Summary
In animal cells the centrosome is commonly viewed as the main cellular structure driving microtubule (MT) assembly into the mitotic spindle apparatus. However, additional pathways, such as those mediated by chromatin and augmin, are involved in the establishment of functional spindles. The molecular mechanisms involved in these pathways remain poorly understood, mostly due to limitations inherent to current experimental systems available. To overcome these limitations we have developed six new Drosophila cell lines derived from Drosophila homozygous mutants for DSas-4, a protein essential for centriole biogenesis. These cells lack detectable centrosomal structures, astral MT, with dispersed pericentriolar proteins D-PLP, Centrosomin and γ-tubulin. They show poorly focused spindle poles that reach the plasma membrane. Despite being compromised for functional centrosome, these cells could successfully undergo mitosis.
Live-cell imaging analysis of acentriolar spindle assembly revealed that nascent MTs are nucleated from multiple points in the vicinity of chromosomes. These nascent MTs then grow away from kinetochores allowing the expansion of fibers that will be part of the future acentriolar spindle. MT repolymerization assays illustrate that acentriolar spindle assembly occurs “inside-out” from the chromosomes. Colchicine-mediated depolymerization of MTs further revealed the presence of a functional Spindle Assembly Checkpoint (SAC) in the acentriolar cells. Finally, pilot RNAi experiments open the potential use of these cell lines for the molecular dissection of anastral pathways in spindle and centrosome assembly.
Key words: Centriole, Mitosis, Spindle, Cytoskeleton, Drosophila, Anastral, Cell lines
Introduction
Cell division is an inherent and essential process in living organisms. Different strategies to partition the genetic material and the cytoplasm into daughter cells have emerged during evolution. In prokaryotes, chromosome separation is mostly based on the attachment of DNA to the plasma membrane. A major evolutionary innovation of eukaryotes for chromosome segregation relies on the assembly of a complex bipolar structure, the mitotic spindle, and the linkage of a specialized region of chromosomes, the kinetochore, to spindle microtubules (MTs), allowing the migration of genetic material to spindle poles.
In metazoans the spindle poles are organized around a pair of small organelles, known as centrioles. These structures were probably initially used as basal bodies in order to generate flagellar axonemes allowing cells to have essential functions such as polarity, sensation, and motion. Only later, at the onset of multi-cellular organization, centrioles were recruited to build true centrosomes functioning as cytoskeleton organizers and cell division organelles. Some phylum such as Metaphyta subsequently lost centrioles. This scenario of evolution has been proposed by Azimzadeh and Bornens (Azimzadeh and Bornens, 2004; Bornens and Azimzadeh, 2007), and subsequently consolidated upon comparative phylogenetic analysis of centrosomal proteins (Azimzadeh and Marshall, 2010; Carvalho-Santos et al., 2010; Hodges et al., 2010). Thus, ancestral eukaryotic cells would rely on functions independent of centrioles for mitotic spindle formation. Remarkably, such primitive cell division mechanisms have been preserved during evolution even after centrioles were co-opted to take part of mitosis.
A recurrent question is the definition of the precise role of centrioles during mitosis in eukaryotes. Centrioles are not essential in at least some eukaryotes, such as most seed plants, which lack this organelle. However, only very few cases are known of natural absence of centrioles in dividing metazoan cells, the most documented situation being female meiosis in the vast majority of animal species. Recently it has been discovered the natural absence of centrioles in dividing cells of the Planarian flatworm Schmidtea mediterranea (Azimzadeh et al., 2012). In addition, mutants affecting centrosome function (Megraw et al., 2001) or centriole duplication (Basto et al., 2006; Bettencourt-Dias et al., 2005) are viable in Drosophila. Even a Drosophila cell line lacking centrioles has been previously established, although the origin of this peculiarity remains obscure (Debec et al., 1982). Finally, in mammalian cells, ablation or destruction of centrioles by laser, microdissection or injection of function-blocking antibodies also support that centrioles are not required for mitotic spindle assembly (Debec et al., 2010; Varmark, 2004; Wilson, 2008). The additional mechanisms allowing organization of a mitotic spindle in the absence of centrosome are beginning to be understood. Studies in Xenopus oocytes extracts revealed that MTs can be nucleated around chromosomes and the bipolar spindle can self-organize through the action of molecular motors like kinesins and dynein (Gatlin and Bloom, 2010; Karsenti and Vernos, 2001; Walczak et al., 1998). Spindles poles are consolidated by cross-linker proteins such as NuMA (Merdes et al., 1996; Merdes et al., 2000) and TPX2 (Wittmann et al., 2000). The small GTPase Ran mediates MT nucleation from chromosomes. Ran is bound to GTP at the surface of the chromosomes and then diffuses in the cytoplasm forming a gradient that spatially regulates MT nucleation and organization (Caudron et al., 2005; Walczak and Heald, 2008). This chromatin/RanGTP pathway appears also to be active in mitotic somatic cells (Kaláb et al., 2006; Ciciarello et al., 2007).
Another contribution to MT nucleation can be found inside the spindle itself. It was already known that a fraction of the γ-tubulin pool is located in the spindle and not only at the centrosomes (Lajoie-Mazenc et al., 1994). Recent studies suggest that many MTs are actually nucleated inside the spindle, producing a MT amplification mechanism for spindle assembly (Lüders et al., 2006; Mahoney et al., 2006; Lüders and Stearns, 2007). This is mediated by augmin, a complex of 8 proteins, which recruit γTuRC along existing spindle MTs and leads to the formation of new MTs, increasing the speed and stability of spindle assembly (Goshima et al., 2007; Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009; Zhu et al., 2009).
It is important to note that these pathways are not really alternative, i.e. they are not backup mechanisms used by cells to compensate for the absence of centrioles, but that they co-exist in a normal cell to accelerate spindle assembly (Lüders and Stearns, 2007; O'Connell and Khodjakov, 2007). The molecular mechanisms responsible for these pathways remain poorly understood and they merit further investigation to discover new partners or even new pathways as they are deregulated in tumor cells. It is well known that most solid tumor cells exhibit extra centrosomes. Ran targets are shown to be overexpressed in various cell types and Ran depletion causes aberrant mitotic spindles and cell death in tumor cell lines while it does not result in loss of cell viability in untransformed cells (Morgan-Lappe et al., 2007; Xia et al., 2008a; Xia et al., 2008b). However, in regular animal cells centrosome activity is dominant over the other pathways, making it difficult to address these questions in normal somatic cultured cells.
In order to better characterize these acentrosomal pathways, we have developed new Drosophila cell lines able to divide without centrioles. In Drosophila, the protein DSas-4 is essential for centriole duplication and the lack of this protein results in the formation of acentriolar somatic cells (Basto et al., 2006). Starting from DSas-4 loss-of-function mutant embryos, we have established 6 immortalized cell lines lacking centrioles. Here we present a first characterization of mitotic spindle assembly in these novel acentriolar cell lines.
These acentriolar cell lines constitute a unique animal somatic cell model to study the mitotic spindle organization independently of centrosomes. In addition these cell lines will be prime candidates for the investigation of the molecular mechanism behind centriole biogenesis.
Results
Establishment of DSas-4 mutant cell lines
DSas-4S2214 mutant flies were balanced over a chromosome bearing an inducible cell lethal transgene, P {hs-hid}, triggering apoptosis upon heat shock stimulation (see Materials and Methods). In order to follow MT dynamics, DSas-4S2214 loss-of-function allele was recombined with the protein trap insertion Jupiter-GFP (Karpova et al., 2006). The Jupiter protein is a small MT associated protein (20 kD) allowing the visualization of MTs during the whole cell cycle. Such recombinants were genotyped by PCR.
We established 132 primary cultures from large batches of dissociated embryos coming from Jup-GFP, DSas-4S2214/TM3 Sb, hs-hid flies (Fig. 1A). Culture protocol is described in more details in Materials and Methods. Most of these primary cultures degenerate after 3–8 weeks (Fig. 1B). In the remaining cases, colonies of undifferentiated cells appear and progressively invade the flask, allowing for a first transfer. In favorable cases transfers became increasingly easy and 11 cell lines where routinely transferred every week in medium supplemented with 10% serum. These lines were named 23, 57, 59, 69, 70, 83, 84, 96, 110, 127, and 131. They differed slightly in phenotype, with a round or a fibroblastic appearance.
Fig. 1. Acentriolar cell lines establishment and characterization.
(A) Different 2–3-week-old primary cultures obtained from dissociated embryos. Cells present various morphologies. Scale bar: 50 µm. (B) Representation of the survival of primary cultures. Most of primary cultures differentiate and stop to grow but some of them spontaneously immortalize. 11 permanent cell lines have been obtained. (C) FACS profiles of a young mixed line (line 23) before (blue) and after (green) heat shock. X-axis: intensity of fluorescence. Y-axis: number of cells. The narrowing of the fluorescence peak reflects the selection against hs-hid genotypes. (D) PCR genotyping of the 11 cell lines. Two couples of primers (AB or BC, see Materials and Methods) allow detection of WT or mutant DSas-4 alleles. From this analysis 6 lines are pure DSas-4−/− (as line 131) and 5 lines are DSas-4+/− (as line 110). (E) Immunofluorescence of control (upper image) and DSas-4−/− no. 70 (lower image) mitotic cells stained with anti-DSas-4 antibody (red). Control cells show a spot of DSas-4 protein at each pole. None of the DSas-4−/− cells present such spots. Blue: DNA, Green: α-tubulin. Scale bar: 5 µm. (F) EM ultrathin section of a control cell showing a centriole (arrow). No centriole has been found upon 1100 sections of DSas-4−/− line 131. Scale bar: 0.2 µm.
FACS monitoring and heat shock treatment of primary cell lines
The GFP profiles of late primary culture cells were followed by FACS. This technique allows the use of a small amount of cell culture to evaluate its genotype. Jupiter-GFP and DSas-4S2214 mutation markers being on the same chromosome, homozygous DSas-4S2214/S2214 cells should display a GFP signal that is 2× more intense than heterozygous DSas-4+/S2214 cells. Most late primary cultures present a fluorescence profile covering both intensities, indicating a mix cell population resulting from the polyclonal origin of the cultures. From the 11 established cell lines, 3 lines derived later towards a pure DSas-4+/S2214 cell population, 6 lines towards a pure DSas-4S2214/S2214 named from now DSas-4−/− and 2 lines stayed mixed according to their GFP profiles. These conclusions were later confirmed by PCR genotyping and immunofluorescence (see below).
We also selected DSas-4−/− homozygous cells by heat shock treatment (90 minutes at 37°C) in order to express the hs-hid transgene and eliminate heterozygous cells for DSas-4S2214. It was not possible to apply directly this treatment on embryos before their dissociation because embryos (even wild-type embryos) are extremely sensitive to heat shock (Dura, 1981; Graziosi et al., 1983). Fig. 1C shows the GFP profile shift after such heat shock treatment on a mixed cell line (line 23). In order to eliminate any wild-type cell from the population we systematically heat shocked the 6 homozygous DSas-4−/− cell lines characterized by FACS analysis.
Identification of the 6 DSas-4−/− cell lines
We genotyped these cell lines by PCR. As control we used the Jupiter cell line obtained from Jupiter-GFP fly stock (Karpova et al., 2006). Fig. 1D shows PCR profiles obtained from cell lines that were either pure DSas-4+/+ (control line Jupiter), pure DSas-4−/− (line 131) or DSas-4+/− (line 110). Within the 11 established cell lines, 6 lines (69, 70, 84, 96, 127, 131) possess only the 500 bp band characteristic of the P element associated with DSas-4S2214 and are therefore purely DSas-4−/−. The 5 other lines (23, 57, 59, 83, 110) present a 500 bp and a 920 bp bands indicating their heterozygosity for DSas-4.
We also tested by immunodetection for the presence or absence of the DSas-4 protein in control and DSas-4−/− cell lines (Fig. 1E). In control cells all the metaphases have spots of DSas-4 protein at the poles (96% with 2 spots, 4% with multiple spots,) and as expected, none of the DSas-4−/− cells presented spots of DSas-4 protein (n = 50 for each line).
Ultrastructural study of DSas-4−/− cells
Due to the fact that centrioles are sub-diffraction structures to be resolved by light microscopy, the ultimate proof of centriole absence requires an ultrastructural analysis. As centrioles are permanent cell organelles, this study can be conducted with cells in interphase. In a control Drosophila cell line (Jupiter), we examined 600 ultrathin sections and found 8 centrioles, which corresponds almost to the same proportion described in previous studies (Debec et al., 1982; Debec and Marcaillou, 1997) (Fig. 1F). In the DSas-4−/− cell line no. 131, we found no centrioles upon 1100 ultrathin cell sections. This is in agreement with a previous ultrastructural study of DSas-4−/− Drosophila third instar larval brain cells (Basto et al., 2006). We therefore conclude that the DSas-4−/− cells lack centrioles.
Morphological analysis of acentriolar mitotic spindles
We then analyzed the overall spindle morphology during mitosis in control and the 6 DSas-4−/− lines by immunofluorescence against α-tubulin. In Jupiter control cells, (Fig. 2A, schematized in Fig. 2A′) the mitotic spindle is bipolar and well polarized (n = 100). During metaphase, the body of the spindle is made of kinetochore fibers (K fibers) as well as many interpolar MTs. Centrosomes are found as a spherical structure at the poles and many astral MTs radiate from the poles. On the contrary, the acentriolar mitotic spindle is quite different in structure (Fig. 2B–F). It has usually a barrel-shaped appearance and it is poorly polarized, with broad poles (Fig. 2C). There are no or only very few astral MTs and no centrosome-like structure can be detected at the poles. In metaphase, the body of the spindle consists of prominent K fibers. Later, during anaphase and telophase interpolar MTs can also be detected. All acentriolar spindles observed were bipolar and anastral. We found, however, a continuum gradient of polarization (Fig. 2B–F, schematized in Fig. 2B′–F′). For example on line 131 29% of spindles are barrel-shaped, 25% are mixed, 17% are not polarized, 15% have a normal polarization, and 14% are hyper polarized (n = 132). The 5 other DSas-4−/− cell lines present the same characteristics with a similar range of variation in spindle shape (data not shown).
Fig. 2. Morphological analysis of acentriolar mitotic spindles.
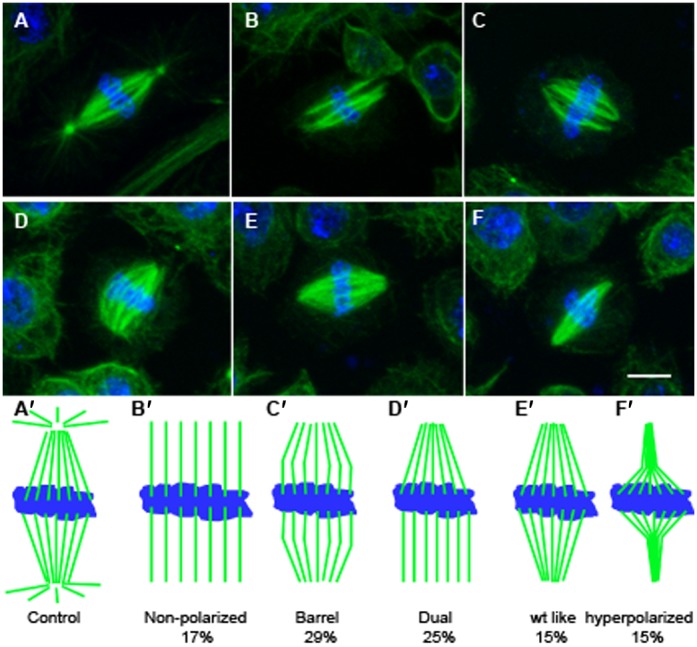
Immunofluorescence on fixed cells. (A) A typical mitotic spindle from control cell, well polarized with many astral MTs. Centrosomes are evidenced at the two poles as a spherical structure. (B–F) Images of the various types of acentriolar spindle morphologies encountered, ranging from non-polarized (B) to extreme polarization (F). The most frequent acentriolar spindle type is represented in (C), with a barrel-shaped appearance. In all cases, there is none or only very few astral MTs and no centrosome-like structure detectable. Blue: DNA, Green: α-tubulin. Scale bar: 5 µm. (A′–F′) Schematic representation of the different mitotic spindles shapes observed either in control (A′) or in the acentriolar line 131 (B′–F′), with percentages of each category (n = 132).
Acentriolar spindles and recruitment of centrosomal proteins
Next, we investigated the recruitment of centrosomal components in DSas-4−/− cell lines (Fig. 3). First, we studied the localization of Centrosomin. As previously described (Megraw et al., 1999; Lucas and Raff, 2007), Centrosomin is essentially recruited to centrosomes during mitosis in wild-type cells (Fig. 3A). In contrast, no Centrosomin was detected at the poles of acentriolar spindles (Fig. 3D). Furthermore we studied the recruitment of γ-tubulin 23C that is massively recruited to the centrosome during mitosis (Debec et al., 1995). In wild-type cells γ-tubulin 23C highlights centrosomes and can also be detected over the spindle itself (Fig. 3B). However, in DSas-4−/− cell lines we failed to detect recruitment of γ-tubulin 23C at the poles although the spindle staining remains (Fig. 3E). In addition, we analyzed the centriolar marker D-PLP (Martinez-Campos et al., 2004) that allows the visualization of centrioles during the whole cell cycle. In wild-type cells D-PLP is detected as one or two spots in interphase and as one dot at each mitotic spindle pole (Fig. 3C). On the contrary, we found no D-PLP staining at the poles during mitosis or interphase in Dsas-4−/− cell lines, (Fig. 3F) but we detected some very small aggregates of D-PLP not localized to the poles, in line with previous observations in the acentriolar cell line 1182 (Moutinho-Pereira et al., 2009).
Fig. 3. Acentriolar spindle poles do not recruit centrosomal components.

Immunofluorescence analysis of control cells Jupiter (upper panel) and acentriolar cells of line 131 (lower panel) for recruitment of PCM proteins. In all cases PCM proteins are strongly recruited at the centrosomes (arrows) in wild-type cells. On the contrary these proteins are dispersed in the cytoplasm in acentriolar cells. Blue: DNA, Green: α-tubulin, White/Red: centrosomal protein. (A,D) Centrosomin staining. (B,E) γ-tubulin 23C. In acentriolar cells the centrosomal staining disappears although a faint spindle staining is conserved. (C,F) D-PLP staining. In (C) (control cells), arrows indicate centriolar structures at the mitotic poles and arrowhead an interphasic centriolar structure. In (F) D-PLP positive granules are found scattered in the cytoplasm of acentriolar cells. Scale bar: 10 µm.
These results demonstrate that in DSas-4−/− cell lines, the centriole and PCM markers were delocalized from the poles and found dispersed in the cytoplasm (although still on the spindle for γ-tubulin). From all these observations we conclude that DSas-4−/− cells do not present a classical MTOC as a consequence of their acentriolar nature.
Modalities of acentriolar mitotic spindle assembly
The results presented above clearly illustrate the absence of centrosomes in DSas-4−/− cell lines; however, these cells are still able to build a spindle allowing chromosome segregation during mitosis. To follow spindle formation in these acentriolar cell lines we used live imaging of Jupiter-GFP (Fig. 4).
Fig. 4. Acentriolar mitotic spindle assembly.
Videomicroscopy of living mitotic cells followed with the Jupiter-GFP fusion protein. (A,C) Control cells (Jupiter line). (B,D) Acentriolar cells (131 line). Scale bar: 10 µm. (A,B) Early steps of mitosis; in wild-type cells the spindle progressively establishes from MTs irradiating from the two centrosomes and the spindle forms according to an “outside-in” process. In acentriolar cells the first mitotic MTs appear from discrete foci in the nuclear space. (C,D) Metaphase–anaphase transition; the wild-type spindle is compact and well polarized with many astral MTs that come in contact with the cell cortex in anaphase. The acentriolar spindle is constituted essentially of robust K-fibers in metaphase, and the extremities of the spindle reach directly the cell cortex in anaphase. Scale bar: 10 µm.
In wild-type cells, live analysis of Jupiter-GFP shows that spindles are progressively established from MTs irradiating essentially from the two centrosomes localized at the antipodes of the nucleus according to an “outside-in” process (Fig. 4A). During metaphase and anaphase (Fig. 4C) the wild-type spindle is compact and well polarized with many astral MTs that come in contact with the cell cortex in anaphase.
In the acentriolar cells (here line 131), due to the absence of centrosomal asters, the initiation of mitosis is particularly difficult to capture. Thus we regularly recorded randomly 20 fields (each representing about 40 interphase cells) of a cell culture overnight by time-lapse microscopy to recover acentriolar prophases. The first step of acentriolar spindle assembly is completely different from the wild-type situation (Fig. 4B). The first mitotic MTs appear from discrete foci close to chromatin. During metaphase, the spindle shows robust K fibers (Fig. 4D) without astral MTs and at the onset of anaphase, the spindle extremities come in contact with the cell cortex, in contrast to control cells. Furthermore, interpolar MTs are more evident from anaphase. Telophase is similar to the wild-type situation with formation of a midbody. In addition, using the live imaging data, we estimated the duration of the different phases of mitosis in both cell types at 23°C. In absence of live labeling of nuclear envelope and chromosomes it was not possible to clearly differentiate the different stages of mitosis. But we can estimate precisely, however, the time for spindle assembly in the two cell types. Spindle assembly in acentriolar cells lasts nearly 3 times (mean of 15 mitoses was 44±2.2 minutes, expressed as s.e.m.) compared to wild-type cells (mean 14±0.7 minutes). The length of the remaining mitotic stages is akin in both cell types. Anaphase is remarkably similar taking place precisely in 8 minutes (8.8±1.1 minutes for line 131 and 8.4±0.6 minutes for control cells). Telophase is also roughly the same in both cell types (about 16 minutes, no accurate measurements available for this stage). Thus the first stages are clearly a critical step for anastral mitosis.
Furthermore, we compared our live imaging observations with fixed cell preparations by counting the number of cells in the different phases of mitosis. Using this approach, we found that more than 30% of mitotic cells were in prophase in acentriolar cell lines (made on lines 70, 127 and 131) whereas in control cells these stages only represent 13% of the mitotic cells.
Kinetochores fibers lead acentriolar spindle assembly
In order to highlight the formation of the acentriolar spindle, we conducted live analysis of MT depolymerisation/repolymerisation assays (Fig. 5A,B). For these experiments, cells were first incubated for 1.5 hours on ice and then allowed to recover at 18°C. Mitotic cells were identified by the presence of condensed chromosomes. In control cells (Fig. 5A), repolymerisation occurs essentially from centrosomes, and lead to large astral MTs. As a consequence of the collapse of mitotic spindle during cold treatment the two centrosomes are usually found closed to the chromosomes. The centrosomes then progressively move away during spindle assembly progression. Despite this apparent centrifugal extension, spindle clearly forms mainly from the centrosomes under an “outside-in” process. The recovery of acentriolar mitotic cells is strikingly different (Fig. 5B). In these conditions we first observe MT repolymerisation close to chromosomes. These MTs then form bundles that progressively elongate in a symmetrical divergent manner from the chromosome mass until a bipolar spindle is formed. Thus acentriolar spindles are clearly re-assembled from the chromosomes following an “inside-out” process.
Fig. 5. Recovery steps of mitotic acentriolar cells after cold MT depolymerisation.
(A,B) Time-lapse sequences of mitotic spindle re-formation at 18°C. Time 0 corresponds to the first image recorded on the microscope (see Materials and Methods). (A) WT cell (Jupiter line). MTs polymerisation occurs essentially from the centrosomes and spindle forms under an “outside-in” process. Centrosomes were first close to the chromosomes and then move away. Scale bar: 5 µm. (B) Acentriolar cell (line 131). Spindle forms from the chromosomes under an “inside-out” mechanism. Scale bar: 5 µm. (C) Immunofluorescence images of acentriolar cells fixed at various times of recovery at 23°C. (a) after 3 minutes of recovery at 23°C, multiples dots of nascent MTs are first observed in the immediate vicinity of the chromosomes. (b) after 6 minutes of recovery elongated bundles of MTs are observed from discrete sites onto the chromosomes. (c) these sites are identified by CID protein as the kinetochores. Inlay: magnified view of c. Blue: DNA, Green: α-tubulin, Red: CID. Scale bar: 10 µm.
We also followed spindle recovery in fixed acentriolar cells. Immediately following incubation on ice, no MTs can be visualized. During recovery, we observed that MT repolymerisation takes place near the chromosomes (Fig. 5C). After 3 minutes of recovery at 23°C multiple MT stubs can be observed in the immediate vicinity of condensed chromosomes (Fig. 5Ca). After 6 minutes of recovery, elongated bundles of MTs are observed from discrete sites on chromosomes (Fig. 5Cb). In order to identify these sites we performed the same experiments after co-staining with CID (centromere identifier). The protein CID (Henikoff et al., 2000) is a histone H3-like protein specifically localized at the centromere during the whole cell cycle and allowing the recruitment of many proteins and establishment of kinetochores (Blower and Karpen, 2001). We found that the chromosomal sites at the base of MT bundles were positive for CID and we identify these sites as the kinetochores (Fig. 5Cc).
We therefore conclude that kinetochores play an essential role in the stabilization of MTs that form in the vicinity of chromosomes resulting in the formation of robust K fibers.
Drosophila acentriolar cell lines as a model system
It has already been shown that Drosophila cell lines are useful tools for genome wide RNA interference (RNAi) and high-throughput screens to dissect various molecular mechanisms of mitosis including spindle assembly (Goshima et al., 2007), centriole coalescence (Kwon et al., 2008) or centriole duplication and centrosome maturation (Dobbelaere et al., 2008). We believe that these acentriolar cell lines represent a new opportunity to study spindle assembly in absence of centrosomes. However, such studies will only be possible if these cell lines have a functional spindle assembly checkpoint (SAC) and respond to RNAi. In addition, the possibility to transfect these cells would be of great interest. We therefore conducted experiments to assess the response of acentriolar cell lines on these three points.
First, we tested the SAC response by quantifying the mitotic index after immune-detection of phospho-Histone 3 epitope (PH3) on random fields after a 5-hour incubation with 2 µM colcemid. Our results show that these acentriolar cell lines have a positive SAC response since the mitotic index increased from 1.4–1.8% of the cell population in normal conditions to 3.2–5.1% after colcemid treatment (Fig. 6A). These results indicate that the absence of centrosomes does not alter the functionality of this checkpoint, and that these acentriolar cell lines can be used in a mitotic blocking assay.
Fig. 6. Acentriolar cells as a model system.
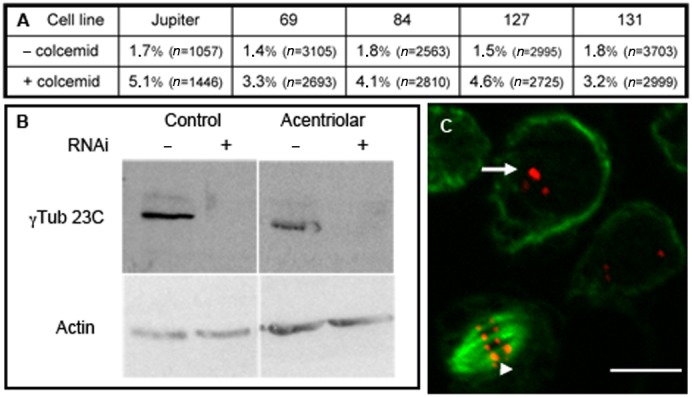
(A) SAC response of control and acentriolar cell lines. Table of the mitotic index (in %) in the different lines in normal conditions of culture or after colcemid treatment (2 µM, 5 hours). Mitotic cells have been visualized by H3P staining on immunofluorescence preparations. (B) Immunoblot showing γ-tubulin 23C depletion after RNAi treatment (lanes +) in control and acentriolar 131 cells. Actin was used as an internal loading control. (C) Live analysis of acentriolar 131 cells transfected with a pMT CIDmCherry plasmid. Interphasic cells show a CID signal in the centromeres (arrow) and mitotic cell is labeled on the kinetochores (arrowhead). Scale bar: 10 µm.
We also performed RNAi experiments using dsRNA specific for γ-tubulin 23C. After 7 days of treatment, γ-tubulin 23C protein level was quantified by Western blot (Fig. 6B) and showed that more than 95% was depleted from both control cells and the acentriolar cell line 131, whereas the depletion was less pronounced for acentriolar cell lines 70 and 127.
We also conducted transfection assays on different acentriolar cells lines using a pMT plasmid bearing a CIDmCherry construct with a blasticidine selection marker. After selection, we easily recovered cell colonies in lines 69, 84 and 131. These cells presented clearly a centromeric mCherry signal in the nucleus during interphase, and on kinetochores during mitosis (Fig. 6C), a result indicating that these three acentriolar cell lines can be used in transfection experiments.
Overall, our results demonstrate that these acentriolar cell lines can be used to study anastral spindle formation and related molecular mechanisms, either through RNAi or drug library high-throughput screening or with a more directed approach with candidate genes or proteins. In addition their capacities to be easily transfected, at least for some of them, complete their qualities as a new biological model.
Discussion
Six new acentriolar cell lines
Here, we present 6 new Drosophila melanogaster cell lines regularly dividing without centrioles. Different protocols have been published to establish either primary or continuous cell lines (Bai et al., 2009; Ueda et al., 2007). However, we here described (see Materials and Methods) an original protocol developed for an heterogeneous population of starting embryos, using the conditional cell lethal transgene P{hs-hid} (Grether et al., 1995).
A Drosophila acentriolar cell line, the 1182-4 cell line, was previously described and analyzed (Debec et al., 1982; Debec and Abbadie, 1989; Moutinho-Pereira et al., 2009; Szöllösi et al., 1986), but the origin of this peculiarity was never elucidated. On the contrary, the origin of these new acentriolar cell lines is clearly defined, the DSas-4 mutation, a key regulator of centriole assembly. DSas-4 is the fly ortholog of C. elegans Sas-4, a large coiled-coil protein functionally associated with the last steps of centriole assembly (Delattre et al., 2006; Kirkham et al., 2003; Leidel and Gönczy, 2003; Pelletier et al., 2006). Fly embryos mutant for DSas-4, which originates from heterozygous mothers progressively lose their centrioles after depletion of the maternal stock of protein (Basto et al., 2006). The persistence of centrioles in the DSas-4 mutant flies is not stated precisely but they probably disappeared during the first or second larval stages. In the case of the DSas-4−/− cell lines, the cells have completed hundreds of cell cycles in the absence of DSas-4 protein.
We investigated spindle assembly during mitosis between control cell lines (Jupiter and S2) and DSas-4−/− cell lines. Our results clearly illustrate that DSas-4−/− cell lines present all the characteristics of an acentriolar state: 1) the mitotic spindles lack or have very few astral MTs, 2) they are usually unfocused, with a high variability in shape, and 3) PCM protein markers (D-PLP, Centrosomin, γ-tubulin 23C) are not recruited to the spindle poles, a result in agreement with centriole function in PCM recruitment and organization into a functional centrosome (Bobinnec et al., 1998; Marshall, 2007). In addition, MT repolymerization after cold treatment never occurs from a focal structure. Overall, these results clearly illustrates that no functional centrosome is present in these DSas-4−/− cell lines. Furthermore, our ultrastructural study on cell line no. 131 confirms the absence of centrioles in DSas-4−/− cells, as previously shown in DSas-4 mutants (Basto et al., 2006). We did not conduct this EM analysis on other DSas-4−/− cell lines, but since these cell lines have the same genotype and the same characteristics by immunofluorescence studies, we consider that most likely none of them present any centriole structure.
The 6 DSas-4−/− lines described in this publication will be available through the DGRC website (https://dgrc.cgb.indiana.edu).
Assembly of anastral mitotic spindles
We have followed mitotic spindle establishment by time-lapse microscopy and on fixed preparations. Overall, our observations show that during anastral spindle formation, the first MTs appeared in the vicinity of the condensing chromosomes. This pattern is very reminiscent of cells depleted for Centrosomin (Mahoney et al., 2006). This is in agreement with the chromatin pathway. The acentriolar mitotic spindle progressively appeared with the growth of robust K fibers. Each chromosome builds its own mini-spindle, exactly as predicted (O'Connell and Khodjakov, 2007). In this process, kinetochores play an essential role. Using MT depolymerisation/repolymerisation experiments and immunostaining against the CID protein we clearly identify kinetochores at the base of growing MT bundles. The exact role of kinetochores in this process remains unknown. We hypothesized that short MTs are first nucleated through the chromatin pathway and then captured by kinetochores at their plus ends allowing their elongation by addition of tubulin dimers at the kinetochore level and translocation due to MT poleward flux (Maiato et al., 2004). Alternatively, kinetochores could directly nucleate MTs. It has been recently shown that the Nup 107–160 complex recruits γ-TuRC at the level of kinetochores promoting MT nucleation and K-fiber assembly; however, this mechanism would produce MTs of opposite polarity (Mishra et al., 2010). This has been shown to occur under particular experimental conditions in S. cerevisiae and MTs nucleated from kinetochores were found to be short-lived (Tanaka et al., 2009). In any event, it is clear that these acentriolar mitotic spindles establish upon an “inside-out” process, contrarily to wild-type cells. It is plausible that the augmin pathway is also active in these cells, and this would contribute to generate new MTs from preexisting spindle MTs. This phenomenon could explain the formation of interpolar MTs in the acentriolar spindles.
We also evaluated the time need for spindle assembly, in wild-type and acentriolar cells. Our results clearly show that the early stages of mitosis (until metaphase) last longer in acentriolar cells than on wild-type ones. These steps are critical for anastral spindle assembly and function. On the contrary the last stages of mitosis are similar in length for both cell types. Despite the fact that it is questionable to present such comparison between cell lines of independent origin, a reasonable hypothesis is that centrosomes would facilitate spindle formation. Furthermore, we noticed that during anaphase B the acentriolar spindle poles goes usually in contact with the plasma membrane. The absence of astral microtubules evidently favors such proximity, but this could also correspond to a real anchoring of the spindle to cell cortex. Surprisingly, nearly all DSas-4−/− mitoses are bipolar.
Acentriolar cell lines bring new research opportunities
The immediate interest of these cell lines will be to study the modalities of anastral spindle assembly in animal somatic cells. We do not intended here to dissect the molecular players involved in this process, but simply to demonstrate that this cellular model will be useful for such studies. Until now, there are only few acentriolar models available. One line of study focuses on oocytes (especially mouse, Xenopus, sea urchin and fly) due to the natural absence of centriole during female meiosis in nearly all the animal species. These models have allowed the identification of the chromatin pathway. However, these models are inappropriate for experiments such as RNAi or pharmacological screens. Another possibility to generate an acentriolar situation is through centriole ablation in a normal cell but this situation is only transitory and induces stress consequences during the procedure (Uetake et al., 2007). Thus, our new cell lines will allow the study of acentriolar mitosis in physiological conditions. It will be possible in the future to transfect these cell lines with markers for organelles and also follow microtubules dynamics during spindle formation using tagged proteins such as EB1.
These acentriolar lines represent a powerful tool to study and screen for new players of anastral spindle assembly, either for the already known chromatin and augmin pathways, or why not for some totally new mechanisms. We showed that these cells respond to RNAi, and are checkpoints competent. Thus it will be possible to test candidate genes specifically affecting mitotic spindle establishment in the acentriolar cells. As SAC activity is maintained in these cells, the depletion of essential proteins could be detected by the variation of the mitotic index, for example using PH3 signal. We can also consider a genome wide screen to identify unknown molecular functions essential for acentrosomal pathways (Goshima et al., 2007).
Furthermore, it may be possible using these acentriolar cell lines to search for molecules affecting specifically the anastral pathways. Indeed, in normal cells both centrosomal and the anastral pathways are effective, but the centrosomal pathway is dominant (Kaláb et al., 2006; Maiato et al., 2004). As mitosis in DSas-4−/− cell lines rely only on anastral pathways it should be theoretically possible to discover drugs affecting mitosis specifically in acentriolar cells.
Many unanswered questions remain concerning the precise function of centrioles in mitotic spindle formation (Debec et al., 2010; Marshall, 2007; Wilson, 2008), cytokinesis (Piel et al., 2001), or during G1/S transition (Wilson, 2008). The acentriolar cell lines we describe here present a great opportunity to develop new strategies and approaches to address the fundamental questions concerning centriole functions and biogenesis.
Materials and Methods
Fly stock
Primary cell cultures are initiated from hundreds of dissociated embryos. As the homozygous DSas-4S2214/S2214 adults flies are sterile (Basto et al., 2006) it is not easy to recover large batches of homozygous mutant embryos from heterozygous parents. The DSas-4S2214 mutation was then balanced over a TM3 Sb chromosome bearing the P(hs-hid) transgene (Bloomington reference 1558 from R. Lehmann). hs-hid is an inducible cell lethal transgene allowing to eliminate cells of undesirable genotypes (DSas-4+/S2214 or +/+). The hid gene (here driven by the heat shock promoter) induces apoptosis through inhibition of IAP (Inhibitor of Apoptosis Protein) who represses caspases (Grether et al., 1995). When this fly stock is submitted to heat shock (90 minutes at 37°C) no Sb adults were recovered.
Cell culture
Batches of embryos (aged from 3 to 14 hours) were collected from heterozygous Jup-GFP, DSas-4S2214/TM3 Sb, P(hs hid) flies raised on food medium plus sterile autoclaved yeast paste. About 500 embryos were rinsed with water then dechorionated and sterilized in 40% Na hypochlorite plus 0.3% Triton for 15 minutes to allow the sterilization of first instar larvae present in the batches. Embryos were rinsed four times with sterile water, and once with culture medium before the dissociation in a glass homogenizer (Tenbroeck type, ref 432–1276 VWR, Fontenay-sous-Bois, France). Embryos dissociation was monitored in order to have cells clusters rather than individuals cells. Cell clusters were let to sediment and the vitellus containing supernatant was discarded. The equivalent of 150 dissociated embryos were placed in a small 12.5 cm2 Falcon tissue culture flask (Becton Dickinson, Le Pont de Chaix, France) with 1.5 ml of culture medium, at 23°C. The culture medium consists of M3 Shields and Sang medium (Sigma–Aldrich, St Louis, MO), with 20% of heat inactivated fetal calf serum (Biowest, Nuaillé, France). These primary cultures differentiated well in numerous cell types and the medium was changed very progressively (by 1/3 then 1/2, at last fully) every week. After immortalization, established cell lines were transferred every week at dilutions ranging from 1/5 to 1/20 with medium containing 10% serum.
Flow cytometry
Living cultured cells were labeled with 1 µg/ml Hoechst 33342 (Sigma–Aldrich, St Louis, MO) and processed for measuring DNA content and GFP fluorescence intensity. Analyses were performed with a Coulter Elite ESP flow cytometer (Beckman Coulter, Fullerton, CA) using an air-cooled argon-ion laser tuned at 488 nm for GFP measurements and a UV (350 nm) water cooled laser for Hoechst 33342 analysis. Doublets were eliminated on the basis of DNA peak versus DNA area signals and 10,000 cells were analyzed after doublets discrimination.
PCR genotyping
The P-element (P{lacW}l(3)s2214) was inserted between basepairs 448 and 449 of the DSas-4 gene; the following primers were designed: a pDSas-4 Fw primer (GAA CGA ATA TAA AAG CAT GC) (Primer A) in the position 5 bp, a pDSas-4 Rev primer (GGA GTT CAT ATC AAT GTT TC) (Primer B) in the position 932 of the DSas-4 gene, and a P1 primer (GAC GAC CTT ATG TTA TTT CA) (Primer C) for the inverted repeats flanking the P-element.
Immunofluorescence analysis
Cells were plated on Concanavalin-A (Calbiochem San Diego, CA) coated glass coverslips for 4 hours. Cells were fixed in cold methanol for 2 minutes at −20°C. The primary antibodies used were: mouse anti-α-tubulin DM1A (1:1000) (Sigma–Aldrich, St Louis, MO) rabbit anti-γ-tubulin T-5168 (1:1000) (Sigma–Aldrich, St Louis, MO), rabbit anti-γ-tubulin 23C (R62, raised in Toulouse lab) (1:1000), rabbit anti-phospho-histone3 (PH3) (1:500) (Upstate, Temecula, CA), rabbit anti-Dsas-4 (1:200) (gift from J. Raff), Chinese hamster anti-D-PLP (1:200) (raised in A.G. lab), and rabbit anti-Cnn (1:2000) (gift from T. Megraw). Secondary antibodies conjugated to FITC, Alexa 488, Alexa 568 (Invitrogen, Camarillo, CA) were used at 1:1000 dilutions.
Images were obtained on a Leica inverted confocal microscope TCS SP5 (Imaging facility Imagoseine, Jacques Monod Institute). Objectives lenses were Leica HCX APO 63×, NA 1.4 and Leica HCX PI APO 100× NA 1.4.
Electron microscopy
Cells were pelleted and fixed for 1 hour with 2% glutaraldehyde/0.5% paraformaldehyde in 0.08 M sodium cacodylate buffer, pH 7.4 (660 mOsmol) then rinsed in cacodylate buffer. Pellets were post-fixed with 2% osmium tetroxyde in distilled water for 1 hour and bulk stained in 1% aqueous uranyl acetate for 45 minutes. Samples were dehydrated in ethanol and embedded in Aradilte Epon. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined using a Tecnai12 Philips electron microscope (Imaging facility, Imagoseine, Jacques Monod Institute).
Live-cell microscopy analysis
For all kinds of living cell microscopy, cells were plated on 32 mm glass coverslips in a special chamber (Jacques Monod Institute, http://www.ijm.fr/en/ijm/facilities/imagoseine) with 2 ml of culture medium.
Videomicroscope (Imaging Facility, Jacques Monod Institute)
Overnight recording of living cells has been performed with a Leica videomicroscope equipped with a high resolution camera (Cool Snap, Photometrics, Tucson, AZ). Objective lens was Leica HCX APO 63× NA 1.2. Images were deconvoluted with the Huygens® Professional 2.7 software (Scientific Volume Imaging, Hilversum, The Netherlands).
Spinning disc (Imaging Facility, Jacques Monod Institute)
Time-lapse recording has been realized with a “spinning disc” multifocal microscope Ultraview CSU10 (PerkinElmer, Waltham, MA) equipped with a high resolution camera Coolsnap HQ (Photometrics, Tucson, AZ). Objective lenses were Leica HCX APO 63× NA 1.4 and HCX PI APO 100× NA 1.4. Images were analyzed with Metamorph 7 software (Molecular Devices, Sunnyvale, CA).
MTs regrowth experiments
Time-lapse recording
Living acentriolar cells were incubated for 1.5 hours in melting ice in a sample microscopy chamber and then directly observed with a confocal spinning disc confocal at 18°C. This relatively low temperature allows a slow spindle recovery without affecting its characteristics. However, the necessary delay (about 2–3 minutes) to place cells under microscope and find mitosis did not allow observation of very first stage of spindle recovery.
Fixed preparations
Living cells were incubated for 2 hours in melting ice, and spindle recovery was performed at 23°C for various times (0, 3, or 6 minutes) before fixation and immunofluorescence.
Transfection assays
Transfections have been realized using the Effecten kit (Quiagen, Hilden, Germany) following the instructions of the manufacturer. 2×105 cells were transfected with 1 µg of pMT Cid-mCherry plasmid (construct from H. Maiato lab). Stable transformed cells were selected after 15 days in 25 µg/ml blasticidine (Invitrogen, Camarillo, CA).
RNAi depletion of γ-tubulin
2×106 cells were incubated for 1 hour in culture medium without serum and with 10 µg of dsRNA specific for γ-tubulin 23C, corresponding to nucleotides 18–722 relative to its start codon. Afterwards complete culture medium was added. The fourth day, the same treatment was applied again. The seventh day, cells were harvested and analyzed by immunofluorescence and Western blot. The DNA templates were generated by PCR from cDNA clone LD 40196 (γ-tubulin 23C), using PCR primers that contained the T7 RNA polymerase minimal promoter sequence. Then PCR products were transcribed in vitro with the RiboMAX large-scale RNA production system T7 (Promega, Madison, WI). After a treatment with RQ1 DNase (Promega, Madison, WI), RNAs were extracted with phenol-chloroform and chloroform, then precipitated with ethanol, and finally resuspended in diethyl pyrocarbonate-treated water, and annealed.
Immunoblotting
Cells were lysed in (25 mM Tris, pH 7.7, 0.1% Triton X-100, 1 mM EDTA, 0.1 M NaCl) with protease inhibitor cocktail (1:100) (Roche, Meylan, France). Proteins were quantified by the Bradford method (Bio-Rad, Hercules, CA). 40 µg of proteins were separated by 10% bis-acrylamide-SDS gel electrophoresis and transferred on nitrocellulose membrane. The membrane was immunoblotted with primary antibody (rabbit anti-γ-tubulin 23C R62 (1:1000) (Toulouse lab) or mouse anti-actin T-5168 (1:5000) (Sigma–Aldrich, St Louis, MO) and with goat anti-rabbit or anti-mouse secondary antibody coupled to peroxydase (1:5000) (Invitrogen, Camarillo, CA).
Acknowledgments
The authors would like to thank Claudio Sunkel for the help with the generation of fly stocks. We thank for their essential help the members of the IJM imaging facility: L. Baba-Aïssa, X. Baudin, C. Chamot, V. Contremoulins, A. Jobart-Malfait, M. Jouve-San Roman and T. Piolot. We are grateful to the following colleagues for providing reagents and Drosophila stocks: Renata Basto, Monica Bettencourt-Dias, Tim Megraw and Jordan Raff. We thank Jay Gopalakrishnan and Andreas Merdes for critical reading of the manuscript. This work was funded by grants ANR “Blanche” grant Cymempol, Blan06-3-139786, ARC grant SL220100601358 and “Ligue Contre le Cancer” grant RS11/75-34. Work in the laboratory of Andreas Merdes is funded by grants from ARC (4720XP023OF), Centre National de la Recherche Scientifique, and Pierre Fabre Laboratories. Work in the laboratory of H.M. is funded by grants PTDC/SAU-GMG/099704/2008 and PTDC/SAU-ONC/112917/2009 from Fundação para a Ciência e a Tecnologia of Portugal (COMPETE-FEDER), the Human Frontier Research Program and the 7th framework program grant PRECISE from the European Research Council. We acknowledge the Portuguese–French cooperation program PESSOA (Egide PHC 20027YD) (MAE and FCT) for traveling support throughout this project.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Azimzadeh J., Bornens M. (2004). The centrosome in evolution. Centrosomes In Development And Disease (ed. Nigg E A.), pp. 93–122 Weinheim: Wiley-VCH. [Google Scholar]
- Azimzadeh J., Marshall W. F. (2010). Building the centriole. Curr. Biol. 20, R816–R825 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J., Wong M. L., Downhour D. M., Sánchez Alvarado A., Marshall W. F. (2012). Centrosome loss in the evolution of planarians. Science 335, 461–463 10.1126/science.1214457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Sepp K. J., Perrimon N. (2009). Culture of Drosophila primary cells dissociated from gastrula embryos and their use in RNAi screening. Nat. Protoc. 4, 1502–1512 10.1038/nprot.2009.147 [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. (2006). Flies without centrioles. Cell 125, 1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bettencourt–Dias M., Rodrigues–Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3, 730–739 10.1038/35087045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L. M., Rieder C. L., Eddé B., Bornens M. (1998). Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143, 1575–1589 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M., Azimzadeh J. (2007). Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 607, 119–129 10.1007/978-0-387-74021-8_10 [DOI] [PubMed] [Google Scholar]
- Carvalho–Santos Z., Machado P., Branco P., Tavares–Cadete F., Rodrigues–Martins A., Pereira–Leal J. B., Bettencourt–Dias M. (2010). Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426 10.1242/jcs.064931 [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. (2005). Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 309, 1373–1376 10.1126/science.1115964 [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Lavia P. (2007). Spatial control of mitosis by the GTPase Ran. Cell. Mol. Life Sci. 64, 1891–1914 10.1007/s00018-007-6568-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec A., Abbadie C. (1989). The acentriolar state of the Drosophila cell lines 1182. Biol. Cell 67, 307–311. [PubMed] [Google Scholar]
- Debec A., Marcaillou C. (1997). Structural alterations of the mitotic apparatus induced by the heat shock response in Drosophila cells. Biol. Cell 89, 67–78 10.1016/S0248-4900(99)80082-3 [DOI] [PubMed] [Google Scholar]
- Debec A., Szöllösi A., Szöllösi D. (1982). A Drosophila melanogaster cell line lacking centriole. Biol. Cell 44, 133–138. [Google Scholar]
- Debec A., Détraves C., Montmory C., Géraud G., Wright M. (1995). Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J. Cell Sci. 108, 2645–2653. [DOI] [PubMed] [Google Scholar]
- Debec A., Sullivan W., Bettencourt–Dias M. (2010). Centrioles: active players or passengers during mitosis? Cell. Mol. Life Sci. 67, 2173–2194 10.1007/s00018-010-0323-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gönczy P. (2006). Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844–1849 10.1016/j.cub.2006.07.059 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura J. M. (1981). Stage dependent synthesis of heat shock induced proteins in early embryos of Drosophila melanogaster. Mol. Gen. Genet. 184, 381–385 10.1007/BF00352509 [DOI] [PubMed] [Google Scholar]
- Gatlin J. C., Bloom K. (2010). Microtubule motors in eukaryotic spindle assembly and maintenance. Semin. Cell Dev. Biol. 21, 248–254 10.1016/j.semcdb.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D. (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziosi G., de Cristini F., di Marcotullio A., Marzari R., Micali F., Savoini A. (1983). Morphological and molecular modifications induced by heat shock in Drosophila melanogaster embryos. J. Embryol. Exp. Morphol. 77, 167–182. [PubMed] [Google Scholar]
- Grether M. E., Abrams J. M., Agapite J., White K., Steller H. (1995). The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9, 1694–1708 10.1101/gad.9.14.1694 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Platero J. S., van Steensel B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716–721 10.1073/pnas.97.2.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M. E., Scheumann N., Wickstead B., Langdale J. A., Gull K. (2010). Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Pralle A., Isacoff E. Y., Heald R., Weis K. (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A. (2006). Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton 63, 301–312 10.1002/cm.20124 [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. (2001). The mitotic spindle: a self-made machine. Science 294, 543–547 10.1126/science.1063488 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Müller–Reichert T., Oegema K., Grill S., Hyman A. A. (2003). SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., Thery M., Pellman D. (2008). Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22, 2189–2203 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie–Mazenc I., Tollon Y., Detraves C., Julian M., Moisand A., Gueth–Hallonet C., Debec A., Salles–Passador I., Puget A., Mazarguil H.et al. (1994). Recruitment of antigenic gamma-tubulin during mitosis in animal cells: presence of gamma-tubulin in the mitotic spindle. J. Cell Sci. 107, 2825–2837. [DOI] [PubMed] [Google Scholar]
- Lawo S., Bashkurov M., Mullin M., Ferreria M. G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J. R., Hegemann B.et al. (2009). HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- Leidel S., Gönczy P. (2003). SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Lucas E. P., Raff J. W. (2007). Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila Centrosomin. J. Cell Biol. 178, 725–732 10.1083/jcb.200704081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U. K., Stearns T. (2006). GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Mahoney N. M., Goshima G., Douglass A. D., Vale R. D. (2006). Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16, 564–569 10.1016/j.cub.2006.01.053 [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C. L., Khodjakov A. (2004). Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167, 831–840 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. (2007). What is the function of centrioles? J. Cell. Biochem. 100, 916–922 10.1002/jcb.21117 [DOI] [PubMed] [Google Scholar]
- Martinez–Campos M., Basto R., Baker J., Kernan M., Raff J. W. (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. (1999). The Centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829–2839. [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Kao L. R., Kaufman T. C. (2001). Zygotic development without functional mitotic centrosomes. Curr. Biol. 11, 116–120 10.1016/S0960-9822(01)00017-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J. D., Cleveland D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W. C., Cleveland D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R. K., Chakraborty P., Arnaoutov A., Fontoura B. M., Dasso M. (2010). The Nup107-160 complex and γ-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12, 164–169 10.1038/ncb2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan–Lappe S. E., Tucker L. A., Huang X., Zhang Q., Sarthy A. V., Zakula D., Vernetti L., Schurdak M., Wang J., Fesik S. W. (2007). Identification of Ras-related nuclear protein, targeting protein for Xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 67, 4390–4398 10.1158/0008-5472.CAN-06-4132 [DOI] [PubMed] [Google Scholar]
- Moutinho–Pereira S., Debec A., Maiato H. (2009). Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol. Biol. Cell 20, 2796–2808 10.1091/mbc.E09-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell C. B., Khodjakov A. L. (2007). Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 120, 1717–1722 10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Müller–Reichert T. (2006). Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., Bornens M. (2001). Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553 10.1126/science.1057330 [DOI] [PubMed] [Google Scholar]
- Szöllösi A., Ris H., Szöllösi D., Debec A. (1986). A centriole-free Drosophila cell line. A high voltage EM study. Eur. J. Cell Biol. 40, 100–104. [PubMed] [Google Scholar]
- Tanaka K., Chang H. L., Kagami A., Watanabe Y. (2009). CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Ueda R., Ui–Tei K., Roberts J., Cherbas L. (2007). Standard protocol for establishing cell lines from Drosophila embryos. CGB Technical Report 2007-04. The Center for Genomics and Bioinformatics, Indiana University, Bloomington, Indiana, USA 10.2506/cgbtr-200704 [DOI] [Google Scholar]
- Uehara R., Nozawa R. S., Tomioka A., Petry S., Vale R. D., Obuse C., Goshima G. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 106, 6998–7003 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J. J., English C. N., La Terra S., Khodjakov A., Sluder G. (2007). Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176, 173–182 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H. (2004). Functional role of centrosomes in spindle assembly and organization. J. Cell. Biochem. 91, 904–914 10.1002/jcb.20013 [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. (2008). Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265, 111–158 10.1016/S0074-7696(07)65003-7 [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Vernos I., Mitchison T. J., Karsenti E., Heald R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903–913 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Wilson P. G. (2008). Centriole inheritance. Prion 2, 9–16 10.4161/pri.2.1.5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., Vernos I. (2000). Tpx2, a novel Xenopus map involved in spindle pole organization. J. Cell Biol. 149, 1405–1418 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F., Lee C. W., Altieri D. C. (2008a). Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 68, 1826–1833 10.1158/0008-5472.CAN-07-5279 [DOI] [PubMed] [Google Scholar]
- Xia F., Canovas P. M., Guadagno T. M., Altieri D. C. (2008b). A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell. Biol. 28, 5299–5311 10.1128/MCB.02039-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Fang K., Fang G. (2009). Microtubule amplification in the assembly of mitotic spindle and the maturation of kinetochore fibers. Commun. Integr. Biol. 2, 208–210 10.4161/cib.2.3.7875 [DOI] [PMC free article] [PubMed] [Google Scholar]





