Summary
Aerobic glycolysis in transformed cells is an unique metabolic phenotype characterized by a hyperactivated glycolytic pathway even in the presence of oxygen. It is not clear if the onset of aerobic glycolysis is regulated by mitochondrial dysfunction and, if so, what the metabolic windows of opportunity available to control this metabolic switch (mitochondrial to glycolytic) landscape are in transformed cells. Here we report a genetically-defined model system based on the gene-silencing of a mitochondrial complex I subunit, NDUFS3, where we demonstrate the onset of metabolic switch in isogenic human embryonic kidney cells by differential expression of NDUFS3. By means of extensive metabolic characterization, we demonstrate that NDUFS3 gene silencing systematically introduces mitochondrial dysfunction thereby leading to the onset of aerobic glycolysis in a manner dependent on NDUFS3 protein levels. Furthermore, we show that the sustained imbalance in free radical dynamics is a necessary condition to sustain the observed metabolic switch in cell lines with the most severe NDUFS3 suppression. Together, our data reveal a novel role for mitochondrial complex I subunit NDUFS3 in regulating the degree of mitochondrial dysfunction in living cells, thereby setting a “metabolic threshold” for the observation of aerobic glycolysis phenotype within the confines of mitochondrial dysfunction.
Key words: Mitochondrial complex I, NDUFS3, Metabolic switch, Aerobic glycolysis, Reactive oxygen species, Mitochondrial dysfunction
Introduction
Cell transformation often entails alterations in energy metabolism to accommodate adaptation in cellular functions as well as to reprogram the interactions within the cellular environment (Tennant et al., 2009). Emerging trend in metabolic profiling of healthy and disease-related tissues has fundamental implications in not only understanding the metabolic origins of the disease phenotype but also in designing effective strategies for prevention as well as treatment (Brandon et al., 2006; Unwin et al., 2003; Wallace, 1999; Wang et al., 2009). In a normal metabolic paradigm, cellular energy demands are primarily met by oxygen-dependent mitochondrial metabolism of carbohydrates, fatty acids and amino acids in almost all cell types while oxygen-independent glycolysis accounts for only a fraction of cellular energy supply. Global propagation of this bioenergetics landscape, from single cells to tissues to the organs, may critically depend on metabolic thresholds that would in turn determine the limits of proper functioning of various tissues. For instance, in the extreme case of hypoxia, anaerobic glycolysis provides for cellular survival, owing to mitochondrial impairment in the absence of oxygen (Hanahan and Weinberg, 2000; Semenza, 2007). A counter-intuitive biochemical phenotype often observed in transformed cancer cells is their increased propensity to utilize glycolytic pathway even in the presence of oxygen. This observation led to a hypothesis in the 1950s that this feature (“aerobic glycolysis”) could stem from acute mitochondrial dysfunction in the cancer cells. In fact, the glycolytic upregulation in tumors has been shown to correlate with proliferative advantages, increased aggressiveness and poor chemosensitivity (Chen et al., 2007; Gogvadze et al., 2010; Hu et al., 2012; Mathupala et al., 2009; Vander Heiden et al., 2009; Warburg, 1956). Despite the fact that this hypothesis has not been proved unanimously, the feature has been long exploited in clinical imaging modalities such as positron emission tomography where tumor imaging contrast is usually achieved by the preferential uptake of isotope-labeled glucose analogs by the tumors (Fischman, 2008). Based on emerging experimental evidences, the metabolic shift from mitochondrial to glycolytic phenotype is no longer viewed as a compromise in cellular ATP as was originally speculated but more as a basis of connecting the increased glycolytic precursors to biosynthetic pathway for rapid proliferation in cancer cells (Vander Heiden et al., 2009). Our laboratory has been involved in addressing the question of metabolic switch (i.e. mitochondrial pathway to aerobic glycolysis pathway) from the mitochondrial point of view and more specifically by investigating the role of mitochondrial complex I in facilitating the metabolic switch in transformed cancer cells.
Mitochondrial complex I is the critical entry point of mitochondrial electron transport chain. Electron donor, NADH, from the TCA cycle is being oxidized by this huge multi-subunit complex I, which then initiates the electron transport. In this context, it is still not clear if mitochondrial dysfunction has a causative role in the observed metabolic switch or if the observed mitochondrial ramifications are simply a consequence of cellular adaptation to an increased glycolytic phenotype (Bellance et al., 2009; Brandon et al., 2006; Nijtmans et al., 2002; Pelicano et al., 2009). A major bottleneck in addressing this vital question is the absence of a genetically defined model system for systematic modulation of mitochondrial function and to dissect the various modes of achieving the metabolic switch in transformed cells. An immediate significance of such a model system will be the possibility to design effective strategies for controlled perturbation of mitochondrial function as well as the possibility of therapeutic modules for targeting critical sites of mitochondrial network. Earlier, we reported that the protein expression of a mitochondrial complex I precursor subunit NDUFS3 was significantly higher in invasive breast carcinoma tissues obtained from clinical patient specimens as compared with normal breast tissues (Suhane et al., 2011). We further discovered that this aberrant expression of NDUFS3 correlated with hypoxic/necrotic regions of the tumor tissues indicating a correlation with tumor aggressiveness. It is intriguing that one of the subunits in the complex-I chain is deregulated in breast cancer. It is unclear if the subunit NDUFS3 has an independent function in regulating metabolic features of transformed cells. We hypothesized that differential expression of NDUFS3 might yield insights into the role of mitochondrial complex I assembly/function in energy metabolism of transformed cells. To test this hypothesis, we established stable cell lines (transformed human embryonic kidney cells) by shRNA-mediated gene silencing NDUFS3 expression and by characterizing the metabolic phenotypes in these isogenic cells. Our data indicate that differential NDUFS3 expression profoundly affects the overall mitochondrial function and enable the cells to undergo metabolic switch (mitochondrial to aerobic glycolysis). The observed metabolic switch phenotype is reversible in cell lines with modest suppression of NDUFS3 levels whereas it is irreversible in cell lines with the most severe suppression of NDUFS3 levels. Our experiments further point out to a critical role that the reactive oxygen species (ROS) status plays in facilitating this metabolic switch. In the absence of any genetically-defined model system for studying the role of mitochondrial complex I function in transformed cells, this study provides the first such evidence based on a clinically-relevant mitochondrial complex I subunit, NDUFS3 function.
Materials and Methods
Generation of stable cell lines expressing shRNA constructs
Human embryonic kidney (HEK-293T) cells were maintained in DMEM medium (1 g/l glucose) supplemented with 10% fetal bovine serum and antibiotics. The shRNA set (5 clones) containing lentiviral pLK0.1 vectors were purchased from Thermo Scientific (catalog no. RHS4533; Openbiosystems, Brookfield, WI, USA). Each shRNA against NDUFS3 target gene was packaged individually in HEK-293T cells using VSV-G envelop pseudotyping. We also included a pLKO.1 vector containing non-specific shRNA as control (sequence: CAA CAA GAT GAA GAG CAC CAA). The packaged lentiviral particles were concentrated by ultracentrification at 30,000 RPM at 4°C for 90 minutes in a Beckman Coulter Optima LE-80K ultracentrifuge with a SW32 Ti swing bucket rotor. The pelleted viral particles were resuspended in 100 µl of DMEM. The viral titer in transducing units was measured in HEK-293T cells. 10-fold serially diluted viruses were inoculated onto the cells in each well of a 96 well plate. Forty eight hours after inoculation, puromycin antibiotic (5 µg/ml) was added to each well. 3–4 days post antibiotic selection, puromycin resistant cell clones were counted and transducing units per ml of each viral vector was calculated. For establishing cell lines constitutively expressing shRNA, the cell lines were plated in a 48-well plate. Individual viral vectors were inoculated onto cells in each well. The next day viral inoculum was replaced with fresh cell culture medium. Stable cell lines were established followed by puromycin antibiotic selection.
Quantitative real-time PCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to instruction of manufacturer. The amount and the integrity of RNA were assessed by measurement of absorbance at 260 and 280 nm. Total RNA was reverse transcribed into first strand cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer. Quantitative real-time PCR was performed with SsoFast EvaGreen (Bio-Rad Laboratories, Inc.). Primer sequences (Integrated DNA Technologies, Skokie, IL, USA) were as follows: NDUFS3 forward, 5′-GCT GAC GCC CAT TGA GTC TG-3′ and reverse 5′-GGC CAG GTG AAT ATG TTT AG-3′; HK2 forward, 5′-AGA TTG AGA GTG ACT GCC TG-3′ and reverse 5′-ACA GTG CAC ACC TCC TTA AC-3′; RN18S1 rRNA forward, 5′-CTT AGA GGG ACA AGT GGC G-3′ and reverse 5′-ACG CTG AGC CAG TCA GTG TA-3′. CT value was normalized to the CT value of 18S ribosomal 1 (RN18S1) rRNA in the same sample. Relative expression changes of mRNA between shNDUSF3 expressed cells and control shRNA expressed cells were calculated by using ΔΔCT method. The comparisons of mRNA expression levels were conducted using the ΔΔCT method, where the ΔΔCT was the difference in the ΔCT values between two samples and 2−ΔΔCT represents the fold change in mRNA expression.
Immunoblotting and immunofluorescence
Cells were lysed in radio-immunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS) (Sigma–Aldrich, St. Louis, MO, USA) containing protease inhibitor cocktail and phosphatase inhibitor cocktail 2. Cell lysates were centrifuged at 15,000 g for 10 minutes at 4°C, and protein concentrations were determined by DC protein assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Western blot analysis was performed according to the guidelines of Trans-Blot Turbo Transfer system protocol. In brief, 30 µg of total proteins were heated for 5 minutes at 95°C, and then separated on 4–20% SDS-polyacrylamide gel and electrotransferred to nitrocellulose membrane. Visualizing protein band was performed according to the guidelines of SNAP i.d. standard protocol (EMD Millipore, Billerica, MA, USA). Membranes were blocked in 0.25% of non-fat dry milk in Tris-buffered saline-Tween 20 (TBS-T) buffer, and then incubated with the following primary antibodies: 1:2000 mouse monoclonal anti-NDUFS3 (ab110246, Abcam, Cambridge, MA, USA), 1:2000 mouse monoclonal anti-NDUFA9 (ab14713, Abcam, Cambridge, MA, USA), 1:1000 rabbit monoclonal anti-HK2 (no. 2867, Cell Signaling Technology, Inc., Dancers, MA, USA), 1:1000 rabbit polyclonal anti-PKM2 (no. 3198, Cell Signaling Technology, Inc., Dancers, MA, USA). The following four rabbit, polyclonal antibodies were purchased from Abgent Inc., San Diego, CA, USA: NDUFS1 (Ap5678c, 1:100), NDUFS2 (Ap9769c, 1:100), NDUFS4 (Ap6932b, 1:100) and NDUFS8 (Ap12552c, 1:100). The Mitoprofile OxPhos westernblot cocktail (ab110411, Abcam, Cambridge, MA, USA) contained a stoichiometric mixture of five mouse monoclonal antibodies specifically targeting the following subunits: complex I subunit NDUFB8 (20 kDa), complex II subunit (30 kDa), complex III subunit Core 2 (47 kDa), complex IV subunit II (24 kDa) and ATP synthase subunit alpha (55 kDa). After washing with TBS-T, membranes were incubated with peroxidase conjugated secondary antibody in TBS-T. Blots were washed and hybridization signals were measured by enhanced chemiluminescence detection system using Luminata Forte Western HRP Substrate (EMD Millipore, Billerica, MA, USA).
For immunofluorescence experiments, HEK Ctrl and NDUFS3-deficient cell lines were seeded on 12 mm glass coverslips in 24 well plates (80,000–120,000 cells/well) for 24 hours. Next day, cells were washed with PBS twice followed by fixation with 4% paraformaldehye/PBS for 20 minutes at RT and then cells were washed three times with PBS. Fixed cells were heated in antigen-retrieval buffer (100 mM Tris, 5% (w/v) urea, pH 9.5) at 95°C for 10 minutes followed by permeabilization with 0.1% Triton X-100/PBS for 15 minutes at RT and then washed three times with PBS. 10% goat serum in PBS was used for blocking for 1 hour at RT and then overnight incubation was done with mouse NDUFS3 (1:150, catalog no. ab110246; Abcam, Cambridge, MA, USA) and Hexokinase 2 (1:1600, catalog no. 2867; Cell Signaling Technology, Inc., Dancers, MA, USA) at 4°C. Next day, cells were washed three times with 1% goat serum in PBS at RT for 10 minutes each wash and then incubated with appropriate secondary antibody in 10% goat serum in PBS (Rabbit Alexafluor488, 1:400, catalog no. A11034; Alexafluor594, 1:400, catalog no. A11032; Life Technologies (Invitrogen), Grand Island, NY, USA) at RT for 2 hours and then washed three times with 1% goat serum in PBS for 10 minutes each wash at RT. Cells were counterstained with 300 ng/ml DAPI in 1% goat serum (catalog no. D3571; Life Technologies (Invitrogen), Grand Island, NY, USA) for 10 minutes and then washed with PBS for 1 minute. Coverslips were mounted with prolong DAPI (catalog no. P36935; Life Technologies (Invitrogen), Grand Island, NY, USA) as described earlier (Suhane and Ramanujan, 2011). Images were acquired in a Leica SP5 confocal microscope with spectral detector.
Metabolic characterization
Flow cytometry analysis
Flow cytometry analysis of live cells was performed in FACScan flow cytometer (BD Biosciences) with appropriate labeling mix in the cell population: 100 µM 2NBDG (Life Technologies (Invitrogen), Grand Island, NY, USA; glucose uptake), 2.5 µM DCFDA (Life Technologies (Invitrogen), Grand Island, NY, USA, hydrogen peroxide levels), 2.5 µM MitoSox Red (Invitrogen, superoxide levels), 200 nM TMRM (Life Technologies (Invitrogen), Grand Island, NY, USA, mitochondrial membrane potential). Apoptotic cell population was determined by labeling live cells with a polarity sensitive indicator of viability and apoptosis (10 µl/ml working concentration; pSIVA-IANBD, Imgenex, San Diego, CA, USA) 30 minutes in PBS with Ca2+ before flow cytometry.
Live cell kinetics imaging
Real-time metabolic response kinetics was monitored in live cells in Delta T chambers (Bioptechs, Butler, PA, USA) and imaging with a two-photon excitation microscope (Leica SP5 MP) built around a femtosecond pulsed laser (Mai Tai, Spectra Physics, Santa Clara, CA, USA). A typical time lapse consisted of collecting images (acquisition time ∼3 seconds/image) every 20 seconds for about 40 minutes.
Lactate measurements
Typically 4 million cells of control and the shRNA clones were plated in 100 mm dishes in complete medium. Extracellular lactate generation was measured using a commercial Lactate Plus analyzer (Sports Resource, USA) by allowing the cell cultures to grow for 36–48 hours in 100 mm petri dishes in complete DMEM medium. Cumulative lactate content was measured at the end of the incubation period. The lactate concentration of the complete medium without cells was found to be 1.4 mM and this value was subtracted from the experimental values measured for the three cell lines before normalizing with respect to the total cell number in each case and the cell culture duration. The final values of lactate generation rate (millimole/million cells/hour) were used to compare the differences between the control and the NDUFS3-deficient ShRNA clones.
Oxygen consumption measurements
Mitochondrial oxygen consumption [pO2] data were obtained with a clark-type oxygen microelectrode (Strathkelvin Instruments, Scotland) in a closed-cell respirometry design. The probe electrodes were calibrated with 5% sodium thiosulfite solution (0% oxygen) and mammalian ringer solution (100% oxygen ∼230 µmol/l) at 38°C. All measurements were performed either in PBS or cell culture medium. Oxygen consumption rate was then calculated from the raw pO2 data as described earlier (Suhane and Ramanujan, 2011).
Amplex Red assay
Supporting measurements for evaluating ROS status in the HEK control and the NDUFS3-deficient shRNA clones were carried out by using Amplex Red assay (catalog no. A22188; Life Technologies (Invitrogen), Grand Island, NY, USA). Briefly this assay relies on the enzymatic activity of hydrogen peroxide/peroxidase pair measured by the Amplex Red reagent (10-acetyl-3-7-dihydroxyphenoxazine). In the presence of peroxidase, the Amplex Red reagent reacts with H2O2 in a 1:1 stoichiometry to produce the red-fluorescent oxidation product, resorufin (571 nm excitation; 585 nm emission). HEK Control and shRNALow and shRNAHigh cells were plated in 12 well plates (300,000 cells/well). After 36 hours of culture, the cells were trypsinized and resuspended in 20 µl KRPG buffer and then added to the 100 µl of the reaction mixture. The reaction mixture consisted of 50 µM Amplex Red Reagent and 0.1 U/ml HRP in Krebs-Ringer phosphate (145 mM NaCl, 5.7 mM sodium phosphate, 4.86 mM KCl, 0.54 mM CaCl2, 1.22 mM MgSO4, 5.5 mM glucose in deionized water, pH 7.35). After incubating the mixture for 30 minutes at room temperature, resorufin fluorescence was measured in a microplate reader. The system was calibrated with hydrogen peroxide standards prior to the experiment to obtain a standard curve. The final readings were converted to the net generated H2O2 values based on this standard curve. All experiments were done in triplicates.
Mitochondrial complex I enzyme activity assay
Cultured cells were washed three times with cold PBS followed by lysis with sonication in cold PBS and then centrifuged at 10,000 g for 10 minutes at 4°C. Supernatant was collected and protein concentration was determined. Complex I activity was measured by following the decrease in absorbance due to oxidation of NADH to NAD at 360 nm. Absorbance measurements were carried out in a microplate reader (Infinite 200 Pro, Tecan Group Ltd, Switzerland). Instrument linearity was first checked with serial dilution of NADH stocks (50–200 µg NADH) and the data acquisition time was optimized to minimize photobleaching artifacts. The reaction mixture contained 25 mM potassium phosphate, pH 7.4, 5 mM MgCl2, 5 mM KCN, 2.5 mg BSA, 100 µM NADH and 400 µM ubiquinone. 200 µg of cell lysates of HEK control, shRNALow and shRNAHigh cell lines were first added to independent wells in the microplate reader along with the above reagents (total reaction volume 100 µl) and the absorbance kinetics measurement was initiated. After monitoring absorbance from these wells for ∼4 minutes (20-second intervals), 2 µM rotenone was added to the three wells to monitor the rotenone-sensitive absorbance kinetics. Initial rates of NADH oxidation before and after rotenone addition were computed by linear regression analysis. Rotenone-sensitive complex I activity was then calculated by subtracting the rotenone-insensitive component from the total activity (before rotenone addition) and is presented as complex I specific enzyme activity per mg protein per minute. The data presented in this paper are a representative of three independent experiments.
Statistics
Data presented are mean±s.e. from at least three independent experiments unless otherwise mentioned. Statistical significance was estimated based on the Student's t-test (P<0.05).
Results
Generation of HEK cell lines with differential NDUFS3 expression
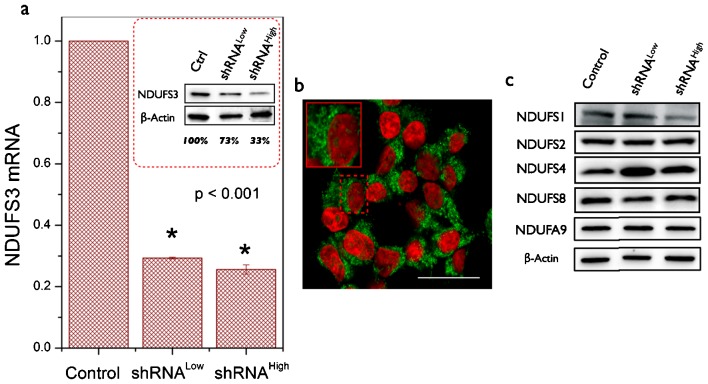
Human embryonic kidney (HEK-293T) cells were transduced with five lentiviral constructs as described earlier. Real-time PCR measurements revealed that all clones except one showed significant suppression of NDUFS3. This observation of partial reduction in NDUFS3 levels in different clones provided an unique opportunity to evaluate the distinct role of NDUFS3 stoichiometry in determining the metabolic phenotypes resulting from the differential suppression of NDUFS3 in the transformed cells. For the sake of clarity and in order to demonstrate the role of NDUFS3 expression level in the observed metabolic phenotype, in this paper we have chosen to present results of metabolic characterization obtained from only two shRNA clones. Fig. 1 shows the relative mRNA levels in the control and the NDUFS3-deficient shRNA clones. Immunoblotting of cell homogenates further revealed that showed robust suppression at the protein level (Control: 100%; shRNALow: 73%; and shRNAHigh: 33%). The observed discrepancy between mRNA levels and the protein levels in the shRNA clones is not uncommon given the fact that starting with a certain mRNA level, multiple regulatory steps could control final protein expression levels. Since the protein level is physiologically relevant for interpreting the metabolic phenotype in cells, all the results presented in this paper refer to these two cell lines with aforementioned suppression levels. High resolution imaging of single cells clearly indicated the mitochondrial distribution of NDUFS3 in all the cell lines as shown in the representative Fig. 1b. Currently available mitochondrial complex I assembly models suggest that NDUFS3 is one of the earliest precursor subunits (Koopman et al., 2010). A preliminary search for potential interacting partners of NDUFS3 yielded two candidates, NDUFS2 and NDUFS8, which are in the same Fe–S cluster of mitochondrial complex I (http://www.hprd.org). Immunoblotting for these two proteins in the control and NDUFS3-deficient cell lines showed no detectable changes due to NDUFS3 suppression. However, two other Fe–S cluster subunit levels (NDUFS1 and NDUFS4) were found to be altered in NDUFS3-deficient cell lines (Fig. 1c). In order to verify that the NDUFS3 suppression did not significantly alter the assembly of later stages of mitochondrial complex I thereby altering the overall function of this complex, we monitored the protein levels of a downstream assembly subunit NADH dehydrogenase [ubiquinone] 1 alpha sub complex subunit 9 (NDUFA9) in the control and the two NDUFS3-deficient cell lines (Fig. 1c). NDUFS3 suppression did not significantly alter the NDUFA9 levels in these cell lines.
Fig. 1. Generation of human embryonic kidney (HEK) cells with selective gene silencing of mitochondrial NDUFS3.
(a) Relative mRNA expression level of NDUFS3 in control and the two NDUFS3-deficient (shRNALow and shRNAHigh) cells as measured by real-time PCR assay. The data are shown after normalizing with the expression of 18S rRNA in each cell line. The inset shows a representative immunoblot result obtained from cellular homogenates. NDUFS3 band intensities were normalized with respect to those of beta actin bands in each case and shown as the percentage of the control cell line. The two clones that are described in this paper had protein expression levels as follows: shRNALow: 73% (27% suppression) and shRNAHigh: 33% (67% suppression). (b) Representative high-resolution confocal image of HEK control cell lines shown here were paraformaldehyde fixed and labeled with primary antibody against NDUFS3 (mouse-NDUFS3) and a fluorescent Alexa594 conjugated secondary antibody (anti-mouse-Alexa594) for immunofluorescence visualization. Similar mitochondrial distribution of NDUFS3 (shown in green) and nuclear DAPI (shown in red) was seen in all the three cell lines. Scale bar = 20 μm. (c) Representative immunoblot results of four Fe–S cluster subunits (NDUFS1, NDUFS2, NDUFS4 and NDUFS8) as well as that of a late assembly subunit NDUFA9 probed in cellular homogenates to verify the effect of NDUFS3 suppression on the related complex I subunit expression levels.
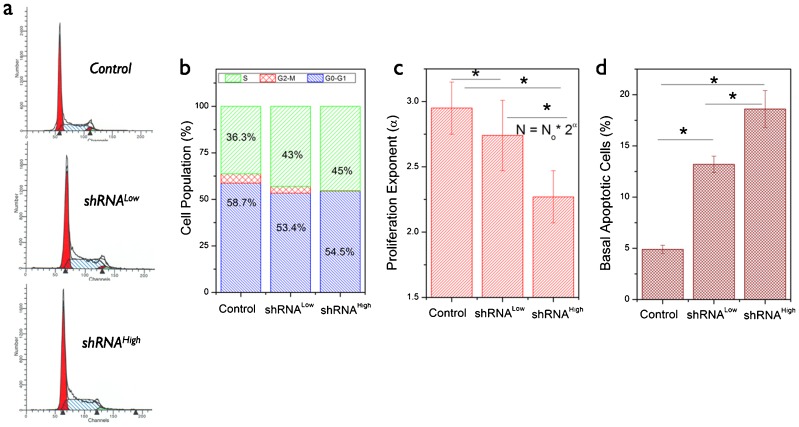
Under the same cell culture conditions, the shRNALow and shRNAHigh cells showed reduced growth rates as compared with the control cells. Cell cycle analysis in these three cell lines is summarized in Fig. 2a,b. We observed that both the stable cells had relatively larger S-phase (DNA synthesis phase) population in comparison with the control cell lines. However, flow cytometry analysis of cell proliferation (using CFSE labeling) in these three cell lines showed a reduction in proliferation exponent (Fig. 2c). We hypothesized that such a reduction in proliferative potential might be related to increase in DNA-synthesis population in the shRNA cells, which could be reconciled by monitoring the basal apoptotic activity in these cells. Using a sensitive apoptosis probe in living cells, we further confirmed that the NDUFS3 shRNALow and shRNAHigh cells have higher basal apoptotic activity (2-fold and 3-fold, respectively) than control cell lines, which accounted for the apoptosis-related increase in S-phase (DNA fragmentation) population.
Fig. 2. Characterization of cell growth and proliferation features in HEK cells.
(a) Flow cytometry based cell cycle analysis measured by labeling the fixed cells with propidium iodide (PI). Cell cycle analysis was performed by ModFit software (BD Biosciences, San Jose, CA, USA) and the cell populations in G0–G1, S and G2–M phases are given in b. (c) Cell proliferation as measured by flow cytometry after labeling with the CFSE probe and culturing for 72 hours. Cell proliferation exponent indicates the differential cell growth/proliferation characteristics in the NDUFS3-deficient cell lines as compared with the control cell lines. (d) Basal apoptotic rate was measured by labeling live cells with pSIVA-IBND that labels the exposed phosphatidyl serine in early stages of apoptosis in each cell line. (* denotes significant difference, P<0.05).
NDUSF3 silencing induces mitochondrial dysfunction in HEK cells
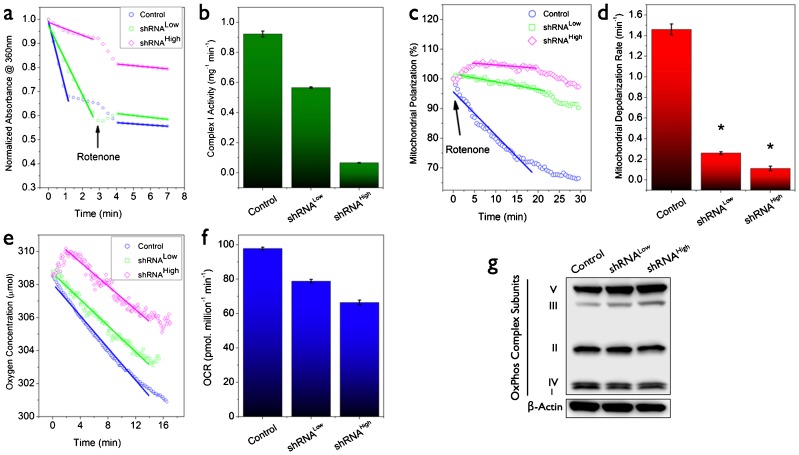
Mitochondrial electron transport chain function critically depends on proper assembly and function of multiple subunits and disrupting this balance by modulating the expression level of one of the precursor subunits such as NDUFS3 is expected to affect the mitochondrial function. Since our goal in this study was to evaluate the effects of such a controlled perturbation of the net mitochondrial function, we developed a set of sensitive assays for evaluating the effects of silencing NDUFS3 expression: expression levels of OxPhos complex systems, mitochondrial depolarization, oxygen consumption and oxygen consumption rate (OCR). Fig. 3a,b show representative mitochondrial complex I activity in the control and the NDUFS3-deficient cell lines. The parental cells displayed relatively higher NADH oxidation rate indicating robust complex I activity whereas NDUFS3 suppression systematically reduced mitochondrial complex I activity in shRNALow and shRNAhigh cells. A sensitive indicator of mitochondrial function is the mitochondrial membrane potential, which reports the potential difference between the mitochondrial matrix and the intermembrane space during metabolic activity. Any perturbation to mitochondrial function will affect the membrane potential and the integrity of mitochondrial function can be tested by the rate of mitochondrial membrane depolarization (collapse of the membrane potential) in living cells. In order to compare the integrity of mitochondrial function in the control and the two stable cells, we labeled the live cells with membrane permeable mitochondrial membrane potential marker (TMRM) as described in Materials and Methods. As established in earlier studies (Genova et al., 1997; Ramanujan and Herman, 2007), we employed mitochondrial complex I inhibitor (100 nM rotenone) as a perturbation reagent, to elicit real-time metabolic responses in TMRM-labeled live cells and monitored the kinetics by two-photon excitation microscopy (Ramanujan and Herman, 2008; Ramanujan et al., 2008; Ramanujan et al., 2005). The rotenone-dependent mitochondrial depolarization kinetics was observed to be drastically reduced in the shRNALow and shRNAHigh cells in contrast to the rapid depolarization kinetics in the control cell lines (Fig. 3c,d). A semi-quantitative analysis of initial decay rates showed that the complex I-inhibition-induced mitochondrial depolarization rates were as follows: control cells (1.44±0.03)×min−1; shRNALow cells (0.28±0.03)×min−1; and shRNAHigh cells (0.10±0.02)×min−1. In order to verify these mitochondrial effects at the overall functional level in live cells, oxygen consumption measurements in NDUFS3 shRNALow and shRNAHigh cells showed that there was a net reduction in oxygen consumption rate (Fig. 3e,f) in these cell lines as compared with the control cells. These results, together with the mitochondrial membrane depolarization data, further confirmed that NDUFS3 silencing induced mitochondrial dysfunction in live cells in a manner dependent on the NDUFS3 expression level. Fig. 3g shows immunoblotting result where we simultaneously monitored the representative subunits of all the five complexes of the mitochondrial oxidative phosphorylation (OxPhos) system. NDUFS3 suppression was correlated with a concomitant upregulation of complex III and V subunits revealing compensatory effects and/or mitochondrial stress.
Fig. 3. Measurement of mitochondrial parameters in the NDUFS3-deficient cell lines.
(a) Mitochondrial complex I enzyme activity as measured by the rate of oxidation of NADH (substrate) to NAD+ by the complex I enzyme in the total cell lysate. Absorbance at 340 nm (NADH absorption maximum) was monitored after adding the substrate (NADH) in the presence of the cell lysate and after ∼4 minutes, 2 µM rotenone (complex I inhibitor) was added to monitor the rotenone-sensitive NADH oxidation rates. Complex I specific NADH oxidation rates are summarized in b. In consensus with the mitochondrial dysfunction scenario, the shRNA clones showed much delayed mitochondrial membrane depolarization rates as compared with the control cell lines under the perturbation of mitochondrial complex I inhibitor (100 nM rotenone). Rotenone-sensitive mitochondrial depolarization is an established means of monitoring mitochondrial integrity or mitochondrial dysfunction as in the case of the NDUFS3-deficient cell lines (c,d). Concomitant reduction in overall mitochondrial oxygen consumption in the NDUFS3-deficient cells further confirm that these cell lines have impaired mitochondrial function induced by NDUFS3 suppression (e,f). Expression levels of representative subunits from each of the mitochondrial oxidative phosphorylation (OxPhos) complexes in control HEK cells and the NDUFS3-deficient isogenic cell lines as assayed by using a OxPhos cocktail described in Materials and Methods. As can be seen there was a significant upregulation of complex III and V subunits in the shRNA clones indicating compensatory adaptive increase in mitochondrial OxPhos subunits and/or mitochondrial stress (g).
NDUFS3-silencing induced mitochondrial dysfunction exacerbates aerobic glycolysis phenotype
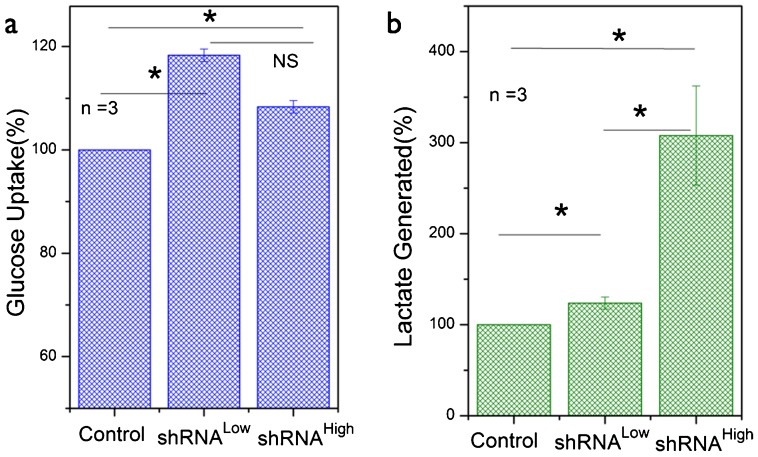
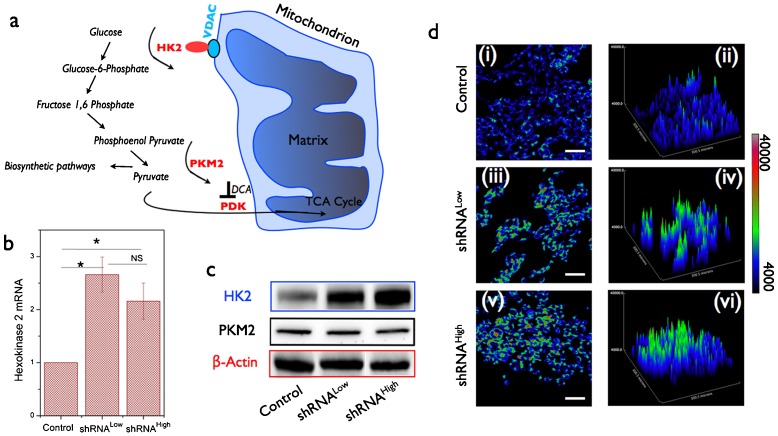
As described in the earlier section, there is still no clarity on the causative role of mitochondrial dysfunction in the aerobic glycolysis phenotype observed in transformed cells. In the present study where we have established a genetically defined model system of NDUFS3 silencing, we sought out to evaluate the role of resulting mitochondrial dysfunction in the observation of aerobic glycolysis. Fig. 4a shows the average (three independent experiments) glucose uptake rate in the three cell lines under investigation. Both the shRNALow and shRNAHigh cells displayed a modest increase in glucose uptake as compared with the control cells. An established hallmark of aerobic glycolysis is the increased rate at which lactate is generated owing to the fact that a larger proportion of pyruvate is converted to lactate rather than entering the mitochondrial TCA cycle (Suhane and Ramanujan, 2011). As can be seen from Fig. 4b, shRNAHigh cell line displayed nearly 3-fold higher lactate generation rate as compared with control cell lines while shRNALow clone showed only a modest increase. This is an intriguing finding given the fact that both the NDUFS3-deficient cell lines displayed a reduction in mitochondrial function. It is important to note here that, besides glucose metabolism, the cellular lactate generation could result from other metabolic pathways such as glutaminolysis. Since the major focus of this study is to address the role of mitochondrial dysfunction in the onset of aerobic glycolysis, no further attempts were taken to characterize these alternate metabolic pathways and we utilized lactate generation rate as a surrogate marker of aerobic glycolysis phenotype. From the molecular point of view, earlier studies have shown that aerobic glycolysis phenotype was accompanied by upregulation of a key glycolytic enzyme hexokinase 2 (HK2) that is mitochondrial outer membrane-bound in almost all cell types (Mathupala et al., 2009). HK2 catalyzes the first committed step of glycolysis (Fig. 5a). Real-time PCR and immunofluorescence measurements revealed an increase in this enzyme level in shRNALow and shRNAHigh cells, both at the mRNA and the protein levels supporting the observed increase in aerobic glycolysis phenotype (Fig. 5b–d). A possible reason for an increase in aerobic glycolysis in transformed cells is to divert the glycolytic precursors to biosynthetic pathway by inhibiting the conversion of phosphoenol pyruvate to pyruvate (catalyzed by M2 isoform of pyruvate kinase, PKM2) (Christofk et al., 2008) (Fig. 5a). In our case, we did not find any observable difference in PKM2 levels in all the three lines studied, thereby confirming no direct role in biosynthetic pathway (Fig. 5c). This is in accordance with the cell cycle/proliferation data (Fig. 2), further demonstrating that the observed increase in aerobic glycolysis phenotype did not give the NDUFS3-deficient cells any proliferative advantage.
Fig. 4. Mitochondrial dysfunction in NDUFS3-deficient cell lines induces aerobic glycolysis in a NDUFS3-dependent manner.
(a) Average glucose uptake rate and (b) average lactate generation rate in the HEK control and NDUFS3-deficient cells demonstrate the onset of aerobic glycolysis in the latter. Data shown are mean±s.e. from three independent experiments and were normalized respect to the values for the control cell lines. Statistical significance was estimated for dataset with a difference in mean values is within P<0.05 threshold. (* denotes significant difference, P<0.05 and NS denotes Not Significant).
Fig. 5. Aerobic glycolysis phenotype observed in NDUFS3-deficient cell lines is substantiated by aberrant hexokinase 2 expression.
(a) Schematic of critical molecular players involved in routing glycolytic oxidation product pyruvate to mitochondrial metabolism. Hexokinase 2 (HK2) catalyzes the first committed step of glycose phosphorylation thereby initiating the conversion of 6-carbon glucose to 3-carbon pyruvate. The terminal step of pyruvate formation is catalyzed by the rate-limiting enzyme, pyruvate kinase M2 (PKM2). Hyperactive PKM2 could inhibit the pyruvate formation thereby diverting the glycolytic precursors to biosynthetic pathway. (b) Relative mRNA levels of hexokinase 2 were found to be significantly higher in both the shRNALow and shRNAHigh cell lines, which is further confirmed at the protein level (c). (* denotes significant difference, P<0.05 and NS denotes Not Significant). Immunofluorescence labeling of cells plated on glass coverslips was carried out with anti-HK2 antibody and Alexa 488-tagged secondary antibody and subsequent fluorescence imaging of these cells revealed a clear upregulation of HK2 in NDUFS3-deficient cell lines (d(i),(iii),(v)). Also shown are the surface plots demonstrating a drastic increase in HK2 expression levels in NUDFS3-deficient cell lines in accordance with immunoblotting results (d(ii),(iv),(vi)). No significant difference was observed in the PKM2 level (c) thereby suggesting no differences in biosynthetic machinery are operative in the NDUFS3-deficient cell lines in comparison with the control HEK cells. Scale bars = 20 μm.
Aerobic glycolysis observed in the NDUFS3-deficient cells is regulated by the ROS imbalance
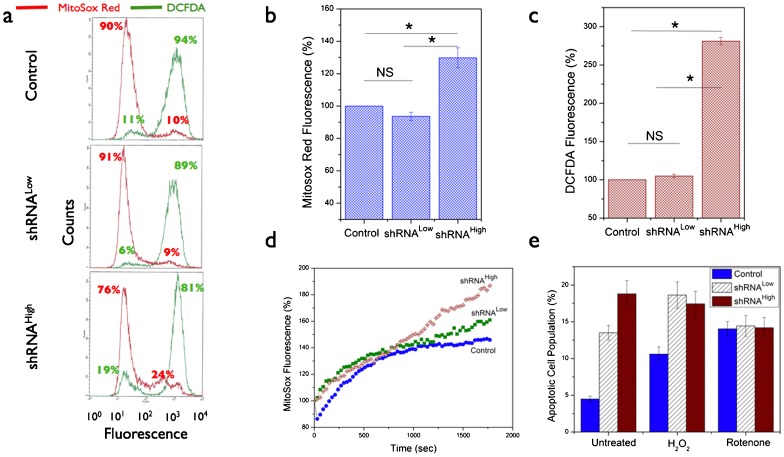
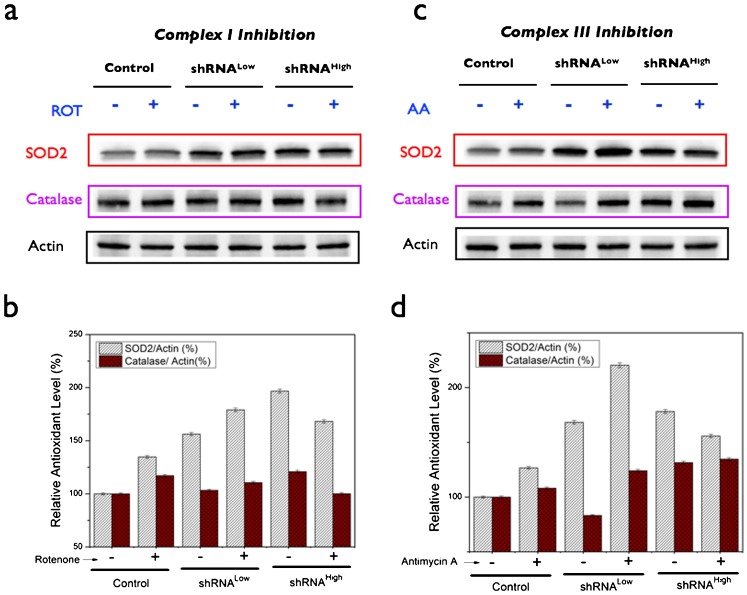
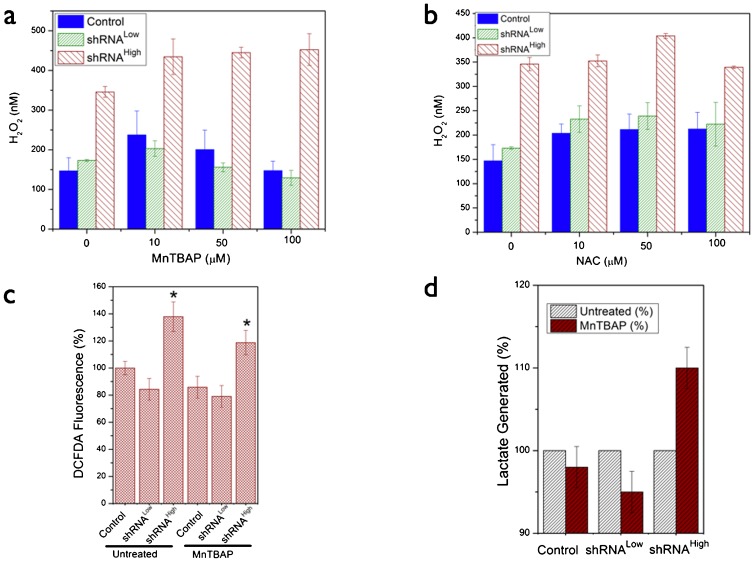
In order to further characterize the metabolic effects of NDUFS3-silencing and to understand the putative mechanisms of the observed metabolic switch (mitochondrial to aerobic glycolysis), we carried out extensive measurements of free radical dynamics in these cell lines as summarized in Figs 6–8. Flow cytometry analysis of basal status of reactive oxygen species (ROS) indicated that the shRNAHigh cells displayed a significantly higher population (24%) of cells having a higher superoxide signal as compared with the control and the shRNALow cells (∼10%). Superoxide-selective (Mitosox labeling) and hydrogen peroxide-selective (DCFDA labeling) signals were significantly higher only in the shRNAHigh cells but not in shRNALow cells (Fig. 6b,c). This was further substantiated by real-time metabolic responses in live cells as measured by the two-photon excitation imaging of superoxide generation after a brief exposure to mitochondrial complex I inhibitor (100 nM rotenone). As shown in Fig. 6d, shRNAHigh cells displayed a sustained increase in superoxide-selective Mitosox red signal in contrast to the control and shRNALow cells. An increase in net apoptotic cells was observed upon exposure of the cells to exogenous oxidative stress (Fig. 6e). In order to further understand if the observed metabolic responses to oxidative stress is reflected in the antioxidant status of the cells, we measured the levels of two major antioxidant enzymes, SOD2 (mitochondrially localized superoxide dismutase 2 catalyzing the conversion of superoxide to hydrogen peroxide) and catalase (cytosolic enzyme converting hydrogen peroxide to hydroxyl radical) under the conditions of complex I inhibition (100 nM rotenone, 48 hours) and complex III inhibition (100 nM Antimycin A, 48 hours). The rationale for this design was to investigate the role of two known sites of ROS generation (complex I and complex III) in contributing to the ROS phenotype in the three cell lines (Ramanujan and Herman, 2007). Fig. 7 summarizes these results. shRNALow and shRNAHigh cells displayed a higher basal levels of antioxidant status as compared with the control cell lines indicating a constitutive mitochondrial dysfunction stemming from NDUFS3 suppression in these cell lines. However, under the various perturbation conditions as described above, shRNALow and shRNAHigh cells displayed opposite trends of antioxidant status (Fig. 7b,d). Integrity of mitochondrial matrix and electron transport chain requires that in response to oxidative stress, antioxidant machinery will correspondingly increase in order to nullify the stress as was observed in the control and the shRNALow cells. On the other hand, shRNAHigh cells displayed an opposite trend. This anomalous feature can be reasoned as either due to the irreversible loss of mitochondrial integrity as a whole and/or degradation of antioxidant proteins in these cell lines. To test these possibilities, we treated the cells with pharmacological agents that mimic cellular antioxidants (Fig. 8). Cell permeable superoxide mimetic (MnTBAP) treatment indeed had an effect on reducing the net cellular ROS signal in a concentration-dependent manner as measured by Amplex Red assay (Fig. 8a) and flow cytometry analysis (Fig. 8d) in the control and the shRNALow cells. On the other hand, this antioxidant mimetic did not have any significant effect on the ROS levels of shRNAHigh cells. Furthermore, cytosolic glutathione precursor, N-acetyl cysteine (NAC) did not show any concentration-dependent reduction in ROS level (Fig. 8b) indicating the source of oxidative stress is predominantly of mitochondrial origin. More importantly, the superoxide mimetic MnTBAP treatment was shown to reduce the net lactate generation in control and the shRNALow cells but not in shRNAHigh cells further supporting the conclusion that sustained ROS is not only a sufficient but a necessary condition for the observation of aerobic glycolysis in the shRNAHigh cells. Together these data point to a scenario in shRNALow cells where the mitochondrial dysfunction induced by the partial silencing of NDUFS3 was below a metabolic threshold necessary to cause a “constitutively active” metabolic switch from mitochondrial to aerobic glycolysis. On the other hand, in shRNAHigh cells, a greater loss of NDUFS3 led to a, plausibly, irreversible status of mitochondrial dysfunction where sustained ROS imbalance was just not sufficient to exacerbate the aerobic glycolysis phenotype but also necessary to sustain this phenotype even in the presence of exogenous antioxidant mimetics.
Fig. 6. Basal reactive oxygen species (ROS) data in the NDUFS3-deficient cell lines.
(a) Representative flow cytometry histograms showing the basal superoxide (red lines) and hydrogen peroxide (green lines) signals as measured by labeling the live cells with superoxide selective Mitosox Red (2.5 µM, 20 minutes, 37°C) and hydrogen peroxide-selective DCFDA (2.5 µM, 20 minutes, 37°C) probes. (b,c) Statistical analyses of steady-state ROS status in the control and NDUFS3 deficient cell lines obtained from at least three independent experiments (P<0.001 and NS denotes Not Significant). (d) Live cell kinetics imaging analysis of real-time superoxide generation rate measured in live cells pre-labeled with 2.5 µM Mitosox Red. The time-lapse imaging was initiated after adding 100 nM rotenone (mitochondrial complex I inhibitor) to monitor the superoxide generation kinetics. Fluorescence values in each case were normalized with respect to the fluorescence at t = 0 seconds and presented as percentage change in fluorescence for the three cell lines studied. These kinetic profiles were representative of three independent experiments under identical imaging conditions. (e) Apoptotic cell population determined under exogenous generic ROS inducer, hydrogen peroxide (25 nM H2O2, 14 hours, 37°C) showed similar response trend in the control and shRNALow cell lines whereas the shRNAHigh cell lines had constitutively higher apopotic cell death even without treatment further substantiating the effects of sustained oxidative stress in these cell lines. Mitochondrial complex I inhibitor (100 nM rotenone, 14 hours, 37°C) was also used in this experiment to demonstrate that apoptotic rates could be regulated by ROS status independent of the source of oxidative stress.
Fig. 7. Antioxidant status in the control and the NDUFS3-deficient cell lines under various perturbation conditions.
Basal antioxidant levels were measured under three specific conditions of ROS induction. Mitochondrial complex I and complex III sites are known to be critical for mitochondrial ROS generation and inhibition of these complexes is anticipated to elicit antioxidant responses in cells with intact mitochondrial function. Antioxidant enzyme levels (superoxide dismutase, SOD2 and catalase) measured under (a,b) mitochondrial complex I inhibition (100 nM rotenone, 48 hours) and (c,d) mitochondrial complex III inhibition (100 nM Antimycin A, 48 hours). Please refer to the main text and Fig. 5a for more details.
Fig. 8. Effects of exogenous antioxidant mimetics on ROS status in the control and NDUFS3-deficient HEK cells.
(a,b) Representative ROS mesurements as assayed by Amplex Red reagent, which reports the net hydrogen peroxide signals. Cells were treated with denoted concentrations of superoxide mimetic MnTBAP and glutathione precursor, N-acetyl cysteine for 36 hours. Corroborative flow cytometry measurements are shown in c. (d) Average (3 independent experiments) lactate generation rate measured in the control and NDUFS3-deficient cell lines in the presence of antioxidant mimetic 10 µM MnTBAP to determine the causative role of ROS status in influencing the observed metabolic switch in the NDUFS3-deficient cells. Lactate values in untreated case were the reference values (100%) for each cell line.
Discussion
The major goal of this study was to establish a genetically defined model system for studying the role of mitochondrial complex I in modulating metabolic switch in transformed cells. Alterations in energy metabolism have been known to be the common denominator for various disease models including diabetes, obesity, cancer and aging. Owing to the central importance of mitochondrial metabolism in cell survival and apoptosis, any fundamental insight into the various mechanisms by which mitochondrial dysfunction dictates the onset of disease phenotype can have high significance. In this context, the present study addresses a fundamental question of how subtle mitochondrial dysfunctions have causative role in the metabolic switch in transformed cells. As the term “switch” implies, it will be intriguing to understand if the switch from predominantly mitochondrial to predominantly glycolytic, even in the presence of aerobic conditions, can be reversed or reprogrammed. Answering this question has huge clinical potential as well, since it is then possible to devise strategies to modulate the metabolic switch and plausibly revert the so-called “dysfunctional mitochondria” to normal-like mitochondrial status. We believe that the present study is an important step towards this direction where we have unraveled the role of a mitochondrial complex I subunit NDUFS3 expression in regulating the ROS-mediated aerobic glycolysis in transformed isogenic HEK cells.
First of all, our extensive metabolic characterization of the NDUFS3-deficient cells revealed that it is possible to systematically induce mitochondrial dysfunction by gene silencing NDUFS3 expression, which is known to be one of the precursor subunits in the mitochondrial complex I assembly (Janssen et al., 2006; Koopman et al., 2010; Vogel et al., 2007). Currently efforts are underway in our laboratory for identifying other critical subunits in the complex I by which it is possible to exert control on the mitochondrial complex I assembly process. Although the dependence of mitochondrial function on this critical subunit (NDUFS3) is intuitive, there have been no attempts to establish a model system without the complications of hypoxia, as is done in the present study. Furthermore, the mitochondrial dysfunction observed in the NDUFS3-deficient cells seems to exacerbate the aerobic glycolysis thereby confirming the role of mitochondrial complex I modulation in metabolic switch in the transformed cells. Even though NDUFS3 suppression did not directly alter the potential NDUFS3-interacting partners (NDUFS2 and NDUFS8) as well as the late assembly subunit NDUFA9 (Fig. 1c), we can not exclude the possibility that the observed metabolic phenotype in NDUFS3-deficient cell lines could stem from a compromised complex I assembly. Our observations of reduced complex I enzyme activity (Fig. 3a) and alterations in other Fe–S cluster subunits (Fig. 1c) support this possibility. On the other hand, despite the fact that the two cell lines (shRNALow and shRNAHigh) investigated in this study displayed similar mitochondrial dysfunction in terms of complex I enzyme activity, oxygen consumption, and the OxPhos subunit expression, the exacerbated aerobic glycolysis phenotype was only observed to be permanent (irreversible) in the cell lines with most sustained ROS imbalance (shRNAHigh cells). This points to an important fact that there may exist a “metabolic threshold” for NDUFS3 stoichiometry in the overall mitochondrial complex I assembly/function. One interesting approach to delineate the distinct roles of metabolic regulatory function and the complex I assembly function of NDUFS3 (and other pertinent complex I subunits) will be by means of rationally-designed small molecule probes specifically targeted to NDUFS3 assembly sites. By this approach, one could exert a controlled perturbation of complex I assembly process and monitor its effects on the metabolic switch phenotype. Currently available crystal structure models are limited to prokaryotic complex I. In the absence of any human complex I structure, many groups have modeled the human subunit based on the available prokaryotic structure with the caveat that human and prokaryotic complex I vary widely in subunit composition, number and complexity (Koopman et al., 2010; Vogel et al., 2007). A more detailed investigation of the human complex I crystal structure, in future, could lead to a better understanding of NDUFS3 and other critical complex I subunits in human health and disease phenotypes.
Finally, our study has clearly demonstrated that the observed mitochondrial dysfunction is a sufficient condition for the observation of aerobic glycolysis phenotype but the necessary condition for sustaining the aerobic glycolysis (or metabolic switch) in these cell lines is the sustained ROS imbalance as shown by the assays described in this paper. Our data are in accordance with a recent study where it was demonstrated that reactive oxygen species can promote aerobic glycolysis in chronic leukemia cell lines (Lu et al., 2012). In addition to establishing the basis for the ROS-mediated aerobic glycolysis, our study highlights a plausible answer to the original question namely the maneuverability of metabolic switch phenotype within the limits of mitochondrial dysfunction. An alternate way to consolidate these observations will be by development of a model system with inducible expression of NDUFS3 (and other critical subunits of mitochondrial complex I) to precisely determine the molecular basis of mitochondrial reprogramming in transformed cells. In conclusion, we believe that we have laid a novel foundation for studying mitochondrial complex I dysfunction in transformed cells. Since mitochondrial metabolism is a fundamental pathway in almost all cell types, we envision that the ideas described in this paper can be easily applied to a variety of metabolic disorders that encompass mitochondrial dysfunction.
Acknowledgments
We gratefully acknowledge financial support from Susan G. Komen for the Cure foundation (Career Catalyst Research Award no. KG090239), National Cancer Institute/National Institutes of Health (ARRA Stimulus Award no. R21-CA124843), Donna and Jesse Garber Foundation award and American Cancer Society Inc. (Research Scholar Award RSG-12-144-01-CCE) (all to V.K.R). We thank Dr Bruce Gewertz and Dr Leon Fine for their intramural support and encouragement.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Bellance N., Lestienne P., Rossignol R. (2009). Mitochondria: from bioenergetics to the metabolic regulation of carcinogenesis. Front. Biosci. 14, 4015–4034 10.2741/3509 [DOI] [PubMed] [Google Scholar]
- Brandon M., Baldi P., Wallace D. C. (2006). Mitochondrial mutations in cancer. Oncogene 25, 4647–4662 10.1038/sj.onc.1209607 [DOI] [PubMed] [Google Scholar]
- Chen Z., Lu W., Garcia–Prieto C., Huang P. (2007). The Warburg effect and its cancer therapeutic implications. J. Bioenerg. Biomembr. 39, 267–274 10.1007/s10863-007-9086-x [DOI] [PubMed] [Google Scholar]
- Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
- Fischman A. J. (2008). PET imaging of brain tumors. Cancer Treat. Res. 143, 67–92 10.1007/978-0-387-75587-8_3 [DOI] [PubMed] [Google Scholar]
- Genova M. L., Bovina C., Marchetti M., Pallotti F., Tietz C., Biagini G., Pugnaloni A., Viticchi C., Gorini A., Villa R. F.et al. (1997). Decrease of rotenone inhibition is a sensitive parameter of complex I damage in brain non-synaptic mitochondria of aged rats. FEBS Lett. 410, 467–469 10.1016/S0014-5793(97)00638-8 [DOI] [PubMed] [Google Scholar]
- Gogvadze V., Zhivotovsky B., Orrenius S. (2010). The Warburg effect and mitochondrial stability in cancer cells. Mol. Aspects Med. 31, 60–74 10.1016/j.mam.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2000). The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu W., Chen G., Wang P., Chen Z., Zhou Y., Ogasawara M., Trachootham D., Feng L., Pelicano H.et al. (2012). K-rasG12V transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 22, 399–412 10.1038/cr.2011.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R. J., Nijtmans L. G., van den Heuvel L. P., Smeitink J. A. (2006). Mitochondrial complex I: structure, function and pathology. J. Inherit. Metab. Dis. 29, 499–515 10.1007/s10545-006-0362-4 [DOI] [PubMed] [Google Scholar]
- Koopman W. J., Nijtmans L. G., Dieteren C. E., Roestenberg P., Valsecchi F., Smeitink J. A., Willems P. H. (2010). Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid. Redox Signal. 12, 1431–1470 10.1089/ars.2009.2743 [DOI] [PubMed] [Google Scholar]
- Lu W., Hu Y., Chen G., Chen Z., Zhang H., Wang F., Feng L., Pelicano H., Wang H., Keating M. J.et al. (2012). Novel role of NOX in supporting aerobic glycolysis in cancer cells with mitochondrial dysfunction and as a potential target for cancer therapy. PLoS Biol. 10, e1001326 10.1371/journal.pbio.1001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala S. P., Ko Y. H., Pedersen P. L. (2009). Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 19, 17–24 10.1016/j.semcancer.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans L. G., Artal S. M., Grivell L. A., Coates P. J. (2002). The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci. 59, 143–155 10.1007/s00018-002-8411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H., Lu W., Zhou Y., Zhang W., Chen Z., Hu Y., Huang P. (2009). Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res. 69, 2375–2383 10.1158/0008-5472.CAN-08-3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujan V. K., Herman B. A. (2007). Aging process modulates nonlinear dynamics in liver cell metabolism. J. Biol. Chem. 282, 19217–19226 10.1074/jbc.M700572200 [DOI] [PubMed] [Google Scholar]
- Ramanujan V. K., Herman B. A. (2008). Nonlinear scaling analysis of glucose metabolism in normal and cancer cells. J. Biomed. Opt. 13, 031219 10.1117/1.2928154 [DOI] [PubMed] [Google Scholar]
- Ramanujan V. K., Zhang J. H., Biener E., Herman B. (2005). Multiphoton fluorescence lifetime contrast in deep tissue imaging: prospects in redox imaging and disease diagnosis. J. Biomed. Opt. 10, 051407 10.1117/1.2098753 [DOI] [PubMed] [Google Scholar]
- Ramanujan V. K., Jo J. A., Cantu G., Herman B. A. (2008). Spatially resolved fluorescence lifetime mapping of enzyme kinetics in living cells. J. Microsc. 230, 329–338 10.1111/j.1365-2818.2008.01991.x [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2007). HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J. Bioenerg. Biomembr. 39, 231–234 10.1007/s10863-007-9081-2 [DOI] [PubMed] [Google Scholar]
- Suhane S., Ramanujan V. K. (2011). Thyroid hormone differentially modulates Warburg phenotype in breast cancer cells. Biochem. Biophys. Res. Commun. 414, 73–78 10.1016/j.bbrc.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhane S., Berel D., Ramanujan V. K. (2011). Biomarker signatures of mitochondrial NDUFS3 in invasive breast carcinoma. Biochem. Biophys. Res. Commun. 412, 590–595 10.1016/j.bbrc.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant D. A., Durán R. V., Boulahbel H., Gottlieb E. (2009). Metabolic transformation in cancer. Carcinogenesis 30, 1269–1280 10.1093/carcin/bgp070 [DOI] [PubMed] [Google Scholar]
- Unwin R. D., Craven R. A., Harnden P., Hanrahan S., Totty N., Knowles M., Eardley I., Selby P. J., Banks R. E. (2003). Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3, 1620–1632 10.1002/pmic.200300464 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R. O., Dieteren C. E., van den Heuvel L. P., Willems P. H., Smeitink J. A., Koopman W. J., Nijtmans L. G. (2007). Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 282, 7582–7590 10.1074/jbc.M609410200 [DOI] [PubMed] [Google Scholar]
- Wallace D. C. (1999). Mitochondrial diseases in man and mouse. Science 283, 1482–1488 10.1126/science.283.5407.1482 [DOI] [PubMed] [Google Scholar]
- Wang J., Bai L., Li J., Sun C., Zhao J., Cui C., Han K., Liu Y., Zhuo X., Wang T.et al. (2009). Proteomic analysis of mitochondria reveals a metabolic switch from fatty acid oxidation to glycolysis in the failing heart. Sci. China C Life Sci. 52, 1003–1010 10.1007/s11427-009-0140-2 [DOI] [PubMed] [Google Scholar]
- Warburg O. (1956). On respiratory impairment in cancer cells. Science 124, 269–270. [PubMed] [Google Scholar]