Abstract
Vaccination is generally considered to be the most effective method of preventing infectious diseases. All vaccinations work by presenting a foreign antigen to the immune system in order to evoke an immune response. The active agent of a vaccine may be intact but inactivated (‘attenuated’) forms of the causative pathogens (bacteria or viruses), or purified components of the pathogen that have been found to be highly immunogenic. The increased understanding of antigen recognition at molecular level has resulted in the development of rationally designed peptide vaccines. The concept of peptide vaccines is based on identification and chemical synthesis of B-cell and T-cell epitopes which are immunodominant and can induce specific immune responses. The accelerating growth of bioinformatics techniques and applications along with the substantial amount of experimental data has given rise to a new field, called immunoinformatics. Immunoinformatics is a branch of bioinformatics dealing with in silico analysis and modelling of immunological data and problems. Different sequence- and structure-based immunoinformatics methods are reviewed in the paper.
Keywords: T-cell epitopes, major histocompatibility-binding prediction, immunoinformatics
2. Introduction
The word ‘vaccination’ was used for first time by Edward Jenner in 1796 to describe the injection of smallpox vaccine [1]. Louis Pasteur developed the concept through his innovative work in microbiology. Now, vaccination is the administration of antigenic agents applied to stimulate the immune system of an individual and to develop adaptive immunity to a disease. Vaccines can ameliorate, or often even prevent, the effects of infection. Vaccination is generally considered to be the most effective method of preventing infectious diseases [2], and the efficacy of vaccination has been extensively studied and verified [3–5]. The administration of some vaccines is conducted after the patient has already been infected by the pathogen. Vaccination conducted after exposure to smallpox, within the first 3 days, is reported to attenuate the disease considerably, and administration up to a week after exposure is able to provide some protection from disease, or may ease its severity [6]. Also, a multi-stage tuberculosis vaccine has recently been developed to confer protection after the exposure to the pathogen [7]. There are numerous vaccine examples, including experimental ones against AIDS, cancer and Alzheimer's disease. The core mechanism behind all the vaccinations is the ability of the vaccine to initiate an immune response in a quicker fashion than the pathogen itself.
The purpose of every vaccination is to present a particular antigen or set of antigens to the immune system in order to evoke a relevant immune response. The main active component of a vaccine may be inactive, but still intact (attenuated bacteria or viruses), or purified components of the pathogen that are known to induce immune reaction.
3. Types of vaccines
3.1. Inactivated vaccines
This type of vaccine consists of virus particles grown in cell culture and inactivated by applying high temperature or chemicals such as formaldehyde. The viral particles are unable to replicate because they are destroyed, but the capsid proteins of the virus have remained intact enough to be recognized and used by the immune system in order to induce a response. If properly produced, the vaccine is not a threat; however, if the inactivation is not performed successfully, active infectious particles can be administered together with the vaccine. Additional booster shots are often needed in order to secure the immune response, because the properly produced vaccine cannot reproduce inside the host.
3.2. Live attenuated vaccines
The attenuated vaccines contain live virus particles with low levels of virulence. They have retained their ability to slowly reproduce, and thus they remain a continuous source of antigen for a certain period after the first vaccination, reducing the need of booster shots to keep the antigen levels sufficiently high. Such vaccines are produced by passing virus in cell cultures, in animals or at suboptimal temperatures, allowing selection of less virulent strains or by mutagenesis, or targeted deletions in genes required for virulence [8–10].
3.3. Subunit vaccines
Subunit vaccines use only the antigenic components that best stimulate the immune system, instead of dealing with the entire micro-organism. The fact that the subunit vaccine content is mainly represented by the essential antigens reduces the chances of adverse reactions to the vaccine. A subunit vaccine introduces an antigen to the immune system without involving any viral particles. The number of antigens in subunit vaccine can range from 1 to 20 or more. Of course, the identification of the most promising antigens to stimulate the immune system is often a time-consuming process, and can be very difficult. Subunit vaccines are often known for causing weaker antibody responses in comparison with the other vaccine classes. One of the most successful subunit vaccines is the hepatitis B vaccine containing the surface antigen HbsAg [11,12].
3.4. Virus-like particles
Virus-like particle (VLP) vaccines are comprised only of viral proteins that take part in the assembly of the virus structure. They have the ability to self-assemble into virus resembling the particles from which they were derived without the presence of the viral nucleic acid, which makes them simply non-pathogenic [13,14]. By contrast with the subunit vaccines, VLPs usually have higher immunogenicity owing to their multi-valent and highly repetitive structure. VLPs have been produced from a broad range of viruses that belong to Retroviridae, Flaviviridae and Parvoviridae families. Vaccines against viruses such as human papillomavirus and hepatitis B are VLP-based vaccines that are currently in clinical use [15]. Additionally, a pre-clinical vaccine against chikungunya virus was developed based on the same approach [16]. VLPs are typically produced in a variety of cell cultures, such as mammalian cell lines, insect cell lines, and plant and yeast cells [17].
3.5. Toxoid vaccines
The toxoid vaccines are typical solution for bacteria that secrete harmful metabolites or toxins. It is common to use them when the main reason for discomfort or sickness is a bacterial toxin. Such toxoid vaccines are produced by treating the toxins with formalin, thus inactivating them, and still retaining their structure for further recognition by the immune system. Examples of toxoid vaccines are the vaccines against diphtheria and tetanus.
3.6. DNA vaccines
DNA vaccination is a very new approach for induction of humoral and cellular immune responses to protein antigens by administering genetically engineered DNA. The majority of DNA vaccines are still in the experimental stage, and have been tested in numerous viral, bacterial and parasitic models of disease, and also in a few tumour models. DNA vaccines represent an innovative approach for immunization, bringing a number of advantages over conventional vaccines and giving the possibility of inducing a broader variety of immune response types [18–25]. The risks of DNA vaccines are limited [22]. Several groups demonstrated that cancer vaccines can be effective for the induction of specific immunity against cancer-associated antigens without negative side effects like integration of plasmid DNA into the host genomes or induction of pathogenic anti-DNA antibodies [23–25].
3.7. Peptide vaccines
The improved knowledge of antigen recognition at molecular level has contributed to the development of rationally designed peptide vaccines. The general idea behind the peptide vaccines is based on the chemical approach to synthesize the identified B-cell and T-cell epitopes that are immunodominant and can induce specific immune responses. B-cell epitope of a target molecule can be conjugated with a T-cell epitope to make it immunogenic. The first epitope-based vaccine was created in 1985 by Jackob et al. [26]. They introduced recombinant DNA and express epitopes against cholera in Escherichia coli. Epitope-based vaccines can be constructed for T and B lymphocytes [27,28]. The T-cell epitopes are typically peptide fragments, whereas the B-cell epitopes can be proteins, lipids, nucleic acids or carbohydrates [27–31]. Peptides have become desirable vaccine candidates owing to their comparatively easy production and construction, chemical stability, and absence of infectious potential. The peptide vaccines against various cancers have been developed, and entered phase I and phase II of clinical trials, with satisfactory clinical outcome. The peptide vaccination is commonly being studied for application in both ameliorating and prophylactic immunotherapy [32]. Yet there is more to be improved in order to eliminate obstacles, such as the need for a better adjuvant and carrier or the low immunogenicity. Nonetheless, current efforts are showing much promise in defying these limitations and providing improvements for this approach.
4. T-cell epitopes
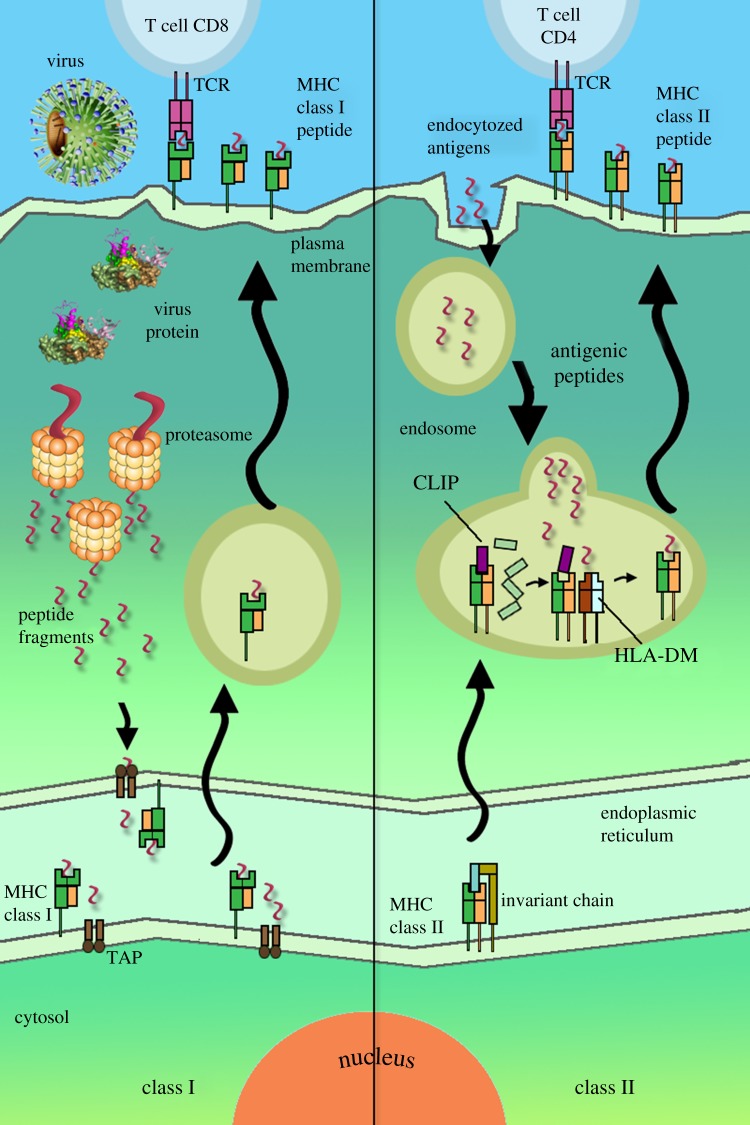
The epitope is recognizable by the immune system part of the antigen, and in particular by antibodies, B cells or T cells. The epitopes may belong to both foreign and self proteins, and they can be categorized as conformational or linear, depending on their structure and integration with the paratope [33]. T-cell epitopes are presented on the surface of an antigen-presenting cell (APC), where they are bound to major histocompatibility (MHC) molecules in order to induce immune response [34]. MHC class I molecules usually present peptides between 8 and 11 amino acids in length, whereas the peptides binding to MHC class II may have length from 12 to 25 amino acids [35]. MHC class II proteins bind oligopeptide fragments derived through the proteolysis of pathogen antigens, and present them at the cell surface for recognition by CD4+ T cells (figure 1). If sufficient quantities of the epitope are presented, the T cell may trigger an adaptive immune response specific for the pathogen. Class II MHCs are expressed on specialized cell types, including professional APCs such as B cells, macrophages and dendritic cells, whereas class I MHCs are found on every nucleated cell of the body [36].
Figure 1.
Antigen-processing pathways in the cell. Left: intracellular pathway. Protein is cleaved into oligopeptides in the proteasome, the peptides enter the endoplasmic reticulum (ER) via TAP protein and bind to MHC class I, and the complex peptide–MHC protein is presented on the cell surface. Right: extracellular pathway. Protein is endocytozed, cleaved into oligopeptides in the endosome, bound to MHC class II protein and presented on the cell surface. In the ER, MHC class II molecules are adjoined to a specific peptide, known as invariant chain (Ii). It blocks the binding cleft of the MHC molecule, thereby preventing the binding of endogenous peptides. In the endosome, the Ii is initially cleaved to CLIP peptide, and is then replaced by an exogenous peptide. The process is facilitated by the HLA-DM molecule.
The recognition of epitopes by T cells and the induction of immune response have a key role for the individual's immune system. Even the slightest deviation from the normal functioning can have a grave impact on the organism. In case of autoimmune disease, the T cells recognize the cells' native peptides as foreign, and attack and eventually destroy the organism's own tissues.
Some viruses, such as human immunodeficiency virus (HIV), hepatitis C, and avian and swine influenza, manage to avoid recognition by the T cell relying on various mutations that effectively alter the amino acid sequences of the proteins encoded by the viral genes [37,38].
Knowledge about the peptide's epitopes has a key role for manufacturing epitope-based vaccines, which, injected into the recipient, can induce immune response. One of the key issues in T-cell epitope prediction is the prediction of MHC binding, as it is considered a prerequisite for T cell recognition. All T-cell epitopes are good MHC binders, but not all good MHC binders are T-cell epitopes.
MHCs are among the most polymorphic proteins in higher vertebrates, with more than 6000 class I and class II MHC molecules listed in IMGT/HLA [39]. Determining the peptide-binding preferences exhibited by this extensive set of alleles is beyond the present capacity of experimental techniques, necessitating the development of bioinformatics prediction methodologies. The most successful prediction methods for T-cell epitopes developed to date have been data-driven. T-cell epitope prediction typically involves defining the peptide-binding specificity of specific class I or class II MHC alleles and then predicting epitopes in silico. Using peptide sequence data, experimentally determined affinity data have been used in the construction of many T-cell epitope prediction algorithms. Such methods include motif-based systems, support vector machines (SVMs) [40,41], hidden Markov models (HMMs) [42–44], quantitative structure–activity relationship (QSAR) analysis [45,46], and structure-based approaches [47].
5. Immunoinformatics
The accelerating growth of bioinformatics techniques and applications along with the substantial amount of experimental data has made a significant impact on the immunology research. This has led to a rapid growth in the field of computation immunology, and a number of immunology-focused resources and software, which help in understanding the properties of the whole immune system, have become available [48]. This has given rise to a new field, called immunoinformatics. Immunoinformatics can be described as a branch of bioinformatics concerned with in silico analysis and modelling of immunological data and problems.
Immunoinformatics research stresses mostly on the design and study of algorithms for mapping potential B- and T-cell epitopes, which speeds up the time and lowers the cost needed for laboratory analysis of pathogen gene products. Using such tools and information, an immunologist can analyse the sequence areas with potential binding sites, which in turn leads to the development of new vaccines. The methodology of analysing the pathogen genome to identify potential antigenic proteins is known as ‘reverse vaccinology’ [49]. This is mainly beneficial because conventional methods need to dedicate time to pathogen cultivation and subsequent protein extraction. Although pathogens grow quickly, extraction of their proteins and then testing of those proteins on a large scale is expensive and time-consuming. Immunoinformatics is capable of reducing time and saving resources for the development of relevant vaccines by revealing virulence genes and surface-associated proteins.
Normally, the investigation of the binding affinity of antigenic peptides to the MHC molecules is the main goal when predicting epitopes. The experimental techniques are found to be difficult and time-consuming, and therefore several in silico methodologies are being created and constantly improved to identify epitopes. The list of approaches includes matrix-driven methods, QSAR analysis, identification of structural binding motifs, protein threading, homology modelling, docking techniques, and design of several machine-learning algorithms and tools. In the past, computational techniques could only identify sequence characteristics, but new improved algorithms and tools are being designed to increase the predictive performance [49]. The methods used for development of prediction models can be divided into structure-based methods that derive information from the three-dimensional structure of the proteins, and sequence-based methods that analyse the amino acid sequence.
5.1. Sequence-based methods
5.1.1. Motif search-based approach
The combination of preferred amino acids at some of the peptide anchor binding positions is called a motif. The motif search is the most outdated, yet the most widely used method for prediction of epitopes [50–53]. The peptide amino acid sequence is searched for motifs by using a motif library [54]. The MHC-binding motifs for a given peptide can be identified by comparison of known binders and non-binders [55]. The motif search approach was used to identify epitopes that bind HLA-DR allele among the proteins expressed by Plasmodium falciparum [56]. EPIPREDICT is another motif-based tool, used for the identification of MHC class II-binding epitopes from proteins involved in the human gluten intolerance [57]. D'Amaro et al. [58] developed the computer program MOTIF, which yields collection of all the known affinity motifs to HLA-A*0201. The program identifies 27 binders when validated against an external test set, and the subsequent experiments confirm that 18 of these peptides exhibit binding affinity with overall accuracy of 61 per cent. Another tool is EpiMer, created at Brown University and used for prediction of HIV-related epitopes [59,60].
One of the widely used epitope prediction tools is SYFPEITHI, which is also based on the motif search approach [54,61]. Similar to the EpiMer approach, SYFPEITHI is used to score the peptides and evaluate their immunogenicity. Numerous experimental in vivo and in vitro assays have been conducted to validate the in silico predictions [62–70].
The accuracy of the motif-based algorithms is about 60–70 per cent, mostly because not all of the binding peptides have recognizable motifs [71]. In many cases, the correlation between the predicted and the experimentally determined affinities is very weak. A study conducted by Andersen et al. [72] compares the affinities predicted by SYFPEITHI and BIMAS binders with the experimentally determined ones from a set of oncogenes and viral proteins. The authors show a large number of wrongly identified false positives, while some of the actual epitopes are predicted as non-binders.
5.1.2. Prediction by artificial neural network
The artificial neural networks (ANNs) provide a convenient method for finding relationships and describing nonlinear data [73]. ANN methods are frequently used by the bioinformatics researchers for solving asthma-related problems [74], to investigate cardiac diseases [75] and drug solubility [76], and for epitope prediction and analysis of MHC haplotypes [77]. When applying for epitope prediction, the peptide length can be highly variable. The sequences included in the training set are usually aligned by assigning a specific anchor position. This is a trivial task when constructing models for MHC I prediction, where the difference in the peptide length is negligible, while it becomes a challenging quest for MHC II, where the length variability is considerably larger.
Nielsen et al. [78] described an improved neural network model to predict T-cell class I epitopes. The NETCTL server [79] (http://www.cbs.dtu.dk/services/NetCTL/) uses a method to integrate the prediction of peptide MHC class I binding, proteasomal C-terminal cleavage and transporter associated with antigen processing (TAP) transport efficiency. It has updated from version 1.0 to 1.2 to improve the accuracy of MHC class I peptide-binding affinity and proteasomal cleavage prediction. NETMHC server v. 3.2 [80] (http://www.cbs.dtu.dk/services/NetMHC) is based on ANN and weight matrices. It has been trained on data from 55 MHC peptides (43 human and 12 non-human) and position-specific scoring matrices for a further 67 HLA alleles. MHC class I molecule motifs are well defined, but the prediction of MHC class II binding peptides is considered harder to achieve, mainly because of the variable length of reported binding peptides, the undetermined core region for each peptide and the number of primary anchor amino acids.
5.1.3. Prediction by support vector machine
The SVM is a computer science concept for a set of supervised learning methods used for data analysis and pattern recognition, developed by Vapnik [81] and commonly used for image and data classification and regression analysis [82]. SVMs belong to the group of the kernel-based approaches [83]. Classically, the SVM takes a set of data and predicts, for each given input, to what type of input class it belongs; therefore, SVM is described as a non-probabilistic binary linear classifier. The SVM model can be represented as two sets of points in space, distributed in a way that the two subsets falling into separate categories are divided by a clear gap that is as wide as possible. The model categorizes the novel data points depending on which side of the gap they fall on.
Another formal description of the SVM method is that it defines a hyperplane or set of hyperplanes in a high- or infinite-dimensional space, which can be used for classification, regression or other purposes. The optimal separation can be achieved by deriving the hyperplane that is positioned at the largest distance from the nearest points belonging to any of the modelled classes. The larger the distance, the more reliable is the model [84].
Nanni [85] demonstrated the use of SVM and SV data description to predict T-cell epitopes. In the case of TAPPRED, Bhasin & Raghava [86], analysed nine features of amino acids to find the correlation between binding affinity and physico-chemical properties. They developed an SVM-based method to predict the TAP binding affinity of peptides, and found cascade SVM to be more reliable. Cascade SVM has two layers of SVMs, and its performance is better than the other available algorithms. It is experimentally determined that the immunoproteasome plays a role in the generation of the MHC class I ligand. Often the computational approach is preferred over experimental analysis for studying and predicting the cleavage specificities of proteasomes. Therefore, a web application called PCLEAVAGE [87] has been developed to predict cleavage sites in antigenic proteins. It uses SVM [88], parallel exemplar-based learning [89] and Waikato Environment for Knowledge Analysis [90].
Sweredoski & Baldi [91] presented COBEPRO, which is a two-step system for the prediction of continuous B-cell epitopes. In the first step, COBEPRO assigns a fragment epitopic propensity score to protein sequence fragments using an SVM. In the second step, it calculates an epitopic propensity score for each residue based on the SVM scores of the peptide fragment in the antigenic sequence. It is incorporated into the SCARTCH prediction suite. However, COBEPRO is not able to find the difference between antigen and non-antigen, and in order to increase the efficacy it should be used with high-throughput technologies.
5.1.4. Hidden Markov models
HMMs were initially described in the second half of the 1960s by Baum et al. [92]. HMMs were first applied for speech recognition in the mid-1970s [93,94]. In the second half of the 1980s, HMMs found their application in the analysis of biological sequences [95], and in particular of DNA sequences. Since then, they have become ubiquitous in the field of bioinformatics [96].
HMM-based approaches are widely used in bioinformatics and proteomics for the prediction of protein sequences with helical secondary structure [97], transmembrane regions [98,99] and protein homology analysis [100]. HMM is also used for sequence alignment [101], and protein family identification by Pfam and SMART [102]. For the purposes of genomics, HMM is used for studying gene splicing [103], phylogenetic tree analysis [104] and gene identification in procariotes [105].
Zhang et al. [106] developed PREDTAP for the prediction of peptide binding to hTAP. They used a three-layer back propagation network with the sigmoid activation function. The inputs were the binary strings, representing nonamer peptide. In addition, they used second-order HMM. The results were both sensitive and specific. Mamitsuka [44] derived HMM-based, high-accuracy models for prediction of peptide-binding affinity to HLA-A*0201 and DR1 proteins. By using Mamitsuka's approach, Udaka et al. [107] derived models for other MHC class I proteins. Brusic et al. [108] also used HMM for binding affinity prediction towards the HLA-A2 family members. The analysis included only the amino acids involved in a direct interaction with the protein. HMM was derived for each allele of the family, and peptides also binding to the other alleles were used as a training set. The test sets comprised peptides binding to the corresponding allele.
Schonbach et al. [109] compare the predictions done by HMM, ANN and quantitative matrices (QMs). Over 500 amino acid sequences of HIV-1 and -2 are scanned for peptides with affinity to A*0201 and B*3501. The ANN model showed high performance for the A*0201 allele, and the HMM was more successful in predicting B*3501 binders. Subsequent experiments showed that 26 per cent of the epitopes were successfully identified by the models based on QMs and ANN.
5.1.5. Prediction by quantitative matrices-driven methods
QMs resemble an extended motif with assigned coefficients for each amino acid at each position in the peptide [110]. In principle, matrix-based epitope prediction can be divided into four steps: first, all possible peptide frames are extracted from a given protein sequence. Second, the corresponding position- and amino acid-specific matrix values are assigned to each residue of a given peptide frame. Next, the side chain values of each peptide are added or multiplied, resulting in the peptide ‘score’. Last, peptides are selected based on their peptide score. Thus, instead of simply counting anchor residues, matrix-based algorithms take into account the relative importance of every amino acid residue in a peptide sequence, as charged by their effect on binding. QMs provide a linear model with easy-to-implement capabilities. Another advantage of using this approach is that it covers a wider range of peptides with binding potential and it gives a quantitative score to each peptide. Their predictive accuracies are also considerable. The capacity to predict HLA class II ligands using QM-based algorithms was first demonstrated for DRB1*0401 molecules [111,112]. These algorithms ranked naturally processed peptides and T-cell epitopes in the top 2–4 per cent of all possible peptide frames of given antigens, even if they owned only one or two anchor residues. More important, however, a correlation between the peptide score and the binding affinity was demonstrated [111], which therefore supports the underlying approximation that a given residue contributes to binding independently of its neighbouring amino acid residues. Later on, many more QM-based algorithms were established, including algorithms for DRB1*0101, DRB1*1501, DRB1*1101, DRB1*0701 and DRB1*0801 molecules. The predictive power of some of these algorithms was validated by a computer simulating the screening of M13 peptide display libraries. QM-based algorithms were used instead of purified HLA-class II molecules to enrich for large class II-binding peptide repertoires [113].
QMs are also applied for the prediction of cleavage sites and are implemented in MAPPP [114]. Similar algorithms are applied for the prediction of linear epitopes of the B lymphocytes. Alix [115] calculates the molecular properties for the 20 common amino acids (side chain flexibility, hydrophilic affinity and accessible surface) and uses these properties for the prediction of potential epitope regions in the proteins that would possibly bind to the B cells.
BIMAS is a T-cell epitope prediction server that implements algorithms based on QM [116]. BIMAS was used for the identification of various potential epitopes [64,70,117,118]. QM was derived from experimental data from the dissociation half-time of the MHC–peptide complexes. The model predicting binding to HLA-A*0201 allele is based on the author's data, and the models for the other alleles are based on the literature data. Servers such as BIMAS and SYFPEITHI are shown to perform well in the prediction of known epitopes, but are accurate enough when screening proteins in search for unknown and novel epitopes [69].
Another QM-based model is EpiMatrix, developed at Brown University [59]. It has been used for the identification of HIV-1 antigens [59,119]. Other similar approaches are implemented in ClustiMer and Conservatrix. ClustiMer identifies promiscuous (for a given HLA superfamily) peptides, and Conservatrix determines unchanged (conserved) regions in the proteins of the mutant pathogens of the same species [120].
Another category of QMs is the position-specific matrices, where the frequency at which the given amino acid appears at a certain position is calculated for binding and non-binding to MHC peptides [121]. Nielsen et al. [78] derive QM for MHC class I and II epitopes accounting for the changes in the Gibbs energy.
Virtual matrix (VM) is another type of QM, created by Sturniolo et al. [122]. VM models the interactions between each amino acid and the pockets of the binding groove. The advantage comes from the applicability of the VM to different alleles that share similar structural characteristics of the binding groove, whereas the QMs are strictly specific to the given allele. TEPITOPE is VM-based and predicts peptides that are HLA-DR binders. TEPITOPE is used for identification of epitopes in the tumour antigen MAGE-3 [123,124]. Another tool using VM is ProPred, created by Singh & Ragava [125], where the profiles of the MHC protein pockets created by Sturniolo served as a foundation for the models.
MHCPred is a sequence-based server using the additive method [126] for developing QMs. The additive method derives QMs using multiple linear regression by partial least-squares (PLS) method. MHCPred was used to design superbinders [127] and to identify the first T-cell epitope binding to HLA-Cw*0102, and originating from HIV proteome [128].
EpiJen is a multi-step algorithm for T-cell epitope prediction. It models the four steps of antigen processing—cleavage in the proteasome, binding to TAP protein, binding to MHC protein and recognition by T cells [129]. For each step, a QM was developed and arranged in a consecutive mode to select only those peptides that will be generated by the proteasome, transported by TAP, bound in MHC and recognized by T cells. In the final set are collected the peptides most probably acting as T-cell epitopes.
VaxiJen predicts immunogenicity of whole proteins. It includes five models derived by PLS-based discriminant analysis, which covers the bacterial, viral, tumour, parasite and fungal kingdoms [130]. The models show accuracy between 70 and 97 per cent.
EpiTOP is a server for MHC class II-binding prediction based on proteochemometrics [131]. Proteochemometrics is a QSAR method specially designed to deal with ligands binding to a set of similar proteins [132]. The structures of the target proteins are described by proper descriptors and enter the X matrix of QSAR. The affinity of a peptide to a particular MHC protein is considered as a function of the structures of both binding peptide and target protein. EpiTOP is among the top three best-working servers for MHC class II-binding prediction [131].
The main drawback of the quantitative models is that they are strongly dependent on the type, number and quality of the data that comprise the training set of peptides. The inclusion of novel data often alters the values upon which the QM is based. Brusic et al. [133] suggest as a prerequisite a threshold value for the derivation of a reliable model to be 150 peptides and the ideal size of training set should reach 600 peptides. However, in reality, most of the alleles are represented by scarce data rarely exceeding more than 50 peptides. This limits the range of applicability for this approach to the alleles that are sufficiently well studied.
5.2. Structure-based methods
The structure-based methods do not solely rely on binding data and sequence information, but rather use the structural information, and use computational methods developed in the field of structural biology for prediction of potentially good binders.
For the MHC molecule to recognize antigenic peptides, geometric and electrostatic complementarities between the receptor and ligand are essential for the formation of a stable complex. Many computational studies that attempt to unravel the rules governing peptide binding to MHC use the sequences of MHC-binding peptides. By aligning the sequences known to bind to a given MHC molecule, residues favouring the binding could be identified along the peptide. The synthesis of this knowledge together with that obtained from crystallographic studies has led to better understanding of the basic principles that guide peptide–MHC recognition [134,135].
5.2.1. Docking of peptides and screening of peptide libraries
Over recent years, many techniques and methods, such as combinatorial peptide library screening and ligand docking, commonly used in the drug design field, have found their application for the purposes of bioinformatics. Davenport et al. [136] generated MHC class II models by evaluating the contribution of a given amino acid to the overall peptide affinity. They took into account how frequently the amino acid is present at a certain position. New peptides exhibiting affinity towards DRB1*0101 were found based on relationships derived from peptide libraries [137]. Screening of peptide libraries was also applied for studying other MHC alleles. Stryhn et al. [138] analysed the peptide specificities of MHC class I binders by using peptide libraries. Stevens et al. [139] used peptide libraries to determine the preferred peptide length for murine MHC alleles. By using the positional screening of combinatorial peptide libraries, Udaka et al. [140,141] characterize the peptides binding to H-Kb Db and Ld alleles. The different amino acids were screened for how frequently they appear at the different positions of the peptides from the training set, and QMs were generated in order to predict the affinity of the peptides from the test set. The accuracy of the predictions reached 80 per cent. Similar studies were conducted by Sung et al. [142] and Nino-Vasquez et al. [143].
Computer-simulated ligand docking is a quick and powerful technique for investigating intermolecular interactions. In general, the purpose of docking simulation is twofold: to find the most probable translational, rotational and conformational juxtaposition of a given ligand–receptor pair and to evaluate the relative binding affinity of the ligand towards its receptor.
Docking is mostly known for its wide application in computer-aided drug design [144]. However, this approach found its application for designing novel peptides exhibiting binding affinity towards MHC. Initially, the docking studies were mainly used for investigation of peptides that bind MHC class I molecules [145,146]. Zeng et al. [147] used residues with different properties (polar, hydrophobic, charged, etc.) by docking them to different positions of the binding groove of the receptor, thus evaluating the most acceptable residues' properties for each position of the potential epitope. Another study [148] uses a genetic algorithm in order to derive QM for A2 and A24 alleles, and peptides with high binding affinity are designed. The peptide structures were modelled and docked to the binding groove. The binding energy was calculated as a sum of the electrostatic and hydrophobic components. After the experimental determination of the peptides' binding affinity, good correlation is observed between the predicted and the experimentally derived values.
Docking is also used for studying peptides binding MHC class II alleles for identification of anchor positions and positions that are solvent-exposed [149]. The interaction between the T-cell receptor and the MHC–ligand complex were also studied via docking [150,151]. Tong et al. [152] develop a novel docking approach that consists of three steps: (i) anchor residue docking; (ii) positioning of the peptide backbone in the binding groove; and (iii) adjustment of the overall positioning of the peptide backbone and the side chains. This approach showed improved accuracy in comparison with the other methods. Liu et al. [67] take into account the flexibility of the MHC proteins during the docking simulation. However, despite the high predictive accuracy, these methods are not feasible for online predictions since the time required for the simulation is unreasonably long. Furthermore, the accuracy of the predictions is highly dependent on the quality of the structural information available for the receptor and the correctly modelled backbone of the ligand.
EpiDOCK is a structure-based server for MHC-binding prediction of peptides using docking score-based QMs (DS-QMs) [153]. It predicts binding to 12 HLA-DR, 6 HLA-DQ and 5 HLA-DP proteins.
5.2.2. Application of threading algorithms
Knowledge-based threading algorithms are used to discriminate the binding and non-binding peptides for particular MHC molecules without relying on previous data. The algorithm usually takes into account the contributions of individual amino acids along the peptide that prompt them to fit into the binding groove of MHC molecule using knowledge-based contact potential [154]. Often, the accurate prediction of peptide structure in the MHC-binding groove is hindered owing to the limited availability of suitable peptide backbone templates. Still, the applicability of the threading algorithm can be extended to a larger number of MHC alleles for the prediction of T-cell epitope by using molecular modelling methods on the peptide–MHC complex. Although the treading is not capable of exact modelling of peptide in the MHC groove, it can verify the probability of a peptide sequence to adopt a particular fold in the MHC groove using binding energy score [155–157].
Adrian et al. [155] studied the MHC complex–peptide interactions, and reveal the significant role played by the peptide's backbone for the overall binder's selection. They also stress the significance of exact knowledge about the ligand's conformation and its impact on the ability to produce more accurate prediction models. They use threading to predict the peptides’ conformations by remodelling them over the existing backbone known from an X-ray study of MHC complexes. The scores used to evaluate the overall binding affinity are additively calculated by summing the individual binding energy score of each amino acid residue at each position [158]. The lower values correspond to higher affinity [156,157].
The drawback of this method is that despite the high level of overlapping between the referent and the tested peptides, some residue side chains tend to be oriented in different directions, and thus worsen the predictability. Additional modelling, however, may improve the predictive accuracy of the model [157].
5.2.3. Binding energy and molecular dynamics
The epitopes can be identified by calculating the change in the free Gibbs energy during the formation of the complex between the ligand and the receptor, which is defined as the difference between the energy of the free and the bound peptide [159,160]. The epitopes can be found by direct comparison of the free energies of two peptides by using scoring functions or molecular dynamics (MD) simulations [161]. MD is used for studying the binding of synthetic peptides [162], MHC peptide–protein complexes [163,164], the role of the water molecules involved in the formation of the peptide–protein complex [165], the interactions between A2 peptides and the receptor's binding groove [161,166], the dissociation of the MHC–peptide complexes [167], and the interactions between the T-cell receptor and the peptide–MHC protein complex [168]. Rognan et al. [163] simulated the binding of six peptides to B*2705 protein and showed the importance of the secondary anchor residues. Lim et al. [169] simulated the interaction between the peptide and HLA-A*0201 protein by using the available X-ray structure. The peptides predicted to have high binding affinity were validated experimentally. In another study, MD is used to identify the contribution of each residue at a given position and the results are used to form a QM for epitope prediction [147]. Analogous MD simulations are performed in order to determine anchor residues for the HLA-A*0217 allele [170]. MD simulations are used for studying peptides binding to DRB1 [171]. Davies et al. [172] built epitope prediction models for MHC class II proteins by using simulated annealing, a common optimization method where the peptide conformation is obtained by rapid increase of the temperature and subsequent recalculation of the protein coordinates by gradually decreasing the temperature at each step. The energy of the resulting complex is derived and used for binding affinity predictions.
Another approach is to derive the binding energy as a difference between the energy of the solvated complex and the energies of the solvated binding partners—peptide and protein receptor. Only the electrostatic and hydrophobic terms are taken into account [173].
Different scoring functions can be used for the evaluation of the interactions between the peptide and the MHC protein. The advantage of this approach is that it delivers more accurate information about which types of interactions govern the stability of the complex [174,175]. Sezerman et al. [159] generate free energy maps describing the binding sites along the binding groove of the MHC class I proteins by using the electrostatic energy, solvation energy and the conformational entropy terms of the amino acid side chains. Froloff et al. [176] calculate the binding energy for eight peptide MHC class I protein complexes based on polar and non-polar interactions. Schapira et al. [173] calculate the binding energy based on three terms—entropic, electrostatic and hydrophobic potentials—and use it for predicting the formation of small protein complexes.
The free energy calculation approach was also applied on peptides binding to HLA-A*0201 [177]. They used an energy evaluation function where the free-binding energy consists of five terms: hydrogen bond energy between the peptide and the receptor, interaction energy between the hydrophobic atoms, entropic loss upon binding, decrease of the binding energy upon interaction between polar and non-polar atoms, and the transition energy required for the transport of an atom between environments with different dielectric constants. For another experiment, Rognan and co-workers [174] used the Fresno method for prediction of the free-binding energy. The training set includes five known binders interacting with HLA-A*0201; there is X-ray data and complex affinity data available for the complexes. Based on the free complex energy, a model is derived to predict the affinity of 26 more binders to the HLA-A*0204 allele that shares significant structure similarities with HLA-A*0201. The study shows, however, that the predictive accuracy is much higher when there is structural information available about the receptor. This approach was used for estimation of the binding energy of peptides binding to A*0201 and B*2705 by using the available X-ray structures [174]. Later on, the Fresno approach is applied to build the peptide MHC–protein complexes via homology modelling and to calculate the binding energy [175]. The main drawback of this method is the amount of time and computational power that it takes to produce results, which makes it inapplicable for online access.
6. Conclusion
Immunoinformatics can effectively leverage computational techniques to deliver effective and utilitarian advantage in the search of new vaccines. It is considered to contribute to vaccine design as the computational chemistry contributes to drug design. Immunoinformatics-based vaccine design is able to achieve effective, cost-efficient development of vaccines or vaccine components.
7. Acknowledgements
The authors thank their colleagues from the Faculty of Pharmacy, the Medical University of Sofia—Ivan Dimitrov, Mariyana Atanasova and Panaiot Garnev—for their contributions in the developing of EpiTOP, AllerTOP and EpiDOCK. I.D. thanks her former colleagues from the Jenner Institute, Oxford University—Darren R. Flower, Pigping Guan, Channa Hattotuwagama and Martin Blythe—for their contributions in the developing of MHCPred, EpiJen and VaxiJen. Part of this work was supported by the National Research Fund of the Bulgarian Ministry of Education and Science (grant no. 02-1/2009).
References
- 1.Lombard M, Pastoret PP, Moulin AM. 2007. A brief history of vaccines and vaccination. Rev. Sci. Tech. 26, 29–48 [DOI] [PubMed] [Google Scholar]
- 2.Hellstrom KE, Hellstrom I. 2003. Novel approaches to therapeutic cancer vaccines. Expert Rev. Vaccines 2, 517–532 10.1586/14760584.2.4.517 (doi:10.1586/14760584.2.4.517) [DOI] [PubMed] [Google Scholar]
- 3.Fiore AE, Bridges CB, Cox NJ. 2009. Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 333, 43–82 10.1007/978-3-540-92165-3_3 (doi:10.1007/978-3-540-92165-3_3) [DOI] [PubMed] [Google Scholar]
- 4.Liesegang TJ. 2009. Varicella zoster virus vaccines: effective, but concerns linger. Can. J. Ophthalmol. 44, 379–384 10.3129/i09-126 (doi:10.3129/i09-126) [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Brewer NT, Rinas AC, Schmitt K, Smith JS. 2009. Evaluating the impact of human papillomavirus vaccines. Vaccine 27, 4355–4362 10.1016/j.vaccine.2009.03.008 (doi:10.1016/j.vaccine.2009.03.008) [DOI] [PubMed] [Google Scholar]
- 6.Mortimer PP. 2003. Can postexposure vaccination against smallpox succeed? Clin. Infect. Dis. 36, 622–629 10.1086/374054 (doi:10.1086/374054) [DOI] [PubMed] [Google Scholar]
- 7.Rupprecht CE, et al. 2010. Use of a reduced (4-dose) vaccine schedule for postexposure prophylaxis to prevent human rabies: recommendations of the advisory committee on immunization practices. MMWR Recomm. Rep. 59, 1–9 [PubMed] [Google Scholar]
- 8.Newman MJ, Livingston B, McKinney DM, Chesnut RW, Sette A. 2002. T-lymphocyte epitope identification and their use in vaccine development for HIV-1. Front. Biosci. 7, d1503–d1515 10.2741/newman (doi:10.2741/newman) [DOI] [PubMed] [Google Scholar]
- 9.Payette PJ, Davis HL. 2001. History of vaccines and positioning of current trends. Curr. Drug Targets Infect. Disord. 1, 241–247 10.2174/1568005014606017 (doi:10.2174/1568005014606017) [DOI] [PubMed] [Google Scholar]
- 10.Smith SG. 1999. The polyepitope approach to DNA vaccination. Curr. Opin. Mol. Ther. 1, 10–15 [PubMed] [Google Scholar]
- 11.Szmuness W, Stevens CE, Harley EJ, Zang EA, Taylor PE, Alter HJ. 1981. The immune response of healthy adults to a reduced dose of hepatitis B vaccine. J. Med. Virol. 8, 123–129 [DOI] [PubMed] [Google Scholar]
- 12.Szmuness W, Stevens CE, Oleszko WR, Goodman A. 1981. Passive–active immunisation against hepatitis B: immunogenicity studies in adult Americans. Lancet 1, 575–577 10.1016/S0140-6736(81)92030-4 (doi:10.1016/S0140-6736(81)92030-4) [DOI] [PubMed] [Google Scholar]
- 13.Adolph KW, Butler PJG. 1976. Assembly of a spherical plant-virus. Phil. Trans. R. Soc. Lond. B 276, 113–122 10.1098/rstb.1976.0102 (doi:10.1098/rstb.1976.0102) [DOI] [PubMed] [Google Scholar]
- 14.Chromy LR, Pipas JM, Garcea RL. 2003. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc. Natl Acad. Sci. USA 100, 10 477–10 482 10.1073/pnas.1832245100 (doi:10.1073/pnas.1832245100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer ME, Blumberg BS, Werner B. 1968. Particles associated with Australia antigen in the sera of patients with leukaemia, Down's Syndrome and hepatitis. Nature 218, 1057–1059 10.1038/2181057a0 (doi:10.1038/2181057a0) [DOI] [PubMed] [Google Scholar]
- 16.Akahata W, et al. 2010. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16, 334–338 10.1038/nm.2105 (doi:10.1038/nm.2105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santi L, Huang Z, Mason H. 2006. Virus-like particles production in green plants. Methods 40, 66–76 10.1016/j.ymeth.2006.05.020 (doi:10.1016/j.ymeth.2006.05.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurunathan S, Klinman DM, Seder RA. 2000. DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol. 18, 927–974 10.1146/annurev.immunol.18.1.927 (doi:10.1146/annurev.immunol.18.1.927) [DOI] [PubMed] [Google Scholar]
- 19.Liu MA. 2003. DNA vaccines: a review. J. Intern. Med. 253, 402–410 10.1046/j.1365-2796.2003.01140.x (doi:10.1046/j.1365-2796.2003.01140.x) [DOI] [PubMed] [Google Scholar]
- 20.Darji A, Guzmán CA, Gerstel B, Wachholz P, Timmis KN, Wehland J, Chakraborty T, Weiss S. 1997. Oral somatic transgene vaccination using attenuated S. typhimurium. Cell 91, 765–775 10.1016/S0092-8674(00)80465-1 (doi:10.1016/S0092-8674(00)80465-1) [DOI] [PubMed] [Google Scholar]
- 21.Paglia P, Medina E, Arioli I, Guzman CA, Colombo MP. 1998. Gene transfer in dendritic cells, induced by oral DNA vaccination with Salmonella typhimurium, results in protective immunity against a murine fibrosarcoma. Blood 92, 3172–3176 [PubMed] [Google Scholar]
- 22.Klinman DM, Takeno M, Ichino M, Gu M, Yamshchikov G, Mor G, Conover J. 1997. DNA vaccines: safety and efficacy issues. Springer Semin. Immunopathol. 19, 245–256 10.1007/BF00870272 (doi:10.1007/BF00870272) [DOI] [PubMed] [Google Scholar]
- 23.Disis ML, Gooley TA, Rinn K, Davis D, Piepkorn M, Cheever MA, Knutson KL, Schiffman K. 2002. Generation of T-cell immunity to the HER-2/neu protein after active immunization with HER-2/neu peptide-based vaccines. J. Clin. Oncol. 20, 2624–2632 10.1200/JCO.2002.06.171 (doi:10.1200/JCO.2002.06.171) [DOI] [PubMed] [Google Scholar]
- 24.Ledwith BJ, et al. 2000. Plasmid DNA vaccines: assay for integration into host genomic DNA. Dev. Biol. 104, 33–43 [PubMed] [Google Scholar]
- 25.Manam S, et al. 2000. Plasmid DNA vaccines: tissue distribution and effects of DNA sequence, adjuvants and delivery method on integration into host DNA. Intervirology 43, 273–281 10.1159/000053994 (doi:10.1159/000053994) [DOI] [PubMed] [Google Scholar]
- 26.Jacob CO, Leitner M, Zamir A, Salomon D, Arnon R. 1985. Priming immunization against cholera toxin and E. coli heat-labile toxin by cholera toxin short peptide-beta-galactosidase hybrid synthesized in E. coli. EMBO J. 4, 3339–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dermime S, Gilham DE, Shaw DM, Davidson EJ, Meziane el K, Armstrong A, Hawkins RE, Stern PL. 2004. Vaccine and antibody-directed T cell tumour immunotherapy. Biochim. Biophys. Acta 1704, 11–35 [DOI] [PubMed] [Google Scholar]
- 28.Meloen RH, Langeveld JP, Schaaper WM, Slootstra JW. 2001. Synthetic peptide vaccines: unexpected fulfillment of discarded hope? Biologicals 29, 233–236 10.1006/biol.2001.0298 (doi:10.1006/biol.2001.0298) [DOI] [PubMed] [Google Scholar]
- 29.Sundaram R, Beebe M, Kaumaya PT. 2004. Structural and immunogenicity analysis of chimeric B-cell epitope constructs derived from the gp46 and gp21 subunits of the envelope glycoproteins of HTLV-1. J. Pept. Res. 63, 132–140 10.1111/j.1399-3011.2003.00113.x (doi:10.1111/j.1399-3011.2003.00113.x) [DOI] [PubMed] [Google Scholar]
- 30.Mahler M, Bluthner M, Pollard KM. 2003. Advances in B-cell epitope analysis of autoantigens in connective tissue diseases. Clin. Immunol. 107, 65–79 10.1016/S1521-6616(03)00037-8 (doi:10.1016/S1521-6616(03)00037-8) [DOI] [PubMed] [Google Scholar]
- 31.Lehner T, Walker P, Smerdon R, Childerstone A, Bergmeier LA, Haron J. 1990. Identification of T- and B-cell epitopes in synthetic peptides derived from a Streptococcus mutans protein and characterization of their antigenicity and immunogenicity. Arch. Oral Biol. 35, S39–S45 10.1016/0003-9969(90)90129-X (doi:10.1016/0003-9969(90)90129-X) [DOI] [PubMed] [Google Scholar]
- 32.Naz RK, Dabir P. 2007. Peptide vaccines against cancer, infectious diseases, and conception. Front. Biosci. 12, 1833–1844 10.2741/2191 (doi:10.2741/2191) [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Honda W. 2006. CED: a conformational epitope database. BMC Immunol. 7, 7. 10.1186/1471-2172-7-7 (doi:10.1186/1471-2172-7-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madden DR. 1995. The three-dimensional structure of peptide–MHC complexes. Annu. Rev. Immunol. 13, 587–622 10.1146/annurev.iy.13.040195.003103 (doi:10.1146/annurev.iy.13.040195.003103) [DOI] [PubMed] [Google Scholar]
- 35.Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. 1996. Crystallographic analysis of endogenous peptides associated with HLA-DR1 suggests a common, polyproline II-like conformation for bound peptides. Proc. Natl Acad. Sci. USA 93, 734–738 10.1073/pnas.93.2.734 (doi:10.1073/pnas.93.2.734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janeway CA. 2001. Immunobiology: the immune system in health and disease. New York, NY: Churchill Livingstone [Google Scholar]
- 37.Letvin NL, Walker BD. 2001. HIV versus the immune system: another apparent victory for the virus. J. Clin. Invest. 107, 273–275 10.1172/JCI12174 (doi:10.1172/JCI12174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirskyj D, Diaz-Mitoma F, Golshani A, Kumar A, Azizi A. 2011. Innovative bioinformatic approaches for developing peptide-based vaccines against hypervariable viruses. Immunol. Cell Biol. 89, 81–89 10.1038/icb.2010.65 (doi:10.1038/icb.2010.65) [DOI] [PubMed] [Google Scholar]
- 39.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SGE. 2011. The IMGT/HLA database. Nucleic Acids Res. 39, D1171–1176 10.1093/nar/gkq998 (doi:10.1093/nar/gkq998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W, Meng XS, Xu QQ, Flower DR, Li T. 2006. Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models. BMC Bioinform. 7, 182. 10.1186/1471-2105-7-182 (doi:10.1186/1471-2105-7-182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan J, Liu W, Xu QQ, Ren Y, Flower DR, Li T. 2006. SVRMHC prediction server for MHC-binding peptides. BMC Bioinform. 7, 463. 10.1186/1471-2105-7-463 (doi:10.1186/1471-2105-7-463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Bickis MG, Wu FX, Kusalik AJ. 2006. Optimally-connected hidden Markov models for predicting MHC-binding peptides. J. Bioinform. Comput. Biol. 4, 959–980 10.1142/S0219720006002314 (doi:10.1142/S0219720006002314) [DOI] [PubMed] [Google Scholar]
- 43.Noguchi H, Kato R, Hanai T, Matsubara Y, Honda H, Brusic V, Kobayashi T. 2002. Hidden Markov model-based prediction of antigenic peptides that interact with MHC class II molecules. J. Biosci. Bioeng. 94, 264–270 [DOI] [PubMed] [Google Scholar]
- 44.Mamitsuka H. 1998. Predicting peptides that bind to MHC molecules using supervised learning of hidden Markov models. Proteins 33, 460–474 (doi:10.1002/(SICI)1097-0134(19981201)33:4<460::AID-PROT2>3.0.CO;2-M) [DOI] [PubMed] [Google Scholar]
- 45.Doytchinova IA, Flower DR. 2003. The HLA-A2-supermotif: a QSAR definition. Org. Biomol. Chem. 1, 2648–2654 10.1039/b300707c (doi:10.1039/b300707c) [DOI] [PubMed] [Google Scholar]
- 46.Doytchinova IA, Walshe V, Borrow P, Flower DR. 2005. Towards the chemometric dissection of peptide–HLA-A*0201 binding affinity: comparison of local and global QSAR models. J. Comput. Aid. Mol. Des. 19, 203–212 10.1007/s10822-005-3993-x (doi:10.1007/s10822-005-3993-x) [DOI] [PubMed] [Google Scholar]
- 47.Wan SZ, Coveney PV, Flower DR. 2005. Molecular basis of peptide recognition by the TCR: affinity differences calculated using large scale computing. J. Immunol. 175, 1715–1723 [DOI] [PubMed] [Google Scholar]
- 48.Brusic V, Petrovsky N. 2005. Immunoinformatics and its relevance to understanding human immune disease. Expert Rev. Clin. Immunol. 1, 145–157 10.1586/1744666X.1.1.145 (doi:10.1586/1744666X.1.1.145) [DOI] [PubMed] [Google Scholar]
- 49.Tomar N, De RK. 2010. Immunoinformatics: an integrated scenario. Immunology 113, 153–168 10.1111/j.1365-2567.2010.03330.x (doi:10.1111/j.1365-2567.2010.03330.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sette A, Buus S, Appella E, Smith JA, Chesnut R, Miles C, Colon SM, Grey HM. 1989. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl Acad. Sci. USA 86, 3296–3300 10.1073/pnas.86.9.3296 (doi:10.1073/pnas.86.9.3296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pamer EG, Harty JT, Bevan MJ. 1991. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature 353, 852–855 10.1038/353852a0 (doi:10.1038/353852a0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suhrbier A, Schmidt C, Fernan A. 1993. Prediction of an HLA B8-restricted influenza epitope by motif. Immunology 79, 171–173 [PMC free article] [PubMed] [Google Scholar]
- 53.Joyce S, Nathenson SG. 1994. Methods to study peptides associated with MHC class I molecules. Curr. Opin. Immunol. 6, 24–31 10.1016/0952-7915(94)90029-9 (doi:10.1016/0952-7915(94)90029-9) [DOI] [PubMed] [Google Scholar]
- 54.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 10.1007/s002510050595 (doi:10.1007/s002510050595) [DOI] [PubMed] [Google Scholar]
- 55.Altuvia Y, Berzofsky JA, Rosenfeld R, Margalit H. 1994. Sequence features that correlate with MHC restriction. Mol. Immunol. 31, 1–19 10.1016/0161-5890(94)90133-3 (doi:10.1016/0161-5890(94)90133-3) [DOI] [PubMed] [Google Scholar]
- 56.Doolan DL, et al. 1997. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity 7, 97–112 10.1016/S1074-7613(00)80513-0 (doi:10.1016/S1074-7613(00)80513-0) [DOI] [PubMed] [Google Scholar]
- 57.Jung G, Fleckenstein B, von der Mulbe F, Wessels J, Niethammer D, Wiesmuller KH. 2001. From combinatorial libraries to MHC ligand motifs, T-cell superagonists and antagonists. Biologicals 29, 179–181 10.1006/biol.2001.0299 (doi:10.1006/biol.2001.0299) [DOI] [PubMed] [Google Scholar]
- 58.D'Amaro J, Houbiers JG, Drijfhout JW, Brandt RM, Schipper R, Bavinck JN, Melief CJ, Kast WM. 1995. A computer program for predicting possible cytotoxic T lymphocyte epitopes based on HLA class I peptide-binding motifs. Hum. Immunol. 43, 13–18 10.1016/0198-8859(94)00153-H (doi:10.1016/0198-8859(94)00153-H) [DOI] [PubMed] [Google Scholar]
- 59.Meister GE, Roberts CG, Berzofsky JA, De Groot AS. 1995. Two novel T cell epitope prediction algorithms based on MHC-binding motifs: comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine 13, 581–591 10.1016/0264-410X(94)00014-E (doi:10.1016/0264-410X(94)00014-E) [DOI] [PubMed] [Google Scholar]
- 60.De Groot AS, Bosma A, Chinai N, Frost J, Jesdale BM, Gonzalez MA, Martin W, Saint-Aubin C. 2001. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine 19, 4385–4395 10.1016/S0264-410X(01)00145-1 (doi:10.1016/S0264-410X(01)00145-1) [DOI] [PubMed] [Google Scholar]
- 61.Dick TP, Stevanovic S, Keilholz W, Ruppert T, Koszinowski U, Schild H, Rammensee HG. 1998. The making of the dominant MHC class I ligand SYFPEITHI. Eur. J. Immunol. 28, 2478–2486 (doi:10.1002/(SICI)1521-4141(199808)28:08<2478::AID-IMMU2478>3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 62.Amicosante M, et al. 2002. Computer-based design of an HLA-haplotype and HIV-clade independent cytotoxic T-lymphocyte assay for monitoring HIV-specific immunity. Mol. Med. 8, 798–807 [PMC free article] [PubMed] [Google Scholar]
- 63.Dong HL, Sui YF, Ye J, Li ZS, Qu P, Zhang XM, Chen GS, Lu SY. 2003. Prediction synthesis and identification of HLA-A2-restricted cytotoxic T lymphocyte epitopes of the tumor antigen MAGE-n. Zhonghua Yi Xue Za Zhi 83, 1080–1083 [PubMed] [Google Scholar]
- 64.Hansson L, Rabbani H, Fagerberg J, Osterborg A, Mellstedt H. 2003. T-cell epitopes within the complementarity-determining and framework regions of the tumor-derived immunoglobulin heavy chain in multiple myeloma. Blood 101, 4930–4936 10.1182/blood-2002-04-1250 (doi:10.1182/blood-2002-04-1250) [DOI] [PubMed] [Google Scholar]
- 65.Wagner C, et al. 2003. Identification of an HLA-A*02 restricted immunogenic peptide derived from the cancer testis antigen HOM-MEL-40/SSX2. Cancer Immun. 3, 18. [PubMed] [Google Scholar]
- 66.Zehbe I, Mytilineos J, Wikstrom I, Henriksen R, Edler L, Tommasino M. 2003. Association between human papillomavirus 16 E6 variants and human leukocyte antigen class I polymorphism in cervical cancer of Swedish women. Hum. Immunol. 64, 538–542 10.1016/S0198-8859(03)00033-8 (doi:10.1016/S0198-8859(03)00033-8) [DOI] [PubMed] [Google Scholar]
- 67.Liu Z, Dominy BN, Shakhnovich EI. 2004. Structural mining: self-consistent design on flexible protein–peptide docking and transferable binding affinity potential. J. Am. Chem. Soc. 126, 8515–8528 10.1021/ja032018q (doi:10.1021/ja032018q) [DOI] [PubMed] [Google Scholar]
- 68.Neumann F, Wagner C, Kubuschok B, Stevanovic S, Rammensee HG, Pfreundschuh M. 2004. Identification of an antigenic peptide derived from the cancer-testis antigen NY-ESO-1 binding to a broad range of HLA-DR subtypes. Cancer Immunol. Immunother. 53, 589–599 10.1007/s00262-003-0492-6 (doi:10.1007/s00262-003-0492-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pelte C, Cherepnev G, Wang Y, Schoenemann C, Volk HD, Kern F. 2004. Random screening of proteins for HLA-A*0201-binding nine-amino acid peptides is not sufficient for identifying CD8 T cell epitopes recognized in the context of HLA-A*0201. J. Immunol. 172, 6783–6789 [DOI] [PubMed] [Google Scholar]
- 70.Ullenhag GJ, Fagerberg J, Strigard K, Frodin JE, Mellstedt H. 2004. Functional HLA-DR T cell epitopes of CEA identified in patients with colorectal carcinoma immunized with the recombinant protein CEA. Cancer Immunol. Immunother. 53, 331–337 10.1007/s00262-003-0441-4 (doi:10.1007/s00262-003-0441-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nussbaum AK, Kuttler C, Tenzer S, Schild H. 2003. Using the World Wide Web for predicting CTL epitopes. Curr. Opin. Immunol. 15, 69–74 10.1016/S0952791502000043 (doi:10.1016/S0952791502000043) [DOI] [PubMed] [Google Scholar]
- 72.Andersen MH, Tan L, Sondergaard I, Zeuthen J, Elliott T, Haurum JS. 2000. Poor correspondence between predicted and experimental binding of peptides to class I MHC molecules. Tissue Antigens 55, 519–531 10.1034/j.1399-0039.2000.550603.x (doi:10.1034/j.1399-0039.2000.550603.x) [DOI] [PubMed] [Google Scholar]
- 73.Beale R, Jackson T. 1990. Neural computing: an introduction. Bristol, UK: Adam Hilger [Google Scholar]
- 74.Tomita Y, Tomida S, Hasegawa Y, Suzuki Y, Shirakawa T, Kobayashi T, Honda H. 2004. Artificial neural network approach for selection of susceptible single nucleotide polymorphisms and construction of prediction model on childhood allergic asthma. BMC Bioinform. 5, 120. 10.1186/1471-2105-5-120 (doi:10.1186/1471-2105-5-120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stefaniak B, Cholewinski W, Tarkowska A. 2004. Prediction of left ventricular ejection fraction in patients with coronary artery disease based on an analysis of perfusion patterns at rest. Assessment by an artificial neural network. Nucl. Med. Rev. Cent. East. Eur. 7, 7–12 [PubMed] [Google Scholar]
- 76.Jouyban A, Majidi MR, Jalilzadeh H, Asadpour-Zeynali K. 2004. Modeling drug solubility in water-cosolvent mixtures using an artificial neural network. Farmaco 59, 505–512 10.1016/j.farmac.2004.02.005 (doi:10.1016/j.farmac.2004.02.005) [DOI] [PubMed] [Google Scholar]
- 77.Bellgard MI, Tay GK, Hiew HL, Witt CS, Ketheesan N, Christiansen FT, Dawkins RL. 1998. MHC haplotype analysis by artificial neural networks. Hum. Immunol. 59, 56–62 10.1016/S0198-8859(97)00231-0 (doi:10.1016/S0198-8859(97)00231-0) [DOI] [PubMed] [Google Scholar]
- 78.Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S, Brunak S, Lund O. 2004. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 20, 1388–1397 10.1093/bioinformatics/bth100 (doi:10.1093/bioinformatics/bth100) [DOI] [PubMed] [Google Scholar]
- 79.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O, Nielsen M. 2005. An integrative approach to CTL epitope prediction. A combined algorithm integrating MHC-I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 35, 2295–2303 10.1002/eji.200425811 (doi:10.1002/eji.200425811) [DOI] [PubMed] [Google Scholar]
- 80.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36(Suppl. 2), W509–W512 10.1093/nar/gkn202 (doi:10.1093/nar/gkn202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vapnik V. 1998. Statistical learning theory. New York, NY: Wiley-Interscience [Google Scholar]
- 82.Ding CH, Dubchak I. 2001. Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics 17, 349–358 10.1093/bioinformatics/17.4.349 (doi:10.1093/bioinformatics/17.4.349) [DOI] [PubMed] [Google Scholar]
- 83.Scholkopf S, Burges CJC, Smola AJ. 1999. Advances in kernel methods: support vector learning. Cambridge, MA: MIT Press [Google Scholar]
- 84.Perner P. 2011. Machine learning and data mining in pattern recognition. In Proc. 7th Int. Conf. MLDM 2011, New York, NY, 30 August–3 September Lecture Notes in Computer Science, vol. 6871. New York, NY: Springer [Google Scholar]
- 85.Nanni L. 2006. Machine learning algorithms for T-cell epitopes prediction. Neurocomputing 69, 866–868 10.1016/j.neucom.2005.08.005 (doi:10.1016/j.neucom.2005.08.005) [DOI] [Google Scholar]
- 86.Bhasin M, Raghava GPS. 2004. Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci. 13, 596–607 10.1110/ps.03373104 (doi:10.1110/ps.03373104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhasin M, Raghava GPS. 2005. Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res. 33(Suppl. 2), W202–W207 10.1093/nar/gki587 (doi:10.1093/nar/gki587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joachims T. 1999. Marking large-scale support vector machine learning practical, pp. 169–84 Cambridge, MA: MIT Press [Google Scholar]
- 89.Cost S, Salzberg S. 1993. A weighted nearest neighbor algorithm for learning with symbolic features. Mach. Learn. 10, 57–78 10.1007/BF00993481 (doi:10.1007/BF00993481) [DOI] [Google Scholar]
- 90.Witten IH, Frank E. 1999. Data mining: practical machine learning tools and techniques with Java implementations, 2nd edn San Francisco, CA: Morgan Kaufman [Google Scholar]
- 91.Sweredoski MJ, Baldi P. 2009. COBEpro: a novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 22, 113–20 10.1093/protein/gzn075 (doi:10.1093/protein/gzn075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baum L, Petrie T, Soules G, Weiss N. 1970. A maximization technique occuring in the statistical analysis of probablistic functions of markov chains. Ann. Math. Stat. 41, 164–171 10.1214/aoms/1177697196 (doi:10.1214/aoms/1177697196) [DOI] [Google Scholar]
- 93.Rabiner LR. 1989. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77, 257–286 10.1109/5.18626 (doi:10.1109/5.18626) [DOI] [Google Scholar]
- 94.Huang X, Jack M, Ariki Y. 1990. Hidden Markov models for speech recognition. Edinburgh, UK: Edinburgh University Press [Google Scholar]
- 95.Bishop M, Thompson E. 1986. Maximum likelihood alignment of DNA sequences. J. Mol. Biol. 190, 159–165 10.1016/0022-2836(86)90289-5 (doi:10.1016/0022-2836(86)90289-5) [DOI] [PubMed] [Google Scholar]
- 96.Durbin R, Eddy SR, Krogh A, Mitchison G. 1999. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge, UK: Cambridge University Press [Google Scholar]
- 97.Delorenzi M, Speed T. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 10.1093/bioinformatics/18.4.617 (doi:10.1093/bioinformatics/18.4.617) [DOI] [PubMed] [Google Scholar]
- 98.Martelli PL, Fariselli P, Krogh A, Casadio R. 2002. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics 18(Suppl. 1), S46–S53 10.1093/bioinformatics/18.suppl_1.S46 (doi:10.1093/bioinformatics/18.suppl_1.S46) [DOI] [PubMed] [Google Scholar]
- 99.Liu Q, Zhu YS, Wang BH, Li YX. 2003. A HMM-based method to predict the transmembrane regions of beta-barrel membrane proteins. Comput. Biol. Chem. 27, 69–76 10.1016/S0097-8485(02)00051-7 (doi:10.1016/S0097-8485(02)00051-7) [DOI] [PubMed] [Google Scholar]
- 100.Qian B, Goldstein RA. 2004. Performance of an iterated T-HMM for homology detection. Bioinformatics 20, 2175–2180 10.1093/bioinformatics/bth181 (doi:10.1093/bioinformatics/bth181) [DOI] [PubMed] [Google Scholar]
- 101.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235, 1501–1531 10.1006/jmbi.1994.1104 (doi:10.1006/jmbi.1994.1104) [DOI] [PubMed] [Google Scholar]
- 102.Bateman A, Haft DH. 2002. HMM-based databases in InterPro. Brief Bioinform. 3, 236–245 10.1093/bib/3.3.236 (doi:10.1093/bib/3.3.236) [DOI] [PubMed] [Google Scholar]
- 103.Cawley SL, Pachter L. 2003. HMM sampling and applications to gene finding and alternative splicing. Bioinformatics 19(Suppl. 2), ii36–ii41 10.1093/bioinformatics/btg1057 (doi:10.1093/bioinformatics/btg1057) [DOI] [PubMed] [Google Scholar]
- 104.Jojic V, Jojic N, Meek C, Geiger D, Siepel A, Haussler D, Heckerman D. 2004. Efficient approximations for learning phylogenetic HMM models from data. Bioinformatics 20(Suppl. 1), i161–i168 10.1093/bioinformatics/bth917 (doi:10.1093/bioinformatics/bth917) [DOI] [PubMed] [Google Scholar]
- 105.Azad RK, Borodovsky M. 2004. Probabilistic methods of identifying genes in prokaryotic genomes: connections to the HMM theory. Brief Bioinform. 5, 118–130 10.1093/bib/5.2.118 (doi:10.1093/bib/5.2.118) [DOI] [PubMed] [Google Scholar]
- 106.Zhang GL, Petrovsky N, Kwoh CK, August JT, Brusic V. 2006. PredTAP: a system for prediction of peptide binding to the human transporter associated with antigen processing. Immunome Res. 2, 3. 10.1186/1745-7580-2-3 (doi:10.1186/1745-7580-2-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Udaka K, Mamitsuka H, Nakaseko Y, Abe N. 2002. Prediction of MHC class I binding peptides by a query learning algorithm based on hidden Markov models. J. Biol. Phys. 28, 183–194 10.1023/A:1019931731519 (doi:10.1023/A:1019931731519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brusic V, Petrovsky N, Zhang G, Bajic VB. 2002. Prediction of promiscuous peptides that bind HLA class I molecules. Immunol. Cell Biol. 80, 280–285 10.1046/j.1440-1711.2002.01088.x (doi:10.1046/j.1440-1711.2002.01088.x) [DOI] [PubMed] [Google Scholar]
- 109.Schonbach C, Koh JL, Sheng X, Wong L, Brusic V. 2000. FIMM, a database of functional molecular immunology. Nucleic Acids Res. 28, 222–224 10.1093/nar/28.1.222 (doi:10.1093/nar/28.1.222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gulukota K, Sidney J, Sette A, DeLisi C. 1997. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 267, 1258–1267 10.1006/jmbi.1997.0937 (doi:10.1006/jmbi.1997.0937) [DOI] [PubMed] [Google Scholar]
- 111.Hammer J, Bono E, Gallazzi F, Belunis C, Nagy Z, Sinigaglia F. 1994. Precise prediction of major histocompatibility complex class II–peptide interaction based on peptide side chain scanning. J. Exp. Med. 180, 2353–2358 10.1084/jem.180.6.2353 (doi:10.1084/jem.180.6.2353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marshall KW, Liu AF, Canales J, Perahia B, Jorgensen B, Gantzos RD, Aguilar B, Devaux B, Rothbard JB. 1994. Role of the polymorphic residues in HLA-DR molecules in allele-specific binding of peptide ligands. J. Immunol. 152, 4946–4957 [PubMed] [Google Scholar]
- 113.Dixon FJ. 1995. Advances in immunology, vol. 66 New York, NY: Academic press [Google Scholar]
- 114.Hakenberg J, Nussbaum AK, Schild H, Rammensee HG, Kuttler C, Holzhutter HG, Kloetzel PM, Kaufmann SH, Mollenkopf HJ. 2003. MAPPP: MHC class I antigenic peptide processing prediction. Appl. Bioinform. 2, 155–158 [PubMed] [Google Scholar]
- 115.Alix AJ. 1999. Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine 18, 311–314 10.1016/S0264-410X(99)00329-1 (doi:10.1016/S0264-410X(99)00329-1) [DOI] [PubMed] [Google Scholar]
- 116.Parker KC, Bednarek MA, Coligan JE. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152, 163–175 [PubMed] [Google Scholar]
- 117.Vonderheide RH, Hahn WC, Schultze JL, Nadler LM. 1999. The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity 10, 673–679 10.1016/S1074-7613(00)80066-7 (doi:10.1016/S1074-7613(00)80066-7) [DOI] [PubMed] [Google Scholar]
- 118.Lu J, Celis E. 2000. Use of two predictive algorithms of the world wide web for the identification of tumor-reactive T-cell epitopes. Cancer Res. 60, 5223–5227 [PubMed] [Google Scholar]
- 119.Schafer JR, Jesdale BM, George JA, Kouttab NM, De Groot AS. 1998. Prediction of well-conserved HIV-1 ligands using a matrix-based algorithm, EpiMatrix. Vaccine 16, 1880–1884 10.1016/S0264-410X(98)00173-X (doi:10.1016/S0264-410X(98)00173-X) [DOI] [PubMed] [Google Scholar]
- 120.Sbai H, Mehta A, DeGroot AS. 2001. Use of T cell epitopes for vaccine development. Curr. Drug Targets Infect. Disord. 1, 303–313 10.2174/1568005014605955 (doi:10.2174/1568005014605955) [DOI] [PubMed] [Google Scholar]
- 121.Reche PA, Glutting JP, Reinherz EL. 2002. Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol. 63, 701–709 10.1016/S0198-8859(02)00432-9 (doi:10.1016/S0198-8859(02)00432-9) [DOI] [PubMed] [Google Scholar]
- 122.Sturniolo T, et al. 1999. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 17, 555–561 10.1038/9858 (doi:10.1038/9858) [DOI] [PubMed] [Google Scholar]
- 123.Manici S, et al. 1999. Melanoma cells present a MAGE-3 epitope to CD4+ cytotoxic T cells in association with histocompatibility leukocyte antigen DR11. J. Exp. Med. 189, 871–876 10.1084/jem.189.5.871 (doi:10.1084/jem.189.5.871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cochlovius B, Stassar M, Christ O, Raddrizzani L, Hammer J, Mytilineos I, Zoller M. 2000. In vitro and in vivo induction of a Th cell response toward peptides of the melanoma-associated glycoprotein 100 protein selected by the TEPITOPE program. J. Immunol. 165, 4731–4741 [DOI] [PubMed] [Google Scholar]
- 125.Singh H, Raghava GP. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics 17, 1236–1237 10.1093/bioinformatics/17.12.1236 (doi:10.1093/bioinformatics/17.12.1236) [DOI] [PubMed] [Google Scholar]
- 126.Guan P, Doytchinova IA, Zygouri C, Flower DR. 2003. MHCPred: a server for quantitative prediction of peptide–MHC binding. Nucleic Acids Res. 31, 3621–3624 10.1093/nar/gkg510 (doi:10.1093/nar/gkg510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Doytchinova IA, Walshe VA, Jones NA, Gloster SE, Borrow P, Flower DR. 2004. Coupling in silico and in vitro analysis of peptide–MHC binding: a bioinformatic approach enabling prediction of superbinding peptides and anchorless epitopes. J. Immunol. 172, 7495–7502 [DOI] [PubMed] [Google Scholar]
- 128.Walshe VA, et al. 2009. Integrating in silico and in vitro analysis of peptide binding affinity to HLA-Cw*0102: a bioinformatics approach to the prediction of new epitopes. PLoS ONE 4, e8095. 10.1371/journal.pone.0008095 (doi:10.1371/journal.pone.0008095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doytchinova IA, Guan P, Flower DR. 2006. EpiJen: a server for multi-step T cell epitope prediction. BMC Bioinform. 7, 131–142 10.1186/1471-2105-7-131 (doi:10.1186/1471-2105-7-131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doytchinova IA, Flower DR. 2007. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 8, 4. 10.1186/1471-2105-8-4 (doi:10.1186/1471-2105-8-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dimitrov I, Garnev P, Flower DR, Doytchinova I. 2010. EpiTOP: a proteochemometric tool for MHC class II binding prediction. Bioinformatics 26, 2066–2068 10.1093/bioinformatics/btq324 (doi:10.1093/bioinformatics/btq324) [DOI] [PubMed] [Google Scholar]
- 132.Lapinsh M, Prusis P, Gutcaits A, Lundstedt T, Wikberg JES. 2001. Development of proteochemometrics: a novel technology for the analysis of drug–receptor interactions. Biochim. Biophys. Acta 1525, 180–190 10.1016/S0304-4165(00)00187-2 (doi:10.1016/S0304-4165(00)00187-2) [DOI] [PubMed] [Google Scholar]
- 133.Brusic V, Schonbach C, Takiguchi M, Ciesielski V, Harrison LC. 1997. Application of genetic search in derivation of matrix models of peptide binding to MHC molecules. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 75–83 [PubMed] [Google Scholar]
- 134.Kellam P, Holzerlandt R, Gramoustianou E, Jenner R, Kwan A. 2008. Immunoinformatics viral bioinformatics: computational views of host and pathogen. In Immunoinformatics: bioinformatic strategies for better understanding of immune function: Novartis Foundation Symp., 254 (eds Bock G, Goode J.), pp. 234–249 Chichester, UK: John Wiley & Sons, Ltd; [PubMed] [Google Scholar]
- 135.Schönbach C, Ranganathan S, Brusic V. 2008. Immunoinformatics. New York, NY: Springer [Google Scholar]
- 136.Davenport MP, Ho Shon IA, Hill AV. 1995. An empirical method for the prediction of T-cell epitopes. Immunogenetics 42, 392–397 10.1007/BF00179401 (doi:10.1007/BF00179401) [DOI] [PubMed] [Google Scholar]
- 137.Fleckenstein B, Kalbacher H, Muller CP, Stoll D, Halder T, Jung G, Wiesmuller KH. 1996. New ligands binding to the human leukocyte antigen class II molecule DRB1*0101 based on the activity pattern of an undecapeptide library. Eur. J. Biochem. 240, 71–77 10.1111/j.1432-1033.1996.0071h.x (doi:10.1111/j.1432-1033.1996.0071h.x) [DOI] [PubMed] [Google Scholar]
- 138.Stryhn A, Pederden LO, Romme T, Holm CB, Holm A, Buus S. 1996. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur. J. Immunol. 26, 1811–1818 10.1002/eji.1830260836 (doi:10.1002/eji.1830260836) [DOI] [PubMed] [Google Scholar]
- 139.Stevens J, Wiesmulles KH, Barker PJ, Walden P, Butcher GW, Joly E. 1998. Efficient generation of major histocompatibility complex class I–peptide complexes using synthetic peptide libraries. J. Biol. Chem. 273, 2874–2884 10.1074/jbc.273.5.2874 (doi:10.1074/jbc.273.5.2874) [DOI] [PubMed] [Google Scholar]
- 140.Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. 1995. Tolerance to amino acid variations in peptides binding to the major histocompatibility complex class I protein H-2Kb. J. Biol. Chem. 270, 24 130–24 134 10.1074/jbc.270.41.24130 (doi:10.1074/jbc.270.41.24130) [DOI] [PubMed] [Google Scholar]
- 141.Udaka K, et al. 2000. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics 51, 816–828 10.1007/s002510000217 (doi:10.1007/s002510000217) [DOI] [PubMed] [Google Scholar]
- 142.Sung MH, Zhao Y, Martin R, Simon R. 2002. T-cell epitope prediction with combinatorial peptide libraries. J. Comput. Biol. 9 527–539 10.1089/106652702760138619 (doi:10.1089/106652702760138619) [DOI] [PubMed] [Google Scholar]
- 143.Nino-Vasquez JJ, Allicotti G, Borras E, Wilson DB, Valmori D, Simon R, Martin R, Pinilla C. 2004. A powerful combination: the use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol. Immunol. 40, 1063–1074 10.1016/j.molimm.2003.11.005 (doi:10.1016/j.molimm.2003.11.005) [DOI] [PubMed] [Google Scholar]
- 144.Vajda S, Camacho CJ. 2004. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 22, 110–116 10.1016/j.tibtech.2004.01.006 (doi:10.1016/j.tibtech.2004.01.006) [DOI] [PubMed] [Google Scholar]
- 145.Sezerman U, Vajda S, Cornette J, DeLisi C. 1993. Toward computational determination of peptide–receptor structure. Protein Sci. 2, 1827–1843 10.1002/pro.5560021105 (doi:10.1002/pro.5560021105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rosenfeld R, Zheng Q, Vajda S, DeLisi C. 1995. Flexible docking of peptides to class I major-histocompatibility-complex receptors. Genet. Anal. 12, 1–21 10.1016/1050-3862(95)00107-7 (doi:10.1016/1050-3862(95)00107-7) [DOI] [PubMed] [Google Scholar]
- 147.Zeng J, Treutlein HR, Rudy GB. 2001. Predicting sequences and structures of MHC-binding peptides: a computational combinatorial approach. J. Comput. Aid. Mol. Des. 15, 573–586 10.1023/A:1011145123635 (doi:10.1023/A:1011145123635) [DOI] [PubMed] [Google Scholar]
- 148.Del Carpio CA, Hennig T, Fickel S, Yoshimori A. 2002. A combined bioinformatic approach oriented to the analysis and design of peptides with high affinity to MHC class I molecules. Immunol. Cell Biol. 80, 286–299 10.1046/j.1440-1711.2002.01090.x (doi:10.1046/j.1440-1711.2002.01090.x) [DOI] [PubMed] [Google Scholar]
- 149.Tzakos AG, Fuchs P, van Nuland NA, Troganis A, Tselios T, Deraos S, Matsoukas J, Gerothanassis IP, Bonvin AM. 2004. NMR and molecular dynamics studies of an autoimmune myelin basic protein peptide and its antagonist: structural implications for the MHC II (I-Au)–peptide complex from docking calculations. Eur. J. Biochem. 271, 3399–3413 10.1111/j.1432-1033.2004.04274.x (doi:10.1111/j.1432-1033.2004.04274.x) [DOI] [PubMed] [Google Scholar]
- 150.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. 2002. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature 418, 552–556 10.1038/nature00920 (doi:10.1038/nature00920) [DOI] [PubMed] [Google Scholar]
- 151.Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. 2003. A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection. Immunity 19, 595–606 10.1016/S1074-7613(03)00269-3 (doi:10.1016/S1074-7613(03)00269-3) [DOI] [PubMed] [Google Scholar]
- 152.Tong JC, Tan TW, Ranganathan S. 2004. Modeling the structure of bound peptide ligands to major histocompatibility complex. Protein Sci. 13, 2523–2532 10.1110/ps.04631204 (doi:10.1110/ps.04631204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Atanasova M, Dimitrov I, Flower DR, Doytchinova I. 2011. MHC class II binding prediction by molecular docking. Mol. Inform. 30, 368–375 10.1002/minf.201000132 (doi:10.1002/minf.201000132) [DOI] [PubMed] [Google Scholar]
- 154.Margalit H, Altuvia Y. 2003. Insights from MHC-bound peptides. Novartis Found. Symp. 254, 77–90 10.1002/0470090766.ch6 (doi:10.1002/0470090766.ch6) [DOI] [PubMed] [Google Scholar]
- 155.Adrian PE, Rajaseger G, Mathura VS, Sakharkar MK, Kangueane P. 2002. Types of inter-atomic interactions at the MHC–peptide interface: identifying commonality from accumulated data. BMC Struct. Biol. 2, 2. 10.1186/1472-6807-2-2 (doi:10.1186/1472-6807-2-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schueler-Furman O, Elber R, Margalit H. 1998. Knowledge-based structure prediction of MHC class I bound peptides: a study of 23 complexes. Folding Des. 3, 549–564 10.1016/S1359-0278(98)00070-4 (doi:10.1016/S1359-0278(98)00070-4) [DOI] [PubMed] [Google Scholar]
- 157.Schueler-Furman O, Altuvia Y, Sette A, Margalit H. 2000. Structure-based prediction of binding peptides to MHC Class I molecules: application to a broad range of MHC alleles. Protein Sci. 9, 1838–1846 10.1110/ps.9.9.1838 (doi:10.1110/ps.9.9.1838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Altuvia Y, Sette A, Sidney J, Southwood S, Margalit H. 1997. A structure-based algorithm to predict potential binding peptides to MHC molecules with hydrophobic binding pockets. Hum. Immunol. 58, 1–11 10.1016/S0198-8859(97)00210-3 (doi:10.1016/S0198-8859(97)00210-3) [DOI] [PubMed] [Google Scholar]
- 159.Sezerman U, Vajda S, DeLisi C. 1996. Free energy mapping of class I MHC molecules and structural determination of bound peptides. Protein Sci. 5, 1272–1281 10.1002/pro.5560050706 (doi:10.1002/pro.5560050706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang C, Cornette JL, Delisi C. 1997. Consistency in structural energetics of protein folding and peptide recognition. Protein Sci. 6, 1057–1064 10.1002/pro.5560060512 (doi:10.1002/pro.5560060512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Meng WS, von Grafenstein H, Haworth IS. 2000. Water dynamics at the binding interface of four different HLA-A2–peptide complexes. Int. Immunol. 12, 949–957 10.1093/intimm/12.7.949 (doi:10.1093/intimm/12.7.949) [DOI] [PubMed] [Google Scholar]
- 162.Scapozza L. 1995. Molecular dynamics and structure-based drug design for predicting non-natural nonapeptide binding to a class I MHC protein. Acta Crystallogr. D Biol. Crystallogr. 51, 541–549 10.1107/S0907444995002678 (doi:10.1107/S0907444995002678) [DOI] [PubMed] [Google Scholar]
- 163.Rognan D, Scapozza L, Folkers G, Daser A. 1994. Molecular dynamics simulation of MHC–peptide complexes as a tool for predicting potential T cell epitopes. Biochemistry 33, 11 476–11 485 10.1021/bi00204a009 (doi:10.1021/bi00204a009) [DOI] [PubMed] [Google Scholar]
- 164.Mata M, Travers PJ, Liu Q, Frankel FR, Paterson Y. 1998. The MHC class I-restricted immune response to HIV-gag in BALB/c mice selects a single epitope that does not have a predictable MHC-binding motif and binds to Kd through interactions between a glutamine at P3 and pocket. D. J. Immunol. 161, 2985–2993 [PubMed] [Google Scholar]
- 165.Petrone PM, Garcia AE. 2004. MHC–peptide binding is assisted by bound water molecules. J. Mol. Biol. 338, 419–435 10.1016/j.jmb.2004.02.039 (doi:10.1016/j.jmb.2004.02.039) [DOI] [PubMed] [Google Scholar]
- 166.Pohlmann T, Bockmann RA, Grubmuller H, Uchanska-Ziegler B, Ziegler A, Alexiev U. 2004. Differential peptide dynamics is linked to major histocompatibility complex polymorphism. J. Biol. Chem. 279, 28 197–28 201 10.1074/jbc.C400128200 (doi:10.1074/jbc.C400128200) [DOI] [PubMed] [Google Scholar]
- 167.Binz AK, Rodriguez RC, Biddison WE, Baker BM. 2003. Thermodynamic and kinetic analysis of a peptide–class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry 42, 4954–4961 10.1021/bi034077m (doi:10.1021/bi034077m) [DOI] [PubMed] [Google Scholar]
- 168.Michielin O, Karplus M. 2002. Binding free energy differences in a TCR–peptide–MHC complex induced by a peptide mutation: a simulation analysis. J. Mol. Biol. 324, 547–569 10.1016/S0022-2836(02)00880-X (doi:10.1016/S0022-2836(02)00880-X) [DOI] [PubMed] [Google Scholar]
- 169.Lim JS, Kim S, Lee HG, Lee KY, Kwon TJ, Kim K. 1996. Selection of peptides that bind to the HLA-A2.1 molecule by molecular modelling. Mol. Immunol. 33, 221–230 10.1016/0161-5890(95)00065-8 (doi:10.1016/0161-5890(95)00065-8) [DOI] [PubMed] [Google Scholar]
- 170.Toh H, Savoie CJ, Kamikawaji N, Muta S, Sasazuki T, Kuhara S. 2000. Changes at the floor of the peptide-binding groove induce a strong preference for proline at position 3 of the bound peptide: molecular dynamics simulations of HLA-A*0217. Biopolymers 54, 318–327 (doi:10.1002/1097-0282(20001015)54:5<318::AID-BIP30>3.0.CO;2-T) [DOI] [PubMed] [Google Scholar]
- 171.Androulakis IP, Nayak NN, Ierapetritou MG, Monos DS, Floudas CA. 1997. A predictive method for the evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins 29, 87–102 (doi:10.1002/(SICI)1097-0134(199709)29:1<87::AID-PROT7>3.0.CO;2-C) [DOI] [PubMed] [Google Scholar]
- 172.Davies MN, Sansom CE, Beazley C, Moss DS. 2003. A novel predictive technique for the MHC class II peptide-binding interaction. Mol. Med. 9, 220–225 10.2119/2003-00032.Sansom (doi:10.2119/2003-00032.Sansom) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schapira M, Totrov M, Abagyan R. 1999. Prediction of the binding energy for small molecules, peptides and proteins. J. Mol. Recognit. 12, 177–190 (doi:10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z) [DOI] [PubMed] [Google Scholar]
- 174.Logean A, Sette A, Rognan D. 2001. Customized versus universal scoring functions: application to class I MHC–peptide binding free energy predictions. Bioorg. Med. Chem. Lett. 11, 675–679 10.1016/S0960-894X(01)00021-X (doi:10.1016/S0960-894X(01)00021-X) [DOI] [PubMed] [Google Scholar]
- 175.Logean A, Rognan D. 2002. Recovery of known T-cell epitopes by computational scanning of a viral genome. J. Comput. Aid. Mol. Des. 16, 229–243 10.1023/A:1020244329512 (doi:10.1023/A:1020244329512) [DOI] [PubMed] [Google Scholar]
- 176.Froloff N, Windemuth A, Honig B. 1997. On the calculation of binding free energies using continuum methods: application to MHC class I protein–peptide interactions. Protein Sci. 6, 1293–1301 10.1002/pro.5560060617 (doi:10.1002/pro.5560060617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rognan D, Lauemoller SL, Holm A, Buus S, Tschinke V. 1999. Predicting binding affinities of protein ligands from three-dimensional models: application to peptide binding to class I major histocompatibility proteins. J. Med. Chem. 42, 4650–4658 10.1021/jm9910775 (doi:10.1021/jm9910775) [DOI] [PubMed] [Google Scholar]