Abstract
Background
The purpose of this study was to determine the functional proteomic characteristics of residual breast cancer and hormone receptor (HR)-positive breast cancer after neoadjuvant systemic chemotherapy, and their relationship with patient outcomes.
Methods
Reverse phase protein arrays of 76 proteins were carried out. A boosting approach in conjunction with a Cox proportional hazard model defined relapse predictors. A risk score (RS) was calculated with the sum of the coefficients from the final model. Survival outcomes and associations of the RS with relapse were estimated. An independent test set was used to validate the results.
Results
Test (n = 99) and validation sets (n = 79) were comparable. CoxBoost revealed a three-biomarker (CHK1pS345, Caveolin1, and RAB25) and a two-biomarker (CD31 and Cyclin E1) model that correlated with recurrence-free survival (RFS) in all residual breast cancers and in HR-positive disease, respectively. Unsupervised clustering split patients into high- and low risk of relapse groups with different 3-year RFS (P ≤ 0.001 both). RS was a substantial predictor of RFS (P = 0.0008 and 0.0083) after adjustment for other substantial characteristics. Similar results were found in validation sets.
Conclusions
We found models that independently predicted RFS in all residual breast cancer and in residual HR-positive disease that may represent potential targets of therapy in this resistant disease.
Keywords: breast cancer, neoadjuvant chemotherapy, residual disease
introduction
More than 209 000 women were diagnosed with breast cancer in 2010 in the USA alone [1]. Neoadjuvant systemic chemotherapy (NST) is associated with tumor down staging, while concurrently allowing in vivo assessment of chemosensitivity. Furthermore, a pathologic complete response (pCR) following NST is considered a surrogate marker for an improved long-term outcome [2–5]. Conversely, patients with residual breast cancer after NST are at increased recurrence risk and may have resistance disease to therapy [3–5]. We have demonstrated a substantial survival benefit for patients who achieved pCR regardless of hormone receptor (HR) status: 5-year overall survival (OS) rates were 96.4% versus 84.5% for patients with and without pCR (P = 0.04) and 5-year recurrence-free survival (RFS) rates were 91.1% versus 65.3%, (P < 0.0001) in the HR-positive group [4]. NST with conventional anthracycline and/or taxane-containing regimens results in pCR in only a minority (8%–31%) of patients [6–8]. Despite the substantial impact of residual disease (RD) after NST on outcomes, no standard and more importantly effective therapy exists. Therefore, there is a critical need to better understand the molecular characteristics of such resistant tumors and to identify novel targets that can be pursued for a more effective, personalized intervention to improve the outcome.
To have a more precise assessment of the consequences of RD, we developed the residual cancer burden (RCB) as a continuous index combining pathologic measurements of the primary tumor and nodal metastases and tested it as an independent predictor of distant relapse-free survival [5]. We have used and validated reverse phase protein arrays (RPPAs) as a quantitative, sensitive, and reproducible proteomic technology as well as a low-cost, sensitive high-throughput platform for marker screening, pathophysiology studies, identification of novel treatment targets, and therapeutic monitoring [9].
Since patients who do not achieve a pCR after NST have a heterogeneous group of tumors, even within a single clinical subtype, there is need for better understanding of their biology, heterogeneity, and treatment sensitivity. More importantly, identifying potential therapeutic targets in RD is a critical need. In this study, we used RPPA to determine the molecular characteristics of residual breast cancer, in general, and in HR-positive tumors after NST, and their relationship with patient outcomes in order to identify potential therapeutic targets.
patients and methods
Two independent tumor sets collected from two different cohorts of patients with RD after NST (n = 99 and 76) for training, and validation sets were included to allow for the generalizability of the risk prediction tool and in particular to reduce biasedness in an overoptimistic direction. All patients were diagnosed with primary breast cancer and treated with uniform taxane and anthracycline-based NST at MD Anderson Cancer Center [12 cycles of weekly paclitaxel or 4 cycles of every 3-week docetaxel, followed by 4 cycles of FAC (5-fluourouracil, doxorubicin, and cyclophosphamide) or FEC100 (5-fluourouracil, epirubicin, and cyclophosphamide)]. All patients with estrogen receptor (ER)- and/or progesterone receptor (PR)-positive disease received adjuvant endocrine therapy with either tamoxifen or an aromatase inhibitor for 5 years or until relapse. Patients were divided into three breast cancer subtypes according to receptor status by immunohistochemistry and/or fluorescent in situ hybridization: HR positive (ER and/or PR positive, HER2 negative), HER2 positive (regardless of HR status), and triple receptor negative (ER, PR, and HER2 negative). For the HR-positive breast cancer subtype, the training and validation sets consisted of 53 and 42 tumors, respectively. The institutional review board approved the laboratory protocol to complete the studies.
reverse phase protein arrays
RPPA was completed independently and at different time points for training and test sets using individual arrays. Protein was extracted from human tumors, and RPPA was carried out as described previously [9, 10]. Lysis buffer was used to lyse frozen tumors by homogenization. Lysates were normalized to 1 µg/µl concentration as assessed by the bicinchoninic acid assay and boiled with 1% sodium dodecyl sulfate. Supernatants were manually diluted in fivefold serial dilutions with lysis buffer. An Aushon Biosystems 2470 arrayer (Burlington, MA) created 1056 sample arrays on nitrocellulose-coated FAST slides (Schleicher & Schuell BioScience, Inc.). Slides were probed with 76 validated primary antibodies (supplementary Table S1, available at Annals of Oncology online) and signal amplified using a DakoCytomation-catalyzed system. Secondary antibodies were used as a starting point for amplification. Slides were scanned, analyzed, and quantified using Microvigene software (VigeneTech Inc., Carlisle, MA) to generate spot signal intensities, which were processed by the R package SuperCurve (version 1.01) [11]. A fitted curve (‘supercurve’) was plotted with the signal intensities on the Y-axis and the relative log2 concentration of each protein on the X-axis using the nonparametric, monotone increasing B-spline model [11]. Protein concentrations were derived from the supercurve for each lysate by curve fitting and normalized by median polish. Protein measurements were corrected for loading as described previously [9, 10]. Antibodies were selected by focusing on markers currently used for breast cancer classification due to their value in treatment decisions (ER, PR, and HER2), targets implicated in breast cancer pathophysiology, and targets implicated in the pathophysiology of other cancer lineages. A final selection of antibodies was also driven by the availability of high quality to pass a strict validation process as previously described [9, 10].
statistical methods
Patient characteristics were tabulated and described by their medians or ranges and compared according to the breast cancer subtype with a chi-square test or Wilcoxon's rank sum test as appropriate. Time to recurrence was measured from the date of diagnosis to the date of recurrence or the last follow-up. Patients who died before experiencing a disease recurrence were censored at their date of death. The median follow-up time was calculated as the median observation time among all patients. The RCB was calculated using the MD Anderson RCB Calculator [12]. Survival outcomes were estimated according to the Kaplan–Meier product limit and compared with log-rank statistic. Cox proportional hazard models were employed to determine the association of risk scores (RSs) after adjustment for other significant patient and disease characteristics. All variables that were significantly associated with RFS (P < 0.05), including breast cancer subtype, nuclear grade, and RCB, were included in the multivariable model. The model selection was based on a backwards selection procedure where all variables of interest were included in a full model and then variables were retained according to their P-values (P < 0.05).
RPPA data analysis
As the first step, both RPPA datasets were variable slope normalized [13] to eliminate batch effects. To train the models (all tumors and HR-positive tumors), for each of the 76 proteins, a univariate CoxPH model was established. A boosting approach with cross validation (R package CoxBoost) was used to develop a robust Cox proportional hazard model and to select RPPA predictors of relapse for all patients and for patients with HR-positive disease. The final combined CoxPH model for the prediction of relapse was established including clinical stage, tumor grade, breast cancer subtype (when all tumors were included), RCB, and selected RPPA proteins. For each model, the C-index was calculated and estimated from 200 randomizations.
The most accurate RPPA set was selected from a series of bootstrapped prediction analyses, whereby different cutoff points were iterated to establish an optimal panel of RPPAs accurately separating the different risk subgroups. Subsequent unsupervised hierarchical clustering analysis was employed as a graphical tool to view the potential clusters obtained from the final multivariable model.
RS were calculated for all patients and patients with HR-positive disease. The sum of the coefficients from the Cox model was the RS for each patient. RS-All was the RS for all tumors, and RS-HR was the RS for HR-positive tumors. Formulas are as follows: RS-All = 0.9528 × CHK1ps345 − 0.7085 × Caveolin1 − 1.2428 × RAB25 and RS-HR = (−2.17) × CD31 + 1.53 × Cyclin E1. To classify validation samples into the specific risk groups, thresholds for receiver operating characteristic (ROC) curves were set for the training set and were subsequently employed for testing. C-index and area under the curve (AUC) were applied to measure the predictive performance and discriminating power of the models.
results
Patient and tumor characteristics for training and validation sets are summarized in Table 1. Training and validation sets were comparable by race, age, stage, and grade. The distributions by breast cancer subtypes: HR positive, HER2 positive, and triple receptor negative were 54%, 15%, and 31%, respectively, in the training set, and 53%, 14%, and 29% in the validation set (P = 0.97). Median follow-up times were 27 (range 7–142) months in the training set and 31 (range 4–145) months in the validation set.
Table 1.
Patients and tumor characteristics
| Training set |
Validation set |
||||
|---|---|---|---|---|---|
| All patients | N = 99 | % | N = 79 | % | |
| Race | |||||
| White | 65 | 65.7 | 50 | 63.29 | |
| Black | 22 | 22.2 | 17 | 21.52 | |
| Hispanic | 10 | 10.1 | 7 | 8.86 | |
| Other | 2 | 3.77 | 5 | 6.33 | 0.5703 |
| Age at diagnosis | |||||
| Minimum | 27 | – | 25 | – | |
| Median | 50 | – | 51 | – | |
| Maximum | 77 | – | 77 | – | 0.7517 |
| Menopausal status | |||||
| Premenopausal | 37 | 37.37 | 30 | 37.97 | |
| Peri/postmenopausal | 62 | 62.63 | 45 | 56.96 | 0.8452 |
| Clinical stage | |||||
| I and II | 48 | 48.48 | 32 | 40.51 | |
| III | 48 | 48.48 | 46 | 58.23 | 0.3038 |
| Nuclear grade | |||||
| 1 | 8 | 8.08 | 2 | 2.53 | |
| 2 | 32 | 32.32 | 19 | 24.05 | |
| 3 | 58 | 58.59 | 55 | 69.62 | 0.1176 |
| Lymphovascular invasion | |||||
| Positive | 62 | 62.6 | 37 | 46.84 | |
| Negative | 37 | 37.4 | 39 | 49.37 | 0.0909 |
| Residual cancer burden | |||||
| I | 1 | 1.01 | 0 | 0 | |
| II | 37 | 37.37 | 5 | 6.33 | |
| III | 57 | 57.58 | 71 | 89.87 | <0.0001 |
| HR positive | N = 53 | % | N = 42 | % | |
| Race | |||||
| White | 41 | 77.4 | 29 | 69.05 | |
| Black | 7 | 13.2 | 6 | 14.29 | |
| Hispanic | 3 | 5.66 | 4 | 9.52 | |
| Other | 2 | 3.77 | 3 | 7.14 | 0.7487 |
| Age at diagnosis | |||||
| Minimum | 30 | – | 31 | – | |
| Median | 50 | – | 51.5 | – | |
| Maximum | 77 | – | 77 | – | 0.597 |
| Menopausal status | |||||
| Premenopausal | 20 | 37.7 | 17 | 40.48 | |
| Peri/postmenopausal | 33 | 62.3 | 24 | 57.14 | 0.8776 |
| Clinical stage | |||||
| I and II | 29 | 54.71 | 19 | 45.24 | |
| III | 21 | 39.62 | 23 | 54.76 | 0.312 |
| Nuclear grade | |||||
| 1 | 8 | 15.1 | 2 | 4.76 | |
| 2 | 27 | 50.9 | 17 | 40.48 | |
| 3 | 17 | 32.1 | 23 | 54.76 | 0.0668 |
| Lymphovascular invasion | |||||
| Positive | 34 | 64.2 | 19 | 45.24 | |
| Negative | 19 | 35.8 | 23 | 54.76 | 0.1020 |
| Residual cancer burden | |||||
| II | 18 | 34.0 | 4 | 9.52 | |
| III | 31 | 54.5 | 38 | 90.48 | 0.0030 |
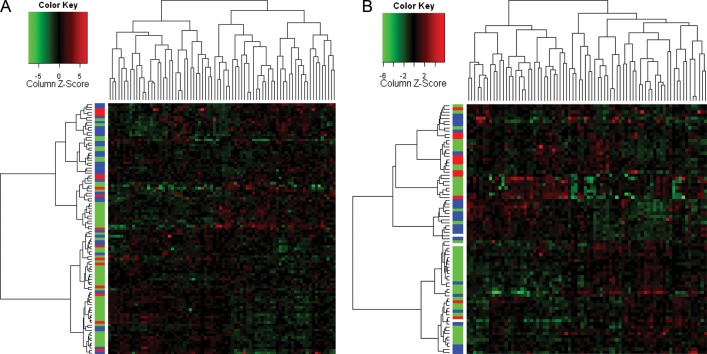
Figure 1 shows the unsupervised clustering of all residual tumors in the training (n = 99) (Figure 1A) and validation (n = 79) (Figure 1B) sets and 76 proteins by the breast cancer subtype. Residual tumors did not cluster together by the breast cancer subtype.
Figure 1.
Unsupervised clustering of all residual tumors training (A) and validation (B) sets and 76 proteins by the breast cancer subtype: hormone receptor positive (green), HER2 positive (red), and triple receptor negative (blue).
unsupervised clustering and survival estimates
all patients
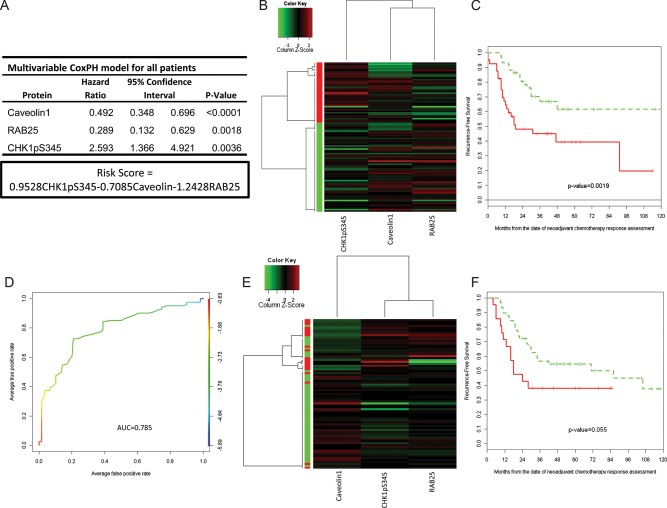
After CoxBoost and stepwise selection, a three-biomarker (CHK1pS345, Caveolin1, and RAB25) CoxPH model was developed as the RPPA relapse predictor model (RS-All = 0.9528 × CHK1pS345 − 0.7085 × Caveolin 1 − 1.2428 × RAB25; Figure 2A). When we applied unsupervised clustering of these three biomarkers to the training set (Figure 2B), patients were split into high and low risk of relapse groups with significantly different 3-year RFS estimates [45.1% (95% confidence interval, CI: 31.7% to 64.3%) versus 66.8% (95% CI: 54.1% to 82.5%), P = 0.002; Table 2 and Figure 2C], and AUC of 0.7476 (Figure 2D). The three protein risk model was then applied to the validation set (n = 79), and patients were again split into high- and low-risk groups (Figure 2E) with near-significant differences in median and 3-year RFS (P = 0.053; Table 2, Figure 2F).
Figure 2.
(A) Multivariable Cox proportional hazard model for the predictive markers and risk score (RS) calculation. (B) Unsupervised clustering of 99 residual breast cancers (training set). Two potential clusters were defined according the independent predictive markers from the multivariable CoxBoost analysis (Caveolin1, Rab25, and CHK1pS345). (C) Recurrence-free survival by clusters in the training set defined a high and a low risk of the recurrence group. (D) ROC curve of the RS in all residual breast cancers (test set). (E) Unsupervised clustering of 79 HR-positive residual breast cancers (validation set). Two clusters characterized by the independent predictive markers from the multivariable CoxBoost analysis (Caveolin1, RAB25, and CHK1pS345). (F) Recurrence-free survival by clusters in the validation set defined a high and a low risk of the recurrence group.
Table 2.
Recurrence-free survival estimates by the risk group in all patients and in patients with hormone receptor (HR)-positive tumors
| N | Number of events | 3-year estimate | 95% confidence interval | P-value | |
|---|---|---|---|---|---|
| Training sets | |||||
| All patients | |||||
| All | 99 | 40 | 58.6 | 48.9–70.3 | |
| High risk clustering | 40 | 23 | 45.1 | 31.7–64.3 | |
| Low risk clustering | 59 | 17 | 66.8 | 54.1–82.5 | 0.002 |
| HR positive | |||||
| All | 53 | 11 | 82.1 | 72.2–93.5 | |
| High risk clustering | 6 | 5 | 16.7 | 2.78–99.7 | |
| Low risk clustering | 47 | 6 | 90.8 | 82.5–99.8 | <0.001 |
| Validation data | |||||
| All patients | |||||
| All | 79 | 37 | 51.8 | 41.6–64.4 | |
| High risk clustering | 21 | 13 | 38.1 | 22.1–65.7 | |
| Low risk clustering | 54 | 24 | 56.7 | 44.9–71.5 | 0.055 |
| HR positive | |||||
| All | 42 | 15 | 70.2 | 57.4–86 | |
| High risk clustering | 5 | 3 | 30 | 6.3–99.9 | |
| Low risk clustering | 37 | 12 | 75 | 62–90.65 | 0.038 |
patients with HR-positive disease
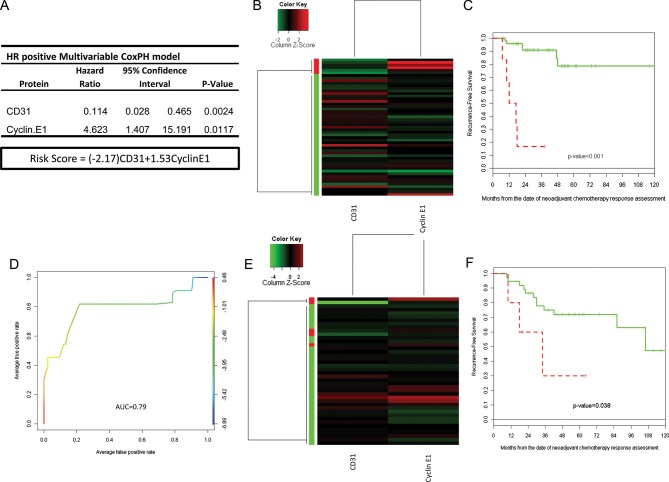
Although our patients received a homogeneous NST regimen, it is well known that breast cancer is a heterogeneous collection of diseases with different biology and therapeutic approaches. We therefore decided to focus further analysis on HR-positive disease. After CoxBoost and stepwise selection, a two-biomarker (CD31 and Cyclin E1) model was considered the optimal RPPA relapse predictor model for HR-positive disease (RS-HR = −2.17 × CD31 + 1.53 × Cyclin E1; Figure 3A). When we applied unsupervised clustering of these two biomarkers to the training set (n = 53; Figure 3B), patients were split into high- and low risk of relapse groups with significantly different 3-year RFS estimates (16.7% [95% CI = 2.78% to 99.7%] versus 90.8% [95% CI = 82.5% to 99.8%], P ≤ 0.001; Table 2 and Figure 3C), with an estimated corresponding AUC of 0.8535 (Figure 3D). The two protein risk model was then applied to the validation set (n = 42), and patients were again split into the high- and low-risk group (Figure 3E) with significant differences in 3-year RFS estimates (P = 0.038; Table 2 and Figure 3F).
Figure 3.
(A) Multivariable Cox proportional hazard model for the predictive markers and risk score (RS) calculation. (B) Unsupervised clustering of 53 HR-positive residual breast cancers (training set). Two potential clusters defined by the independent predictive markers from the multivariable CoxBoost analysis (CD31 and Cyclin E1). (C) Recurrence-free survival by clusters in the training set defined a high and a low risk of the recurrence group. (D) ROC curve of the RS in the HR-positive breast cancers (test set). (E) Unsupervised clustering of 42 HR-positive residual breast cancers (validation set). Two clusters defined by the independent predictive markers from the multivariable CoxBoost analysis (CD31 and Cyclin E1). (F) Recurrence-free survival by clusters in the validation set defined a high and a low risk of the recurrence group.
final multivariable models
Table 3 summarized the Cox proportional hazard models based on stepwise regression with backward elimination for all patients and for patients with HR-positive disease in the training sets. Considering all patients, the RS-All was the only independent predictor of RFS [hazard ratio = 2.09, 95% CI = 1.36–3.21, P = 0.0008] after adjustment for other substantial patient and disease characteristics including breast cancer subtype, clinical stage at diagnosis, nuclear grade, and RCB (a measurement of the volume of the RD at the time of surgery). Considering patients with HR-positive disease only, the RS-HR was the only independent predictor of RFS (hazard ratio = 2.87, 95% CI = 1.31–6.27, P = 0.0083) after adjustment for other substantial patient and disease characteristics including clinical stage at diagnosis, nuclear grade, and RCB. Similar results were found in both validation sets (P = 0.007 and P = 0.018 for all patients and patients with HR-positive disease, respectively).
Table 3.
Multivariable models for recurrence-free survival in all patients and in patients with HR-positive tumors
| Variables | Recurrence-free survival |
||
|---|---|---|---|
| Hazard ratio | 95% confidence interval | P-value | |
| All patients | |||
| Breast cancer subtype | |||
| HR positive | 1.0 | ||
| HER2 positive | 2.58 | 0.97–6.83 | 0.0572 |
| Triple receptor negative | 2.13 | 0.83–5.48 | 0.117 |
| Clinical stage at diagnosis | |||
| I and II | 1.0 | ||
| III | 1.02 | 0.515–2.029 | 0.95 |
| Nuclear grade | |||
| 1 and 2 | 1.0 | ||
| 3 | 1.64 | 0.56–4.79 | 0.362 |
| Residual cancer burden (continuous) | 1.01 | 0.99–1.02 | 0.346 |
| Risk score all patients (RS-All) | 2.09 | 1.36–3.21 | 0.0008 |
| Patients with HR-positive disease | |||
| Clinical stage at diagnosis | |||
| I and II | 1.0 | ||
| III | 0.33 | 0.08–1.43 | 0.1388 |
| Nuclear grade | |||
| 1 and 2 | 1.0 | ||
| 3 | 2.2 | 0.59–8.43 | 0.2405 |
| Residual cancer burden (continuous) | 1.02 | 0.99–1.05 | 0.2211 |
| Risk score HR positive (RS-HR) | 2.87 | 1.31–6.26 | 0.0083 |
discussion
Achieving a pCR after NST correlates with improved disease-free and OS; conversely patients with residual breast cancer after NST are at increased risk for recurrence. Therefore, identifying markers able to better discriminate different prognostic categories, and understanding the biology and the clinical implications of RD may help us identify specific targets and better tailor management. In this study, we used RPPA to determine the molecular characteristics of residual breast cancer, and their relationship with patient outcomes. We found a three protein model that independently predicted RFS risk in all patients and a two protein model that independently predicted RFS risk in patients with HR-positive disease.
There is a growing interest within the scientific community in understanding the biology and characteristics of RD. Guarneri et al. [14] evaluated the effect of NST on tumor biomarker expression and its prognostic role after therapy-induced variation. They observed that NST induced a substantial reduction in the expression of ER, PR, Ki-67, and apoptosis markers and that a Ki-67 ≥15% and nodal positivity after treatment were the predictors of inferior disease-free survival. Our group evaluated whether patients with HER-2 overexpressed tumors had a change in HER-2 status in the RD after trastuzumab-based NST. Despite the small sample size, we observed the loss of HER-2 in one-third of the patients with substantial RD, and this change was associated with poor RFS [15]. Lastly, Creighton et al. [16] used gene expression signatures showing increased expression of mesenchymal markers in residual tumor cells.
To our knowledge, this is the first analysis of RD after NST using functional proteomics. When looking at all tumors, we found a three marker model that predicted RFS. It included CHK1pS345, Caveolin1, and Rab25. CHK1 is the downstream protein kinase central to replication checkpoints. It responds primarily to replication fork abnormalities via Ataxia-Telangiectasia-Rad3-dependent phosphorylation at two sites, Ser345 and Ser317, through which it activates cycle arrest, activates DNA repair, and terminates the checkpoint to resume cell division cycle [17, 18]. A recent study showed that expression levels of CHK1 and FBX6 exhibited an inverse correlation in both cultured cancer cell lines and breast tumors and that defects in CHK1 degradation rendered tumor cells resistant to topoisomerase inhibitors [19]. Targeting DNA repair mechanisms in breast cancer is a potential therapeutic approach being explored in patients with germline DNA repair defects. Similar drugs may be useful for patients with RD and high levels of activated CHK1. RAB25, a member of the RAS superfamily of small GTPases, has been implicated in the pathophysiology of ovarian, breast, and other cancers. It is implicated in endosomal transport and recycling of cell-surface receptors and signaling proteins [20]. In breast cancer 1q amplification, RAB25 gene amplification and mRNA overexpression, measured by array comparative genomic hybridization and quantitative polymerase chain reaction, have been correlated with shorter OS [21, 22]. However, RAB25 mRNA and protein appear to exhibit a subtype-specific pattern of expression with high levels of expression in HR- and HER2-positive tumors, intermediate expression in basal tumors, and low or absent expression in metaplastic cancers [20, 21], suggesting that it may have different roles in breast cancer subtypes explaining, in part, our findings, since over 50% of the tumors were HR-positive and the HER2-positive cancers did not receive trastuzumab. The role of Caveolin1 in tumor biology appears to be multidimensional [21]. Caveolin1 appears to suppress the development of mammary tumors and metastases in vivo, as well as growth and transformation in vitro [23]. Studies reporting a reduction of Caveolin1 in breast cancer compared with normal tissue require cautious interpretation due to its location and technical limitations [24]. The best evidence cited in support of its tumor suppressor role come from a report of somatic inactivating CAV1 mutations in 16% of breast cancers [25] corroborated by an independent group in 35% of HR-positive breast cancer [26]. Among patients harboring mutations, 82% suffered recurrence, compared with 48% without mutations [26]. Our data, although limited by the stromal contamination of RPPA, support these findings, showing Caveolin1 as a predictor of better RFS.
Although our patients received a homogeneous NST regimen, it is well known that breast cancer is a heterogeneous collection of diseases with different biology and therapeutic approaches. We therefore decided to focus our analysis on HR-positive disease. We found a two marker model that predicted RFS. It included Cyclin E1 and CD31. CD31 has been used a marker of angiogenesis; however, no clear predictive or prognostic role in breast cancer has been previously reported.
Cyclins are critical to cell cycle progression, and Cyclins D1 and E1 have been well studied in breast cancer [27–30]. Overexpression of Cyclin E1 has been consistently associated with an increased risk of cancer relapse and death [29, 30]. We previously demonstrated that the levels of Cyclins B1, D1, and E1 were differentially deregulated in breast cancer subtypes and that although Cyclin B1 appeared to be the dominant cyclin associated with poor prognosis in HR-positive breast cancer, coordinated overexpression of Cyclins B1 and E1 was associated with adverse patient outcomes across all breast cancers and specifically in HR positive [31]. In our patients, these findings are interesting since these tumors were selected to be resistant to standard anthracycline/taxane-based chemotherapy (extensive RD), and with the two marker model, tumors were further discriminated into high- and low risk, implying that the high-risk tumors that had high levels of cyclin E1 were also resistant to adjuvant endocrine therapy. Further, PCNA, a proliferation marker for Ki67, was not found to be an independent predictor of outcome in this patient population. Recent work from our group showed that overexpression of low-molecular-weight (LMW) Cyclin E in letrozole-treated postmenopausal patients was associated with poor prognosis [31]. Letrozole treatment-inhibited Cyclin E-CDK2 kinase activity by preventing androstenedione-induced CDK2 increase. LMW Cyclin E bypassed this effect rendering cells resistant to letrozole. Patients with breast cancer whose tumors overexpressed LMW Cyclin E did not respond to therapy with aromatase inhibitors [31]. Ongoing clinical trials are looking at the role of CDK inhibitors in patients with HR-positive and HR-negative breast cancers [32].
Our study has some limitations. RPPA is useful for the objective quantification of protein expression in cell lines and tumor tissues, but does not provide information on intratumoral or cellular localization of the evaluated proteins and thus on the effect of different protein localization patterns on clinical outcomes. Further, and in particular with regard to Cyclin E1, RPPA does not discriminate between the protein products of full length and LMW variants.
In summary, we used RPPA to determine the molecular characteristics of RD after NST, and their relationship with patient's outcome. We found a three protein model that independently predicted RFS risk in all patients and a two protein model that independently predicted RFS risk in patients with HR-positive disease. These models may have value in stratifying patients based on their risk of relapse, and these biological differences may provide potential targets for therapy and help us to guide our adjuvant treatment modalities including novel agents in this resistant disease.
funding
This work was supported in part by 1K23CA121994-01 (AMG), Komen for the Cure Catalyst Award KG090341 (AMG), American Cancer Society Research Scholar Grant 121329-RSG-11-187-01-TBG (AMG), and National Cancer Institute through The University of Texas MD Anderson's Cancer Center support grant (P30 CA016672).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
references
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Kuerer HM, Newman LA, Smith TL, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17:460–469. doi: 10.1200/JCO.1999.17.2.460. [DOI] [PubMed] [Google Scholar]
- 3.Bear HD, Anderson S, Smith RE, et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2006;24:2019–2027. doi: 10.1200/JCO.2005.04.1665. [DOI] [PubMed] [Google Scholar]
- 4.Guarneri V, Broglio K, Kau SW, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006;24:1037–1044. doi: 10.1200/JCO.2005.02.6914. [DOI] [PubMed] [Google Scholar]
- 5.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 6.O'Regan RM, Von Roenn JH, Carlson RW, et al. Final results of a phase II trial of preoperative TAC (docetaxel/doxorubicin/cyclophosphamide) in stage III breast cancer. Clin Breast Cancer. 2005;6:163–168. doi: 10.3816/CBC.2005.n.019. [DOI] [PubMed] [Google Scholar]
- 7.Stearns V, Singh B, Tsangaris T, et al. A prospective randomized pilot study to evaluate predictors of response in serial core biopsies to single agent neoadjuvant doxorubicin or paclitaxel for patients with locally advanced breast cancer. Clin Cancer Res. 2003;9:124–133. [PubMed] [Google Scholar]
- 8.Colleoni M, Viale G, Zahrieh D, et al. Chemotherapy is more effective in patients with breast cancer not expressing steroid hormone receptors: a study of preoperative treatment. Clin Cancer Res. 2004;10:6622–6628. doi: 10.1158/1078-0432.CCR-04-0380. [DOI] [PubMed] [Google Scholar]
- 9.Hennessy BT, Lu Y, Gonzalez-Angulo AM, et al. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu J, He X, Baggerly KA, et al. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–1994. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 12.Residual Cancer Burden Calculator. http://www3.mdanderson.org/app/medcalc/index.cfm?pagename=jsconvert3. (26 April 2012, date last accessed).
- 13.Neeley ES, Kornblau SM, Coombes KR, et al. Variable slope normalization of reverse phase protein arrays. Bioinformatics. 2009;25:1384–1389. doi: 10.1093/bioinformatics/btp174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarneri V, Piacentini F, Ficarra G, et al. A prognostic model based on nodal status and Ki-67 predicts the risk of recurrence and death in breast cancer patients with residual disease after preoperative chemotherapy. Ann Oncol. 2009;20:1193–1198. doi: 10.1093/annonc/mdn761. [DOI] [PubMed] [Google Scholar]
- 15.Mittendorf EA, Wu Y, Scaltriti M, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res. 2009;15:7381–7388. doi: 10.1158/1078-0432.CCR-09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Guntuku S, Cui XS, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Develop. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 19.Merry C, Fu K, Wang J, et al. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2010;9:279–283. doi: 10.4161/cc.9.2.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal R, Jurisica I, Mills GB, et al. The emerging role of the RAB25 small GTPase in cancer. Traffic. 2009;10:1561–1568. doi: 10.1111/j.1600-0854.2009.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng JM, Ding M, Aribi A, et al. Loss of RAB25 expression in breast cancer. Int J Cancer. 2006;118:2957–2964. doi: 10.1002/ijc.21739. [DOI] [PubMed] [Google Scholar]
- 22.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Williams TM, Lee H, Cheung MW, et al. Combined loss of INK4a and caveolin-1 synergistically enhances cell proliferation and oncogene-induced tumorigenesis: role of INK4a/CAV-1 in mammary epithelial cell hyperplasia. J Biol Chem. 2004;279:24745–24756. doi: 10.1074/jbc.M402064200. [DOI] [PubMed] [Google Scholar]
- 24.Sagara Y, Mimori K, Yoshinaga K, et al. Clinical significance of Caveolin-1, Caveolin-2 and HER2/neu mRNA expression in human breast cancer. Br J Cancer. 2004;91:959–965. doi: 10.1038/sj.bjc.6602029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi K, Matsuda S, Machida K, et al. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- 26.Li T, Sotgia F, Vuolo MA, et al. Caveolin-1 mutations in human breast cancer: functional association with estrogen receptor alpha-positive status. Am J Pathol. 2006;168:1998–2013. doi: 10.2353/ajpath.2006.051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshadri R, Lee CS, Hui R, et al. Cyclin DI amplification is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis. Clin Cancer Res. 1996;2:1177–1184. [PubMed] [Google Scholar]
- 28.Bieche I, Olivi M, Nogues C, et al. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86:580–586. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keyomarsi K, Tucker SL, Buchholz TA, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–1575. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 30.Span PN, Tjan-Heijnen VC, Manders P, et al. Cyclin-E is a strong predictor of endocrine therapy failure in human breast cancer. Oncogene. 2003;22:4898–4904. doi: 10.1038/sj.onc.1206818. [DOI] [PubMed] [Google Scholar]
- 31.Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res. 2009;15:3654–3662. doi: 10.1158/1078-0432.CCR-08-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ClinicalTrials.gov. http://www.clinicaltrials.org/ (26 April 2012, date last accessed).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.