BSAC methods for antimicrobial susceptibility testing, version 9.1.
- Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types—induction of transformation by a deoxyribonucleic-acid fraction isolated from Pneumococcus type-III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- Bernstein H, Byers GS, Michod RE. Evolution of sexual reproduction - importance of DNA-repair, complementation, and variation. Amer Nat. 1981;117:537–549. [Google Scholar]
- Bogaert D, Engelen MN, Timmers-Reker AJ, Elzenaar KP, Peerbooms PG, Coutinho RA, et al. Pneumococcal carriage in children in The Netherlands: a molecular epidemiological study. J Clin Microbiol. 2001;39:3316–3320. doi: 10.1128/JCM.39.9.3316-3320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rumke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- Carrolo M, Pinto FR, Melo-Cristino J, Ramirez M. Pherotypes are driving genetic differentiation within Streptococcus pneumoniae. BMC Microbiol. 2009;9:191. doi: 10.1186/1471-2180-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P, Siren J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, McGee L, Rozen DE. Polymorphic competence peptides do not restrict recombination in Streptococcus pneumoniae. Mol Biol Evol. 2010;27:694–702. doi: 10.1093/molbev/msp287. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Linares J, et al. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson CG, Hutchison A, Woodford N, Johnson AP, George RC, Spratt BG. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol. 1999;16:1687–1695. doi: 10.1093/oxfordjournals.molbev.a026082. [DOI] [PubMed] [Google Scholar]
- Ferrandiz MJ, Ardanuy C, Linares J, Garcia-Arenzana JM, Cercenado E, Fleites A, et al. New mutations and horizontal transfer of rpoB among rifampin-resistant Streptococcus pneumoniae from four Spanish hospitals. Antimicrob Agents Chemother. 2005;49:2237–2245. doi: 10.1128/AAC.49.6.2237-2245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujise O, Lakio L, Wang Y, Asikainen S, Chen C. Clonal distribution of natural competence in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2004;19:340–342. doi: 10.1111/j.1399-302x.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- Griffith F. The significance of Pneumococcal types. J Hygiene. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez O, Juan C, Perez JL, Oliver A. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob Agents Chemother. 2004;48:3573–3575. doi: 10.1128/AAC.48.9.3573-3575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L, Beker T. On the evolutionary advantage of fitness-associated recombination. Genetics. 2003;165:2167–2179. doi: 10.1093/genetics/165.4.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Hahn J, Haynes J, Dubnau D. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol. 2001;40:52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Tang J, Connor TR, Corander J. Hyper-recombination, diversity, and antibiotic resistance in Pneumococcus. Science. 2009;324:1454–1457. doi: 10.1126/science.1171908. [DOI] [PubMed] [Google Scholar]
- Havarstein LS, Hakenbeck R, Gaustad P. Natural competence in the genus Streptococcus: Evidence that streptococci can change pherotype by interspecies recombinational exchanges. J Bacteriol. 1997;179:6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Wang JT, Lee WS, Hsueh PR, Shao PL, Chang LY, et al. Serotype competence and penicillin resistance in Streptococcus pneumoniae. Emerg Infect Dis. 2006;12:1709–1714. doi: 10.3201/eid1211.060414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli F, Oggioni MR, Pozzi G. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiol Lett. 2005;252:321–326. doi: 10.1016/j.femsle.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Johnsen PJ, Levin BR. Adjusting to alien genes. Mol Microbiol. 2010;75:1061–1063. doi: 10.1111/j.1365-2958.2010.07075.x. [DOI] [PubMed] [Google Scholar]
- Joloba ML, Kidenya BR, Kateete DP, Katabazi FA, Muwanguzi JK, Asiimwe BB, et al. Comparison of transformation frequencies among selected Streptococcus pneumoniae serotypes. Int J Antimicrob Agents. 2010;36:124–128. doi: 10.1016/j.ijantimicag.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby E, Rainey PB. Exclusion rules, bottlenecks and the evolution of stochastic phenotype switching. Proc Biol Sci. 2011;278:3574–3583. doi: 10.1098/rspb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Cohan FM. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan H, Redfield RJ. Extensive variation in natural competence in Haemophilus influenzae. Evolution. 2009;63:1852–1866. doi: 10.1111/j.1558-5646.2009.00658.x. [DOI] [PubMed] [Google Scholar]
- Mortier-Barriere I, de Saizieu A, Claverys JP, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. The Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- Otto SP. The evolutionary enigma of sex. Am Nat. 2009;174:S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, et al. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- Redfield RJ. Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all. Genetics. 1988;119:213–221. doi: 10.1093/genetics/119.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae. Genetics. 1993;133:755–761. doi: 10.1093/genetics/133.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Do bacteria have sex. Nat Rev Genet. 2001;2:634–639. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- Schoustra S, Rundle HD, Dali R, Kassen R. Fitness-associated sexual reproduction in a filamentous fungus. Curr Biol. 2010;20:1350–1355. doi: 10.1016/j.cub.2010.05.060. [DOI] [PubMed] [Google Scholar]
- Sikorski J, Teschner N, Wackernagel W. Highly different levels of natural transformation are associated with genomic subgroups within a local population of Pseudomonas stutzeri from soil. Appl Environ Microbiol. 2002;68:865–873. doi: 10.1128/AEM.68.2.865-873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmoen H, Knutsen E, Havarstein LS. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Nat Acad Sci USA. 2002;99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. Control of competent state in Pneumococcus by a hormone-like cell product - an example for a new type of regulatory mechanism in bacteria. Nature. 1965;208:155–159. doi: 10.1038/208155a0. [DOI] [PubMed] [Google Scholar]
- Tomasz A, Mosser JL. On the nature of the pneumococcal activator substance. Proc Natl Acad Sci USA. 1966;55:58–66. doi: 10.1073/pnas.55.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends Microbiol. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Vestrheim DF, Gaustad P, Aaberge IS, Caugant DA. Pherotypes of pneumococcal strains co-existing in healthy children. Infect Genet Evol. 2011;11:1703–1708. doi: 10.1016/j.meegid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Vos M. Why do bacteria engage in homologous recombination. Trends Microbiol. 2009;17:226–232. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, Barcus VA, Dowson CG. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DL, Bell JA, Young VB, Wilder SR, Mansfield LS, Linz JE. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology. 2003;149:3603–3615. doi: 10.1099/mic.0.26531-0. [DOI] [PubMed] [Google Scholar]
- Wylie CS, Trout AD, Kessler DA, Levine H. Optimal strategy for competence differentiation in bacteria. PLoS Genet. 2010;6:e1001108. doi: 10.1371/journal.pgen.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Evans BA, Rozen DE. Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc Biol Sci. 2010;277:2991–2999. doi: 10.1098/rspb.2010.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.