Abstract
Copper has an important role in the life cycle of many streptomycetes, stimulating the developmental switch between vegetative mycelium and aerial hyphae concomitant with the production of antibiotics. In streptomycetes, a gene encoding for a putative Sco-like protein has been identified and is part of an operon that contains two other genes predicted to handle cellular copper. We report on the Sco-like protein from Streptomyces lividans (ScoSl) and present a series of experiments that firmly establish a role for ScoSl as a copper metallochaperone as opposed to a role as a thiol-disulphide reductase that has been assigned to other bacterial Sco proteins. Under low copper concentrations, a Δsco mutant in S. lividans displays two phenotypes; the development switch between vegetative mycelium and aerial hyphae stalls and cytochrome c oxidase (CcO) activity is significantly decreased. At elevated copper levels, the development and CcO activity in the Δsco mutant are restored to wild-type levels and are thus independent of ScoSl. A CcO knockout reveals that morphological development is independent of CcO activity leading us to suggest that ScoSl has at least two targets in S. lividans. We establish that one ScoSl target is the dinuclear CuA domain of CcO and it is the cupric form of ScoSl that is functionally active. The mechanism of cupric ion capture by ScoSl has been investigated, and an important role for a conserved His residue is identified.
Keywords: Streptomyces, copper-metallochaperone, cytochrome c oxidase, Sco protein, morphological development, thiol-disulphide reductase activity
2. Introduction
Streptomycetes are soil-dwelling Gram-positive bacteria best known for their ability to produce a bewildering array of secondary metabolites that have antibiotic, antimicrobial and anti-cancer properties [1]. The onset of chemical differentiation (antibiotic production) coincides with the onset of morphological development from a vegetative growth to a reproductive growth phase beginning with aerial mycelium followed by cell division and sporulation. The bioavailability of copper (Cu) has been shown in certain Streptomyces strains to stimulate morphological development and antibiotic production [2–5]. This dependence is restricted to the reproductive growth phase (aerial mycelium and spores), whereas vegetative growth proceeds under strongly Cu-limiting conditions. The Cu proteome of Streptomyces coelicolor has been reported based on in silico methods and contains a large number of cuproproteins and cupro enzymes [6,7]. The stimulatory effect of Cu in development therefore suggests that some Cu-dependent biochemical functions correlate with the complex life cycle of streptomycetes and that cuproproteins and cuproenzymes must play a part. However, their identification and role in development have yet to be fully elucidated [7].
Streptomyces lividans has an even more pronounced dependence on Cu for development than S. coelicolor. The former shares a high level of sequence identity and genome organization with S. coelicolor but the annotated genome has not yet been reported. Therefore, the gene numbering of the annotated S. coelicolor genome database is used in this study in keeping with previous studies of cuproproteins in S. lividans [8,9]. A recent study has highlighted that in the five sequenced and annotated genomes of streptomycetes a conserved unidirectional gene cluster is present that includes three genes (3966, 3965, 3964) predicted to encode cuproproteins (figure 1) [4]. In S. coelicolor, experimental evidence suggests that this cluster has a central role in the utilization of Cu and may therefore have a complex role in the Cu-dependent stimulation of development [4]. The operon has been named the sco operon owing to the presence of a gene encoding for a protein with a CXXXC motif and sequence homology with the Sco/SenC/PrrC protein family. Synthesis of cytochrome c oxidase (Sco) proteins is present in both eukaryotes and prokaryotes, and was first identified, along with other cytochrome c oxidase (CcO) assembly factors, in the mitochondria of yeast and then subsequently in humans where a role as a Cu(I)-metallochaperone in co-factoring the mixed valance di-nuclear CuA site of the aa3-type CcO of the mitochondrial electron transport chain was first proposed [12–14].
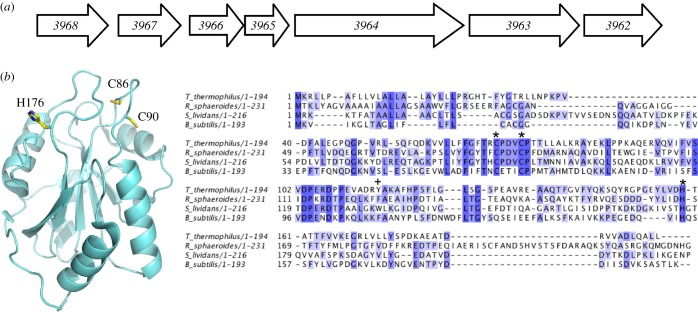
Figure 1.
The sco operon, X-ray crystal structure of apo-ScoBs and multiple sequence alignment of bacterial Sco proteins for which biochemical or structural data have been reported. (a) Schematic of the unidirectional sco operon (3966, 3965 and 3964), including two upstream and two downstream genes found in Streptomyces coelicolor. The coding sequence numbers assigned in the S. coelicolor genome are indicated with the respective annotations obtained from the StrepDB; 3968 (integral membrane protein), 3967 (hypothetical membrane protein), 3966 (Sco/SenC/PrrC-like protein), 3965 (conserved hypothetical protein), 3964 (integral membrane putative Cu transport protein with Cu binding CopC-like and CopD-like domains), 3963 (hypothetical protein) 3962 (prephenate dehydratase involved in Phe biosynthesis). (b) Cartoon representation of the X-ray crystal structure of apo-ScoBs (pdb 1XZO) [10]. The location of the His and Cys residues considered to be involved in Cu(II) coordination are labelled with S. lividans numbering. ClustalW2 sequence alignments [11] of bacterial Sco proteins with completely and partially conserved residues boxed from dark blue to light blue, respectively. The residues involved in Cu(II) coordination are indicated (asterisk) and a plus symbol indicates the position of Trp-132 for S. lividans. The UniprotKB NCBI accession numbers are gi |257157333| (Bacillus subtilis), gi |77386477| (Rhodobacter sphaeroides), gi |198443051| (Thermus thermophilus).
The most extensively studied bacterial Sco protein is that from Bacillus subtilis (ScoBs). This protein has been reported to bind both Cu(I) and Cu(II) in vitro, and a genetic knockout of ScoBs leads to a marked phenotype on the activity of the aa3-type CcO and organism viability [15] suggesting a role in CuA site maturation in this Gram-positive bacteria. An alternative role as a thiol-disulphide reductase has been proposed for bacterial Sco proteins [10,16,17]. Evidence for this comes from an in vitro NMR study using the periplasmic Sco protein form Thermus thermophilus (ScoTt), which was found to be incapable of delivering Cu to the CuA site of CcO [18]. Instead, a role in keeping the Cys residues of the CuA site in the reduced form to facilitate metallation of the CuA site by a newly identified periplasmic Cu(I) chaperone, PCuAC (periplasmic CuA chaperone) [18], was proposed. PCuAC is not present in all bacteria and is absent in B. subtilis, but in S. lividans the 3965 gene, part of the sco operon (figure 1a), is predicted to encode a PCuAC-like protein [7]. The structures of Sco proteins from yeast [19], human [20,21], B. subtilis [10,22] and T. thermophilus [18] all have a fold very similar to the thioredoxin (Trx) superfamily (figure 1b), and, importantly, the CXXXC motif implicated in Cu binding (figure 1b) superimposes on the CXXC redox motif of Trx making a redox role for Sco proteins an attractive proposition. Subsequent studies have reported thiol-disulphide reductase activity of ScoBs and PrrC (a Sco-like protein found in Rhodobacter sphaeroides) [23,24].
A consensus functional role of bacterial Sco proteins has not therefore been reached and it may be that variation exists across species [17]. In this study, we have focused our attention on the Sco-like protein identified in S. lividans (ScoSl), in which the gene (3966) is located in the so-called sco operon (figure 1a) [4]. We report herein on the properties of an N-terminal-truncated form of the 3966 gene product upon addition of cupric ions and assess the redox and thiol-disulphide reductase activity of the CXXXC motif in the absence of Cu. Parallel in vivo studies have been performed and imply a critical role for ScoSl both in the development switch from vegetative mycelium to aerial hyphae and in the activity of the aa3-type CcO at low [Cu]. The development switch is found to be independent of CcO activity, and we present evidence to suggest that a second ScoSl target, requiring Cu for function, is essential for morphological development in S. lividans under low Cu levels. Mechanistic features of Cu(II) loading to ScoSl and the H176A mutation have been studied, which together with the in vivo data leads to new insights into the functional role of ScoSl in S. lividans.
3. Material and methods
3.1. Generation of the Sco and Cox mutants of Streptomyces lividans 1326
The agar media soya flower mannitol (SFM), R5 (complex medium) and Difco nutrient agar (DNA) were prepared according to Kieser et al. [25]. Antibiotics were used in the following final concentrations: apramycin (50 μg μl−1) and thiostrepton (5–20 μg μl−1). Agar plates were incubated at 30°C, and spore stocks were obtained from cultures grown on SFM plates and stored in 20 per cent glycerol at −20°C. The 3966 gene encoding for ScoSl was deleted in S. lividans 1326 (S. lividans 66, stock number 1326 from the John Innes Centre) in a two-step process using the CRE-lox system [26]. First, the gene (nt + 33 to 490) was replaced by homologous recombination with an apramycin-resistance cassette flanked by loxP sites. For this purpose, the upstream flanking region of 3966 (from −2160 to +32) and the downstream flanking region (from +491 to 2471) were amplified from genomic DNA by PCR introducing EcoRI, XbaI and XbaI, HindIII sites, respectively, for cloning purposes. These two fragments and the apramycin-resistance cassette flanked by loxP sites were cloned in the delivery vector pWHM3 that is unstable in Streptomyces [27]. Following protoplast transformation, recombinants that were apramycin-resistant but had lost the vector (thiostrepton-resistance) were isolated. Second, an unstable, plasmid-encoding Cre recombinase was introduced [26]. The Cre recombinase allowed for the excision of the apramycin-resistance cassette on the lox sites. The resulting strain, Δsco, has lost most of the coding sequence of ScoSl and has only a 61 nt scar, including two XbaI sites left in the genome. The Δsco strain was analysed by PCR to confirm the loss of 3966 and vector sequences. The cox::Apra mutant was isolated with a similar protocol replacing the open reading frames of 2155 and 2156 with the apramycin-resistance cassette. The replacement was confirmed by PCR.
3.2. Plasmid construction for complementation
For complementation/over-expression of ScoSl in wild-type (wt) 1326 and Δsco strains of S. lividans, the 3966 gene including 747 nt upstream was cloned in pHJL401 as an EcoRI–BamHI fragment generated by PCR with genomic DNA as template. The 747 nt upstream contains the entire promoter region of 3966 as determined by promoter probing with the redD gene as reporter [28]. The ScoSl H176A mutation was introduced in this construct by fusion PCR of two fragments with a 24 nt overlap. The inserts of all plasmids used in this study were sequenced on both strands. Introduction of the plasmids in S. lividans was carried out by protoplast transformation according to standard protocols.
3.3. In vivo cytochrome c oxidase assay
The in vivo CcO activity was visualized with N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) as substrate essentially according to references [29,30]. The wt and mutant strains were spotted on DNA plates (10 μl containing 1000 spores) and incubated for 24–30 h at 30°C. Spots were overlaid with 10 ml of a 25 mM phosphate buffer pH 7.4 solution containing 20 per cent ethanol, 0.6 per cent agarose, 1 per cent sodium deoxycholate and 10 mg TMPD. CcO activity is detected by the appearance of the blue colour of the oxidized product (indophenol blue) of the TMPD substrate and was recorded by taking digital images every 30 s for 5–10 min.
3.4. Cloning and site-directed mutagenesis of ScoSl for over-expression in Escherichia coli
An N-terminal-truncated version of ScoSl (3966), with nucleotides encoding amino acid residues 1–23 deleted, was amplified from S. lividans DNA and ligated into the NdeI and BamH restriction sites of a pET28a vector (Novagen) to create an N-terminal His6-tagged construct for over-expression in Escherichia coli. The Quikchange (Stratagene) site-directed mutagenesis method was used to create the H176A mutation in the N-terminal-truncated ScoSl. The following mutagenic primers were used, with the nucleotides changed to create the mutation in lower case: 5′-GATCGTCTCGACCgcCGGCACCCAGGTCGTC-3′ and 5′-CGACCTGGGTGCCGgcGGTCGAGACGATC-3′.
3.5. Over-expression and purification of ScoSl and the H176A mutation
ScoSl and H176A were over-expressed in E. coli strain BL21 (DE3) starting from overnight pre-cultures (2 ml 2xYT, 2 μl Kan (50 mg ml−1), 37°C) that were subsequently used to inoculate 750 ml of medium in 2 l flasks. At an OD600 of 0.6, 1 M isopropyl β-d-1-thiogalactopyranoside solution (Melford) was added to give a final concentration of 1 mM and the temperature decreased to 25°C. Cells were harvested after 16 h at 4000 r.p.m. and lysed using an EmulsiFlex-C5 cell disrupter (Avestin) followed by centrifugation at 18 000 r.p.m. for 20 min at 4°C. The clarified supernatant was loaded to a 5 ml Ni2+–NTA Sepharose column (GE Healthcare) equilibrated with buffer A (50 mM Tris/HCl, 500 mM NaCl, 20 mM imidazole, pH 8.0) and eluted with a linear imidazole gradient using buffer B (buffer A with 500 mM imidazole). A single peak at approximately 25 per cent buffer B was eluted from the column, and fractions were pooled and dialysed overnight at 4°C against buffer C (50 mM Tris/HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0). Following dialysis, the N-terminal His6-tag was removed by incubating the protein at room temperature overnight in the presence of 125 U of thrombin (Sigma). The protein/thrombin mixture was reapplied to the Ni2+–NTA Sepharose column (GE Healthcare) and the flow-through collected and concentrated at 4°C using a centrifugal concentrator (Vivaspin) with a 5 kDa cut-off. Concentrated protein was loaded to a G75 Sephadex column (GE Healthcare) equilibrated with buffer D (buffer C with 2 mM dithiothreitol (DTT)). Fractions eluting in the major peak of the G75 column were analysed by 15 per cent SDS–PAGE and those deemed of good purity were concentrated and stored at −20°C until required. Electrospray ionization–mass spectrometry (ESI–MS) using a Micromass Quattro Ultima triple quadrupole instrument operating in the positive ion detection mode was used to determine the mass of the purified samples. Proteins were desalted and exchanged into 1 M ammonium acetate followed by a 1 : 20 dilution with a 50 per cent methanol and 1 per cent formic acid solution.
3.6. Preparation of apo- and Cu(II)-ScoSl for circular dichroism spectroscopy
Apo-proteins were reduced in an anaerobic chamber (DW Scientific, [O2] less than 2 ppm) with 10 mM DTT and desalted (twice) using a PD-10 column (GE Healthcare) into the desired DTT-free buffer. Concentrations of apo-proteins were determined by UV–vis spectroscopy (Varian Cary 50 UV–vis spectrophotometer) using an extinction coefficient of 14 565 M−1 cm−1 at 280 nm [31]. Cu(II)-loaded samples were prepared in 100 mM sodium phosphate, 50 mM NaCl, pH 7.5 starting from reduced apo-ScoSl samples incubated with an excess of Cu(II)SO4 for 30 min. Unbound Cu(II) was removed by desalting (PD-10 column) and concentrating at 4°C using a 5 kDa cut-off centrifugal concentrator (Vivaspin). Samples for circular dichroism (CD) analysis were exchanged into 10 mM KPi, 50 mM KF. Far-UV CD spectra were recorded between 260 and 175 nm and visible spectra between 600 and 300 nm at 20°C on a Chirascan CD spectrophotometer (Applied Photophysics, Leatherhead, UK) equipped with a thermostatic cell holder controlled with a Peltier system. CD spectra were analysed using DichroWeb [32,33] with the programs CDSSTR [34–36] and Contin-LL [37] and the databases 4, 7 [36,38] and SP175 [39].
3.7. Electron paramagnetic resonance spectroscopy
Wilmad SQ electron paramagnetic resonance (EPR) tubes (Wilmad Glass, Buena, NJ, USA) were used and filled with 200 μl of protein solutions (100 mM sodium phosphate, 50 mM NaCl pH 7.5) treated with a sub-stoichiometric amount of Cu(II)SO4 (50 μM protein + 45 μM Cu(II)SO4) and frozen in methanol kept on dry ice. EPR spectra were measured at 10 K on a Bruker EMX EPR spectrometer (X-band) at a modulation frequency of 100 kHz. A spherical high-quality Bruker resonator SP9703 and an Oxford Instruments liquid helium system were used to measure the low-temperature EPR spectra. The EPR spectra of the blank samples (frozen water) were subtracted from the EPR spectra of the protein samples to eliminate the baseline caused by the resonator's walls, quartz insert or quartz EPR tube. The baseline was corrected by subtraction of a polynomial line drawn through a set of points randomly chosen on the baseline.
3.8. Cu(II) binding to apo-ScoSl monitored by UV–vis and fluorescence spectroscopy
A stock solution of 100 mM Cu(II)SO4 was prepared anaerobically and diluted as required. Reduced apo-ScoSl proteins were prepared anaerobically in 100 mM sodium phosphate, 50 mM NaCl, pH 7.5. Titration of a Cu(II) solution of known concentration using a gastight syringe (Hamilton) into 100 μM of reduced apo-protein in an anaerobic quartz cuvette (Hellma) was monitored at 20°C in the UV–vis spectrum. Fluorescence spectroscopy was carried out on an LS 50B fluorimeter (Perkin Elmer). A Trp fluorescence emission spectrum was collected between 300 and 400 nm with excitation at 280 nm. The excitation slit and the emission slit were set at 5 nm, and the change in emission at approximately 330 nm was monitored upon titration of aliquots of a Cu(II) solution into a 5 µM protein solution.
3.9. Stopped-flow kinetics
Kinetic experiments were carried out using an SX20 stopped-flow spectrophotometer (Applied Photophysics) thermostatted at 20°C with a Peltier system. A stock of 100 mM Cu(II)SO4 and reduced apo-proteins (20 mM sodium phosphate 50 mM NaCl, pH 7.5) were prepared. Time courses were taken at 360 and 380 nm with various Cu(II) concentrations (20–1000 µM before mixing) and 10 µM protein with the transients fitted to a two-step exponential function. A point-to-point accumulation was carried out over 320–420 nm with a step of 5 nm using 30 µM protein and 100 µM Cu(II) before mixing. The resulting kinetic data were analysed using the program ‘ProK’ (Applied Photophysics). This program was also used to construct spectra of intermediates using a simple sequential model (e.g. a > b > c). Analysis by singular value decomposition was used to determine the minimum number of species required to reconstruct the original dataset.
3.10. Determination of the mid-point redox potential of apo-proteins
Apo-ScoSl (2.5 μM) and the H176A mutant (2.5 μM) in 100 mM sodium phosphate, 50 mM NaCl, pH 7.5 were oxidized by the addition of 1 mM oxidized DTT (Sigma). The oxidized proteins were titrated with a stock solution of 10 mM reduced DTT (Melford), allowing 10 min for the protein to equilibrate to each new potential. The transition from oxidized to reduced protein was monitored by the increase in Trp fluorescence emission at approximately 330 nm (excitation at 280 nm) measured at 25°C with 5 nm excitation and emission slits. Intensity was corrected for dilution effects. A mid-point reduction potential (Em) was determined by calculating the fraction of ScoSl reduced (fr) at each point in the titration and plotting this as a function of the system potential. The data were then fit to the Nernst equation (3.1)
| 3.1 |
in which Em and n, the number of electrons, were allowed to float. F is the Faraday constant, R is the gas constant, T is the temperature and Eh is the standard reduction potential couple for DTT, which was taken to be −330 mV at pH 7 and 25°C.
3.11. Thiol-reductase activity assay
The turbidimetric insulin assay as outlined by Holmgren [40] was used to monitor thiol-reductase activity. E. coli thioredoxin (Trx) and human insulin were purchased from Sigma and reaction mixtures containing 3 μM of protein and 2.5 mM DTT were incubated in K2HPO4, pH 8 followed by the addition of insulin to a final concentration of 0.17 mM. The absorbance change at 650 nm caused by the precipitation of reduced insulin chains was monitored using a Cary 50 spectrophotometer every 60 s for up to 2 h. Controls were carried out with Trx in the absence of DTT, bovine serum albumin (BSA) in the presence of DTT and insulin with DTT.
4. Results
4.1. ScoSl is over-expressed, folded and has two reactive Cys residues
An N-terminal His6-tag fusion construct in which the first 23 amino acids of ScoSl were absent was prepared. The design of this construct was based on bioinformatics analysis of the ScoSl amino acid sequence in which an N-terminal transmembrane helix or a signal sequence (residues 1–21) for secretion with putative lipidation of Cys-22, which subsequently becomes the N-terminal amino acid upon cleavage by signal peptidase II, was predicted. The purified protein had a mass of 21 265.8 (1.1) Da (predicted mass, 21 264 Da) determined by denaturing ESI–MS. This mass corroborated that the N-terminal fused His6-tag had been successfully removed by thrombin treatment during the purification stages (mass with His6-tag 23 147.9 Da). The visible region of the absorption spectra was featureless with a prominent peak in the UV region at 280 nm. Anaerobic treatment of the purified ScoSl with DTT and subsequent removal by desalting under anaerobic conditions yielded a protein with accessible thiol groups as determined by the reduction of DTNB (5,5′-dithiobis-(2-nitrobenzoic acid) [41]. An average thiol : protein ratio of 1.6 : 1 was consistently determined, which is close to the expected ratio of 2 : 1 based on the presence of two Cys residues in the CXXXC motif (figure 1b). The far-UV CD spectra of reduced and oxidized ScoSl are shown in figure 2a and are typical for a mixed secondary structure protein whereby the α-helical segments are dominant with some distortion in the ‘classical’ α-helical CD shape resulting from β-strand segments. Secondary structure content of these two ScoSl forms was determined using DichroWeb and is reported in table 1.
Figure 2.
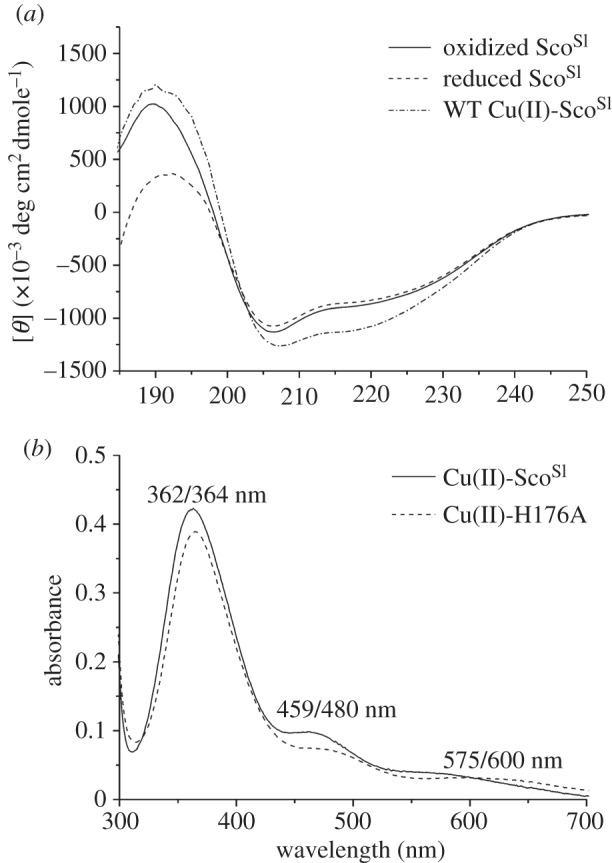
CD and visible absorption spectra of ScoSl and the H176A mutant. (a) Far-UV CD spectra of apo-oxidized (disulphide), apo-reduced (free-thiol) and Cu(II)-loaded ScoSl. (b) Visible absorbance spectra of Cu(II)-loaded ScoSl and the H176A mutant. All spectra were recorded at 20°C, with protein concentrations ranging between 20 and 100 μM.
Table 1.
Output from DichroWeb of average secondary structure content in the different forms of ScoSl used in this study.
| protein | α-helix (%) | β-sheet (% ) | turn (%) | unordered (%) |
|---|---|---|---|---|
| oxidized ScoSl | 27.3 | 17.1 | 20.9 | 34.7 |
| reduced ScoSl | 35.0 | 18.7 | 16.2 | 30.0 |
| Cu(II)-ScoSl | 34.6 | 17.3 | 18.0 | 30.0 |
| oxidized H176A | 29.2 | 15.1 | 19.9 | 35.8 |
| reduced H176A | 27.1 | 18.6 | 20.9 | 33.4 |
| Cu(II)-H176A | 26.1 | 21.0 | 21.7 | 31.2 |
4.2. Reduced apo-ScoSl binds Cu(II)
To determine whether ScoSl has affinity for Cu(II), reduced apo-ScoSl was titrated with a Cu(II) solution. A number of distinct absorption bands in the UV–vis spectrum appear (figure 2b). The λmax are similar, but not identical, to those reported for Cu(II)-ScoBs (354, 450 and 545 nm) and have been attributed to arise from S(Cys)–Cu(II) charge transfer [42]. The absorbance increase at 362 nm plotted as a function of [Cu(II)]/[ScoSl] reveals a steep linear transition with a break point at approximately 1 equivalent of Cu(II) (figure 3a, inset), indicating a 1 : 1 binding stoichiometry with a strong affinity for Cu(II). In the presence of excess EDTA (up to 50-fold), the intensity of the absorption peaks in the UV–vis spectra of Cu(II)-ScoSl remained unchanged over many days. The far-UV CD spectrum of Cu(II)-ScoSl is similar to the apo-reduced or -oxidized forms (figure 2a) and thus consistent with no gross secondary structural change upon Cu(II) binding (table 1). The visible CD spectrum for the Cu(II)-ScoSl complex is shown in figure 3b and has a number of peaks and troughs that probably arise from S(Cys)–Cu(II) charge transfer. A single Trp residue (W132) is present in ScoSl (figure 1b). Excitation of reduced apo-ScoSl at 280 nm results in an emission spectrum with a peak maximum at approximately 330 nm, which upon addition of Cu(II) is quenched (figure 3c, inset). A plot of the emission intensity at 330 nm versus [Cu(II)]/[ScoSl] indicates a 1 : 1 binding stoichiometry (figure 3c) consistent with the data from UV–vis spectroscopy.
Figure 3.
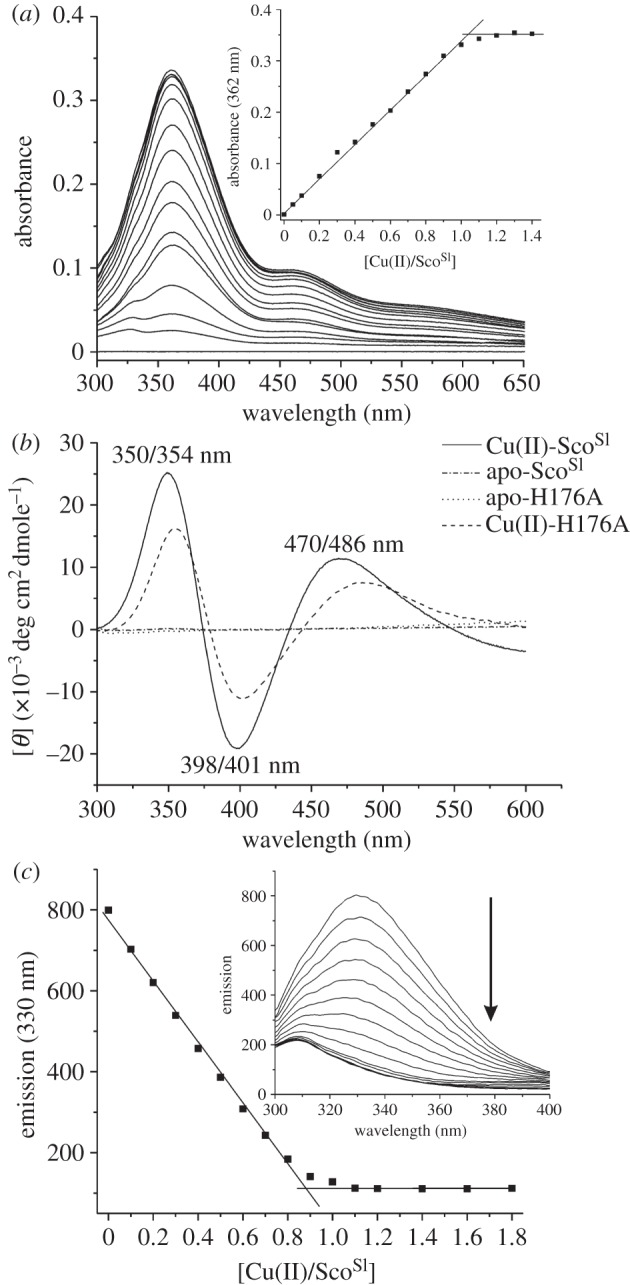
Cu(II) binding to reduced apo-ScoSl. (a) Changes in the UV–vis spectrum (pH 7.5, 20°C) of reduced apo-ScoSl (110 μM) upon stepwise addition of 10 μM Cu(II)SO4. Inset: the change in absorbance at 362 nm plotted as a function of [Cu(II)]/[ScoSl]. (b) Visible CD spectra of Cu(II)-loaded and apo-reduced ScoSl and the H176A mutant. (c) Inset: changes in the Trp emission spectrum (pH 7.5, 20°C) of reduced apo-ScoSl (5 μM) upon titration with 0.5 μM of Cu(II)SO4 with the emission at 330 nm plotted as a function of [Cu(II)]/[ScoSl]. The arrow indicates the direction of the emission change. The stoichiometry of the reaction in both (a) and (c) is indicated by the intersection of the two lines.
4.3. ScoSl possesses a redox active CXXXC motif but not thiol-disulphide reductase activity
To explore a possible role for apo-ScoSl as a thiol-disulphide reductase, the mid-point redox potential (Em) of the Cys residues in the CXXXC motif of apo-ScoSl was first determined by measuring the difference in fluorescence intensity of W132 in the oxidized and reduced forms of apo-ScoSl upon addition of DTT (figure 4a). The data fit to the Nernst equation (3.1) to give an Em value of –280 mV. An insulin precipitation assay with E. coli Trx as a control was used to assess whether ScoSl displayed thiol-disulphide reductase activity [40]. From the data presented in figure 4b, it is clear that Trx displays catalytic activity, whereas the long lag-phase followed by the slow increase in ΔA650 for apo-ScoSl is equivalent to the assay with DTT only and BSA with DTT (figure 4b), i.e. no catalytic activity. This indicates that despite having a redox active CXXXC motif, ScoSl does not possess catalytic thiol-disulphide reductase activity in the apo-form.
Figure 4.
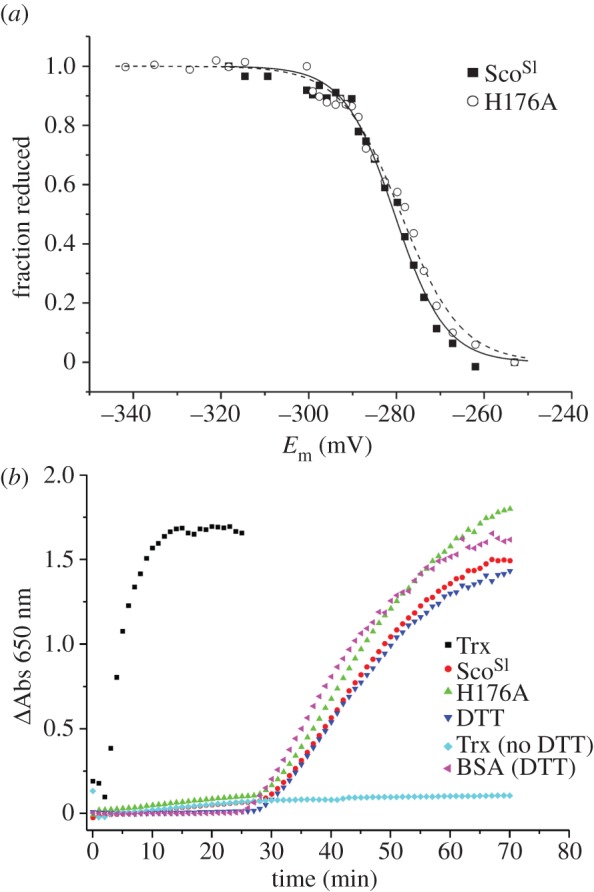
Mid-point redox potential and thiol-disulphide reductase activity of ScoSl and the H176A mutant. (a) Plot of fraction of reduced apo-ScoSl and the H176A mutant calculated from the fluorescence intensity at 330 nm as a function of the cell potential. The solid and dashed lines show the individual fits to the data for the respective proteins using equation (3.1). (b) Insulin reductase activity of E. coli Trx, ScoSl and the H176A mutant in the presence of 2.5 mM DTT, along with a number of controls, DTT only, BSA + DTT and Trx with no DTT. The precipitation of insulin was monitored at 650 nm. ScoSl or the H176A mutant do not show thioredoxin-like activity in this assay.
4.4. ScoSl is critical for morphological development and CcO activity at low exogenous Cu levels
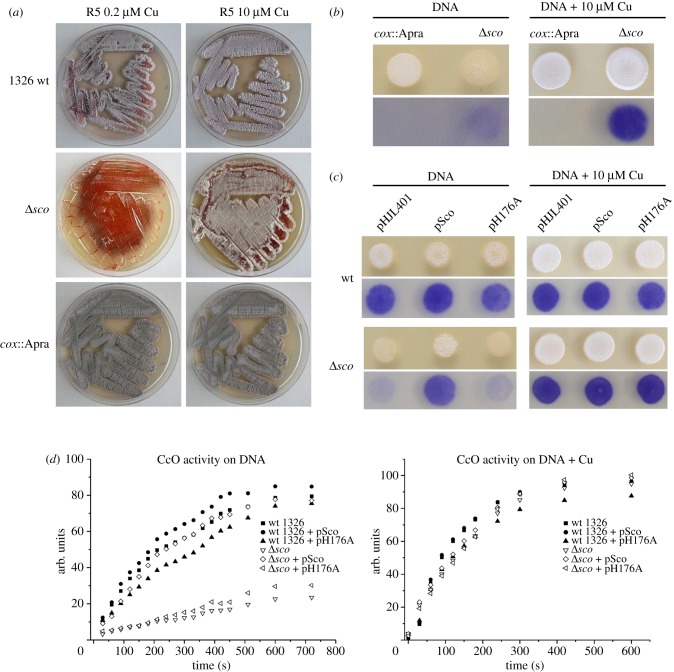
The function of ScoSl was probed by comparing morphological development and CcO activity of wt S. lividans 1326 and Δsco strains. The wt strain shows almost full development, aerial hyphae and spore production on R5 medium, and a clear stimulation of development on R5 supplemented with 10 μM Cu(II) (figure 5a). The Δsco mutant, on the other hand, is not capable of switching from vegetative to aerial growth and spore production on R5 medium (less than 0.2 μM Cu(II)), whereas development is clearly restored upon addition of 10 μM Cu(II) (figure 5a). This demonstrates that ScoSl has a critical role in the onset of aerial hyphae formation at low Cu availability. A similar effect on development of the Δsco strain is seen on DNA medium under low levels of exogenous Cu(II) but development is once more restored on addition of 10 μM Cu(II) (figure 5b). A cox::Apra strain, in which the genes SL2155 and SL2156 encoding for subunits I and II of the aa3-type CcO have been removed, appears to be viable and not defective in development under low or elevated Cu(II) (figure 5a,b). However, development is somewhat delayed compared with the wt on both R5 and DNA medium. CcO activity was tested for in the cox::Apra mutant by using TMPD as substrate. It is apparent from figure 5b that no indophenol blue was observed and this is therefore consistent with the absence of TMPD oxidation by the aa3-type CcO, but also points to the absence of any other Cu/haem oxidases that may be upregulated in the absence of the aa3-type (the bo-type and ba3-type Cu/haem oxidases that do not react with TMPD are absent in S. livdians). In line with a putative function of ScoSl in the assembly of the CuA site, the CcO activity in the Δsco mutant (figure 5b) is strongly reduced and maximal 25 per cent of the activity is found in the wt strain (figure 5c,d). Increasing the Cu(II) concentration in the medium clearly restores CcO activity to wt levels in the Δsco mutant, corroborating the observation with development that ScoSl is also not required for maturation of CcO at elevated Cu(II) levels.
Figure 5.
Morphology and CcO activity in wt and mutant strains of S. lividans. (a) S. lividans 1326 (wt), the Δsco and the cox: :Apra mutants were grown on R5 and R5 supplemented with 10 μM Cu(II) for 5 days. (b) The top two frames show the growth of the cox::Apra and Δsco mutants on DNA medium and DNA supplemented with 10 μM Cu(II). Drops of 10 μl containing 1000 spores were spotted on the plates and incubated for 48 h. The appearance of the white fluffy aerial mycelium and grey spores demonstrates that a strain is capable of full morphological development. The lower two frames show the in vivo detection of CcO activity with TMPD as substrate. The blue colour is the result of TMPD conversion to indophenol blue by CcO. (c) Morphology and CcO activity of wt and Δsco mutant transformed with empty vector (pHJL401), pHJL401 containing sco under its own promoter (pSco) and pHJL401 expressing the H176A mutant (pH176A) on DNA plates and DNA supplemented with 10 μM Cu(II). (d) CcO activity detected by the TMPD assay and recorded by digital imaging of the plates. The ImageJ software (http://imagej.nih.gov/ij/) was used to calculate average pixel intensities (s.d. varied from 1% to 15%) of the indophenol blue stained mycelium as seen in (b) and (c). After correction for background reading, the data were multiplied by −1 and plotted as arbitrary units against time.
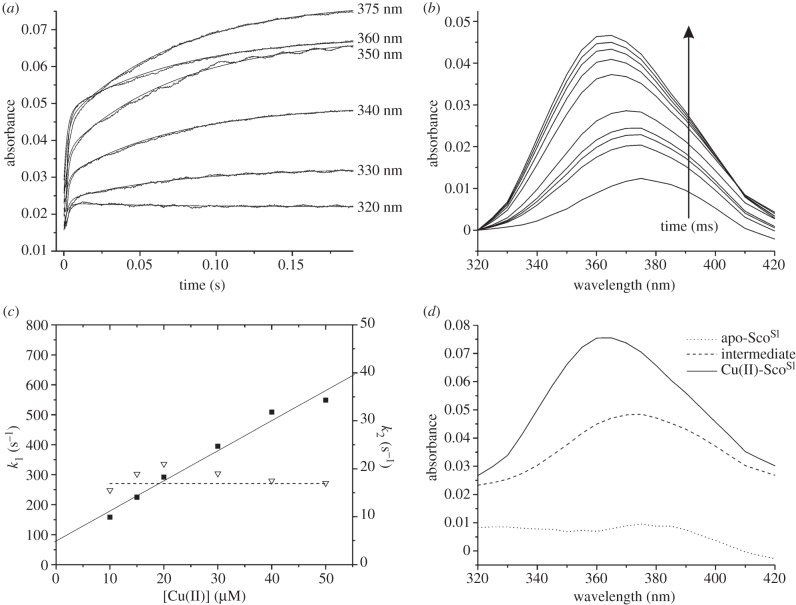
4.5. ScoSl binds Cu(II) rapidly in a biphasic process
On mixing reduced apo-ScoSl with excess Cu(II), a biphasic time course was observed at all wavelengths examined (figure 6a). The rapid phase occupied the first 10 ms after mixing and was followed by a slower phase taking approximately 200 ms to complete (figure 6a). This behaviour suggests that the formation of the final Cu(II)-ScoSl complex from the apo-protein passes through an intermediate. The time-dependent amplitudes of the time courses were used to construct the time-resolved spectra given in figure 6b. The observed rate constant for the faster process is seen to be linearly dependent on [Cu(II)] up to approximately 500 s−1. This phase therefore represents the second-order process in which Cu(II) binds initially to form an intermediate. Fitting of this dependency yields a value of the second-order rate constant (k1) of 1 × 107 M−1 s−1. It is evident that at a [Cu(II)] of zero there is a significant value of the extrapolated pseudo first-order rate constant (79 s−1) (figure 6c). This value may be assigned to the dissociation rate constant (k−1) of Cu(II) from this intermediate. Taken together, these values allow an equilibrium binding constant (Kb) of 1.3 × 105 M−1 (k1/k−1) to be calculated for Cu(II) binding to the intermediate. A similar Kb value was determined from the examination of the amplitudes of the fast process as a function of [Cu(II)] (not shown). The slower kinetic process that leads from the intermediate to the final product showed little dependency on [Cu(II)], maintaining a value of approximately 20 s−1 at all [Cu(II)] used (figure 6c). This behaviour indicates that the step forming the final product from the intermediate involves no further Cu(II) binding but an internal rearrangement within the Cu(II) protein complex. Based on a sequential model with one intermediate, global fitting of the kinetic profiles in figure 6a,b gave rise to the spectral features of the intermediate and the final product as shown in figure 6d. It may be observed that the intermediate has a spectrum distinct from the final complex, having a λmax at 375 nm and a considerably lower extinction coefficient (figure 6d). These spectral features are similar to those reported for the binding of Cu(II) to ScoBs [43]. The mechanism described earlier, namely a rapid second-order binding process leading to a spectrally distinct intermediate that then rearranges to form a final complex, is identical to that proposed for ScoBs [43] but in the case of ScoSl the rate constants are considerably larger.
Figure 6.
Stopped-flow kinetics of Cu(II) binding to ScoSl. (a) Time courses at the indicated wavelengths observed on reacting 15 μM ScoSl with 50 μM Cu(II). (b) Spectra constructed at the following times, 3, 4.5, 6, 16.5, 60, 87, 114, 141, 200 ms, after mixing from the time courses given in (a). (c) First-order rate constants for the ‘fast’ (k1; filled squares) and ‘slow’ (k2; open triangles) phase of Cu(II) binding to ScoSl as a function of [Cu(II)]. The solid lines indicate a fit to the first rapid phase to obtain a second-order rate constant, and the dashed line indicates the independence of the rate of the slower phase (k2) on [Cu(II)]. (d) Spectra obtained from global fitting of the data presented in (a) and (b) to the model a > b > c.
4.6. H176 is a Cu(II) ligand and stabilizes the cupric state
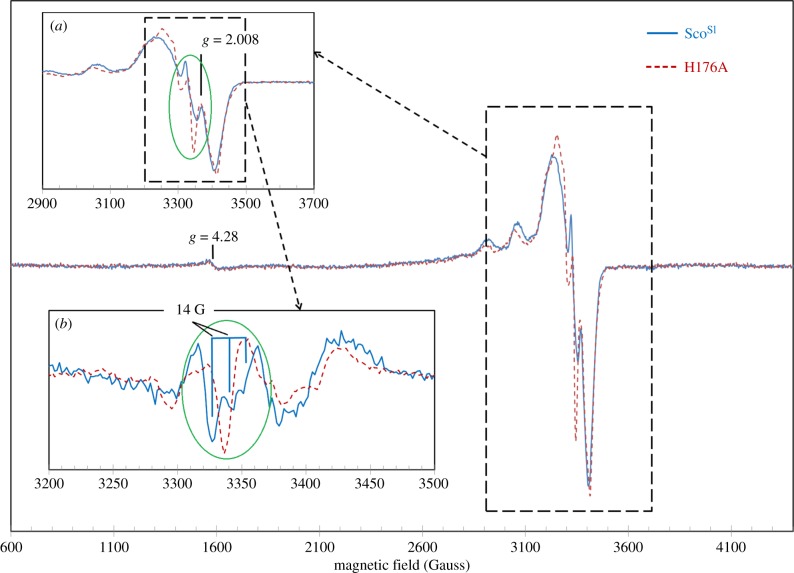
To explore further the mechanistic and functional properties of Cu(II)-ScoSl, the effect of removing a putative first coordination sphere Cu(II) ligand was assessed by creating the H176A mutant. The EPR spectra of Cu(II)-loaded wt and H176A mutant are typical for cupric ions [44] (figure 7). The perfect fit of the baselines, including the g = 4.3 signal from the adventitious ferric iron in rhombic coordination [45], indicates that the differences in the EPR spectra of the two proteins, although small, are significant. An additional hyperfine structure is present in the wt spectrum (figure 7a, inset). This is better observed from the derivatives of the spectra (figure 7b, inset), showing that wt ScoSl has a triplet structure, whereas the mutant H176A has a single line. The separation of these three components in Gauss (approx. 14 G) is consistent with an interaction of the electronic spin with a nitrogen nucleus [46]. Thus, the differences in the EPR spectra of the two proteins can be explained by the presence of an additional N atom in the vicinity of the Cu(II) atom in the wt ScoSl, whereas such an atom is absent in the H176A mutant, strongly suggesting that His176 is a Cu(II) ligand in ScoSl.
Figure 7.
EPR spectra of Cu(II)-loaded ScoSl and the H176A mutant. The superimposed spectra within the dashed line box are shown in greater detail in inset (a). Derivatives by field have been taken of both spectra and inset (b) shows the second derivatives of the EPR absorption. The most rapidly changing part of the spectra, as the field increases, is highlighted by an oval in both first (inset a) and second derivatives (inset b) of the EPR absorption. The latter representation demonstrates that instead of a single peak in the H176A mutant, there is a triplet with a 14 G separation between the lines in the wt protein spectrum. Instrumental conditions were as follows: microwave frequency νMW = 9.466 GHz, microwave power PMW = 3.181 mW, modulation frequency νm = 100 kHz, modulation amplitude Am = 5 G, time constant τ = 81.9 ms, scan rate V = 22.6 G s−1, number of scans per spectrum NS = 1, each spectrum represents an average of three spectra obtained from three independent samples.
The far-UV CD spectra of the reduced and oxidized H176A ScoSl mutant are shown in figure 8a and the deconvolution using DichroWeb reported in table 1. The H176A mutant readily bound Cu(II) with slight shifts in λmax compared with the wt protein (figure 2b). Stoichiometric Cu(II) binding was further corroborated from the decay of the W132 emission in the fluorescence spectrum (figure 8b) and the presence of a visible CD spectrum similar to that of wt ScoSl, but with significant wavelength shifts of the peaks and trough (figure 3b). At Cu(II) stoichiometries of 1 or greater, the absorption bands in the UV–vis spectra spontaneously bleach, and a slight but significant increase in the W132 emission is observed (figure 8b). Similarly, the absorption bands in the visible CD spectrum spontaneously decay with time at Cu(II) stoichiometries of 1 or greater, with a decay rate, kred, of 3.1 × 10−4 s−1 (figure 8c, inset). The observation that the W132 emission does not return to apo-ScoSl intensities suggests that an autoreduction process is occurring resulting in Cu(I)-H176A. Finally, an Em of −279 mV for the CXXXC motif in the H176A mutant was determined (figure 4a), and from the insulin precipitation assay catalytic thiol-disulphide reductase activity was absent for the apo-H176A (figure 4b).
Figure 8.
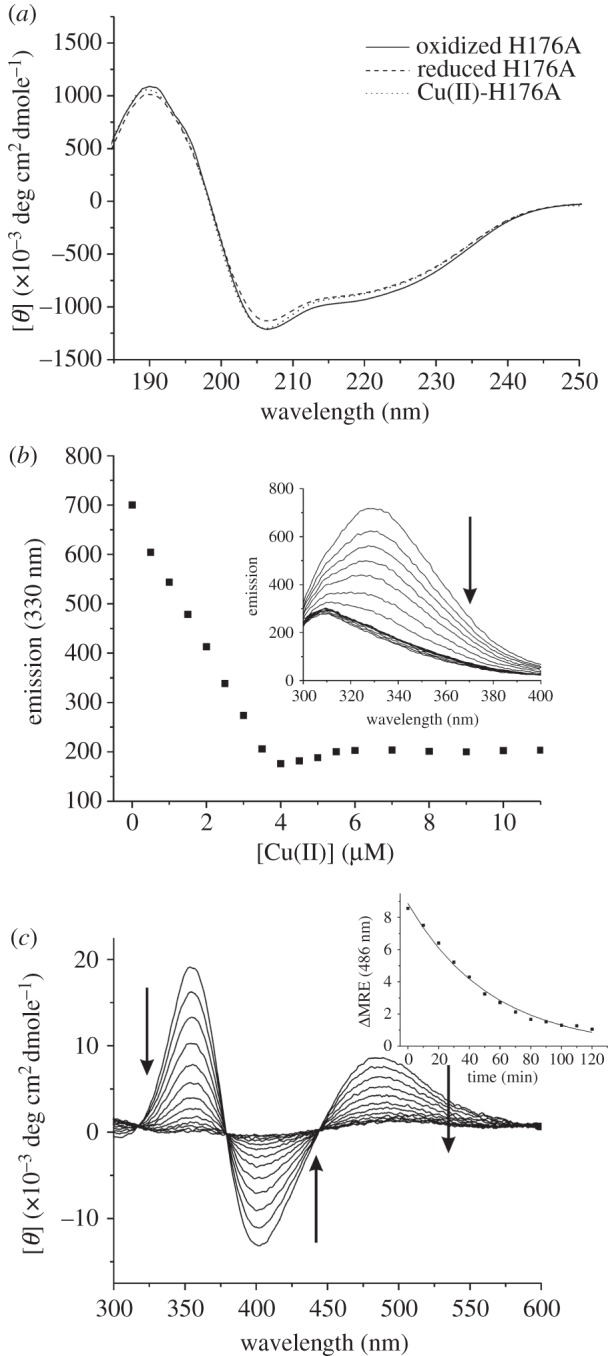
Cu(II) binding to the H176A mutant. (a) Far-UV CD spectra of apo-oxidized (disulphide), apo-reduced (free-thiol) and Cu(II)-loaded H176A. (b) Changes in the Trp emission spectrum of reduced apo-H176A (inset) upon titration with 0.5 μM of Cu(II)SO4 with the emission at 330 nm plotted as a function of [Cu(II)]/[H176A]. The arrow indicates the direction of the emission change up to 1 equivalent of Cu(II), with a slight increase and plateau observed above 1 equivalent of Cu(II). (c) Visible CD spectrum for the H176A mutant following addition of 1 equivalent of Cu(II). The spontaneous decay over time of the visible bands are indicated by arrows, and the inset is an example of a plot of mean residue ellipticity (MRE) at 486 nm versus time fitted to a single exponential decay function to give a decay rate constant (k) for the process. All spectra were recorded at 20°C, with protein concentrations ranging between 5 and 400 μM.
4.7. Complementation of the Δsco strain with pScoSl restores function but pH176A does not
To test the effect of the H176A mutant on development and CcO activity, complementation experiments with low copy number plasmids expressing the wt ScoSl gene (pSco) and the H176A mutant (pH176A) were carried out. For transformation of pSco and pH176A into the 1326 wt strain, no effect on growth or development on DNA at low [Cu] or with 10 μM Cu(II) was observed. However, a small and reproducible effect on CcO activity was observed on DNA without added Cu(II). Figure 5d illustrates that the H176A mutation has a negative effect on CcO activity in the wt strain, whereas the wt Sco protein has a small positive effect. In the Δsco mutant, the development and CcO activity under low Cu(II) concentrations are restored to near wt 1326 levels upon transformation with pSco (figure 5c,d). However, pH176A was not capable of restoring development or CcO activity to wt levels in the Δsco mutant although a small increase in activity is observed in the absence of exogenous Cu(II) (figure 5d). We also note that the time courses for the TMPD assays are multiphasic in the absence of added Cu(II). At 10 μM Cu(II), the development and CcO activity in all Δsco transformants on DNA are restored to wt levels (figure 5c,d), and the TMPD time courses now concur to a single phase (figure 5d). Taken together, these data indicate that the phenotype and reduced CcO activity in the Δsco mutant can be attributed completely to the absence of ScoSl, and that despite the H176A mutant being able to stoichiometrically bind Cu(II) in vitro, it is not capable of supporting wt levels of development and CcO activity at low Cu(II) concentrations in vivo.
4.8. The H176A mutant reveals a spectrally distinct intermediate on binding Cu(II)
The kinetics of Cu(II) binding to the H176A mutant were studied in a similar manner to those of the wt ScoSl and revealed rapid formation of an intermediate that eventually yielded the final complex. Similar analysis to that undertaken for the wt protein shows that the intermediate formed in the H176A mutant is distinctly different in spectral characteristics from that shown in figure 6d, with a λmax now at 365 nm (figure 9a). The initial rapid kinetic process again displayed a Cu(II) concentration dependence but now the pseudo first-order rate constant plateaued at approximately 170 s−1 (figure 9b). Fitting of the initial linear portion of the rate dependence at low [Cu(II)] yields a second-order rate constant (k1_H176A) of 2 × 106 M−1 s−1 and a dissociation rate constant (k−1_H176A) of 30 s−1. Thus, the rates of binding and dissociation appear lower than in wt ScoSl and correspond to a slightly lower Kb for the intermediate complex of 6.7 × 104 M−1. The slow process again shows very little dependence on [Cu(II)], yielding a first-order rate constant (k2_H176A) of approximately 5 s−1, again significantly slower than wt.
Figure 9.
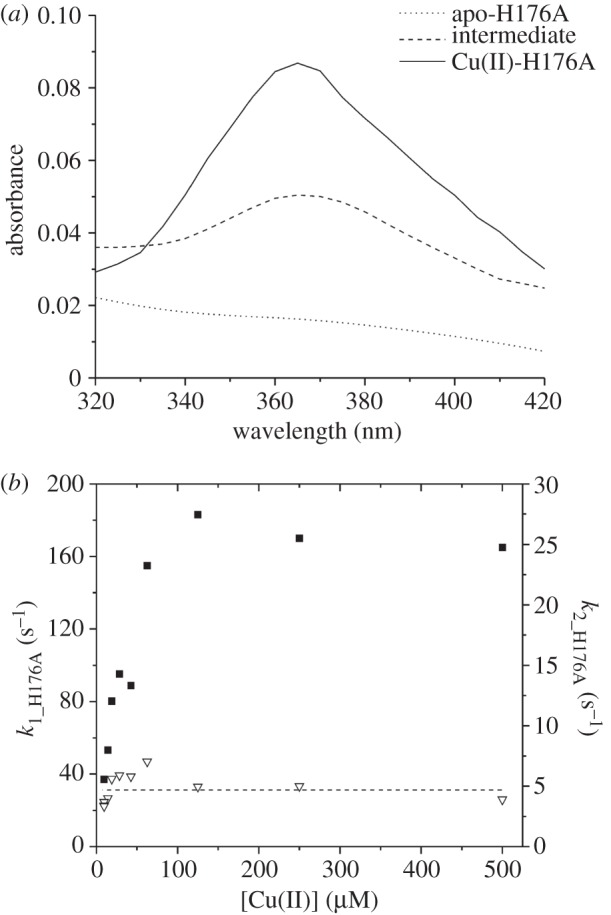
Stopped-flow kinetics of Cu(II) binding to the H176A mutant. (a) Global fitting of time courses at wavelengths from 420 to 320 nm to a sequential model a > b > c. (b) First-order rate constants for the ‘fast’ (k1_H176A; filled squares) and ‘slow’ (k2_H176A; open triangles) phases of Cu(II) binding to the H176A as a function of Cu(II) concentration. The dashed line indicates the [Cu(II)] independence for k2_H176A.
5. Discussion
5.1. The sco operon of Streptomyces lividans encodes a gene for a Cu(II) binding Sco protein
Previous studies with Cu(II)-Sco proteins from human, yeast and B. subtilis [16,20,23,42,47] have identified a type 2 Cu centre, with UV–vis absorption spectra displaying similar features to the red cupredoxin nitrosocyanin that binds Cu(II) in a square pyramidal geometry using two N(His) ligands, a single S(Cys), a solvent water molecule in the basal plane and a coordinating O(Glu) as the axial ligand [48]. The major absorption band in the visible region for Cu(II)-nitrosocyanin (approx. 390 nm) and Cu(II)-Sco (approx. 360 nm) are reported to arise from S(Cys)–Cu(II) charge transfer dominated by σ rather than π orbital interactions as is the case in cupredoxins [49]. The absorption spectra for Cu(II)-ScoSl and the H176A mutant (figure 2b) have the same overall features as Cu(II)-ScoBs [23], the Cu(II)-H135A mutant of ScoBs [50] and the eukaryotic Sco1 proteins form yeast and human [20,47], corroborating that the 3966 gene, part of the sco operon in S. lividans, encodes for a Cu(II) binding Sco protein. From the EPR data (figure 7), a contribution to the Cu(II) coordination sphere from the imidazolate N of His176 is apparent, yet the H176A mutant retains the ability to bind Cu(II) with the same stoichiometry and a similar affinity (derived from kinetic studies) as the wt protein. However, in the absence of the His ligand, the Cu(II)-ScoSl is susceptible to autoreduction to Cu(I). This has also been reported for the H135A mutant of ScoBs but occurs an order of magnitude faster (1 × 10−3 s−1) compared with the H176A ScoSl mutant (3.1 × 10−4 s−1) [50].
5.2. ScoSl is unlikely to function as a thiol-disulphide reductase
An alternative ‘non-Cu’ role for bacterial Sco proteins as thiol-disulphide reductases has been postulated, based on the ability of ScoTt to reduce the Cys residues of the CuA site in CcO, thus facilitating Cu delivery by the periplasmic Cu(I) chaperone PCuAC [18]. To maintain ScoTt in the reduced state competent for redox function, a supply of reducing equivalents in the periplasmic environment is required and thus a redox role has been questioned [51]. Although we have not looked at the same reaction with the S. lividans proteins, namely the reduction of the Cys residues in the CuA domain of CcO by ScoSl, our in vitro assay clearly shows no enzymatic enhancement of thiol-disulphide reductase activity for apo-ScoSl compared with E. coli Trx (figure 4b). Our findings are thus in keeping with results from a similar assay with human Sco1, where thiol-disulphide activity was not detected [20]. An Em of −280 mV for the CXXXC motif of ScoSl has been determined in this study and is comparable to that of the Em for the CXXC motif of Trx (−270 mV). However, these similarities in Em are not sufficient to infer catalytic activity in apo-ScoSl when present in an insulin precipitation assay where Trx clearly possesses thiol-disulphide activity and, therefore, catalytic activity is not inherent on the Em of the disulphide motif in the respective proteins. Furthermore, the Em of the H176A mutant is unaffected, and this finding coupled with the observation that Cu(I) is the more stable oxidation state of the Cu bound H176A has ramifications for interpreting the lack of development and CcO activity in vivo with this mutant (vide infra).
5.3. At low Cu levels morphogenesis and CcO activity require the assistance of Cu(II)-ScoSl
The absence of indophenol blue from the TMPD assay in the cox::Apra strain is consistent with no activity from the aa3-type oxidase (figure 5b). However, in the Δsco mutant indophenol blue is detected, albeit to a much lower level than in the wt strain or upon addition of exogenous Cu(II) (figure 5b–d). Therefore, a basal level of active CcO is present under low Cu and in the absence of ScoSl, suggesting that the CuA site of CcO is metallated and active. Despite the presence of low levels of CcO activity in the Δsco mutant, the developmental switch from vegetative to aerial mycelium clearly does not occur in the absence of ScoSl (figure 5a). The morphological phenotype for the Δsco mutant coinciding with the significantly decreased CcO activity compared with the wt strain is restored upon complementation with pSco (figure 5c,d). This is therefore evidence that the switch from vegetative to aerial growth in S. lividans is dependent on ScoSl with a clear correlation to CcO biogenesis implied. It therefore appears that when ScoSl is present CcO is metallated much more efficiently at low [Cu]. In contrast at elevated [Cu], the development block is lifted, and CcO activity is restored in the Δsco mutant, indicating that under these conditions the role of ScoSl is bypassed with Cu being delivered to the CuA site through another means or by spontaneous self-assembly.
The H176A mutant binds Cu(II) readily, but is susceptible to an autoreduction process that results in the Cu(I)-bound state. The inability of the H176A mutant to restore CcO activity in the Δsco mutant under low [Cu(II)] may therefore be due to the mutant's inability to maintain the Cu(II) oxidation state, with the Cu(I) bound state no longer active in transferring Cu to the CuA site. The absence of CcO activity and development with this mutant further questions the role of the proximal 3965 gene that encodes for a PCuAC-like protein in the co-factoring of the CuA domain. Recent data from R. sphaeroides are in accordance with our data supporting the maturation of the CuA site proceeding only via a Sco-like protein, PrrC, with a PCuAC-like protein having an as yet undefined role [52]. Although not tested in this study, it is conceivable that Cu plays a role in thiol-disulphide redox activity of ScoSl and that the stabilization of the cuprous state by the H176A mutant inhibits any electron donation involving Cu oxidation. Further studies into this role are planned.
5.4. A second Cu(II)-ScoSl target is required for aerial hyphae formation in Streptomyces lividans
As indicated, despite the presence of low levels of CcO activity in the Δsco mutant, the developmental switch from vegetative to aerial mycelium clearly does not occur in the absence of ScoSl (figure 5a). If deleting the sco gene would be equivalent to only strongly reducing the CcO activity, the phenotypes of the Δsco and cox::Apra mutants would be expected to be very similar. However, this is not the case as the cox::Apra mutant is still capable of full development at low [Cu(II)], whereas the Δsco mutant is stalled in the vegetative growth phase (figure 5a). This leads us to speculate that a second ScoSl target is involved in initiation of aerial hyphae formation. This Cu-ScoSl target is likely to be a cuproenzyme and its identification is currently in progress. Furthermore, the observation that a strain lacking the terminal aa3-type oxidase can survive and initiate a full life cycle demonstrates that another terminal oxidase, incapable of reacting with TMPD, can take over respiration (figure 5b). Genes for the terminal quinol bd-type oxidase are present in S. lividans and is thus a good candidate.
5.5. ScoSl captures exogenous Cu(II) rapidly
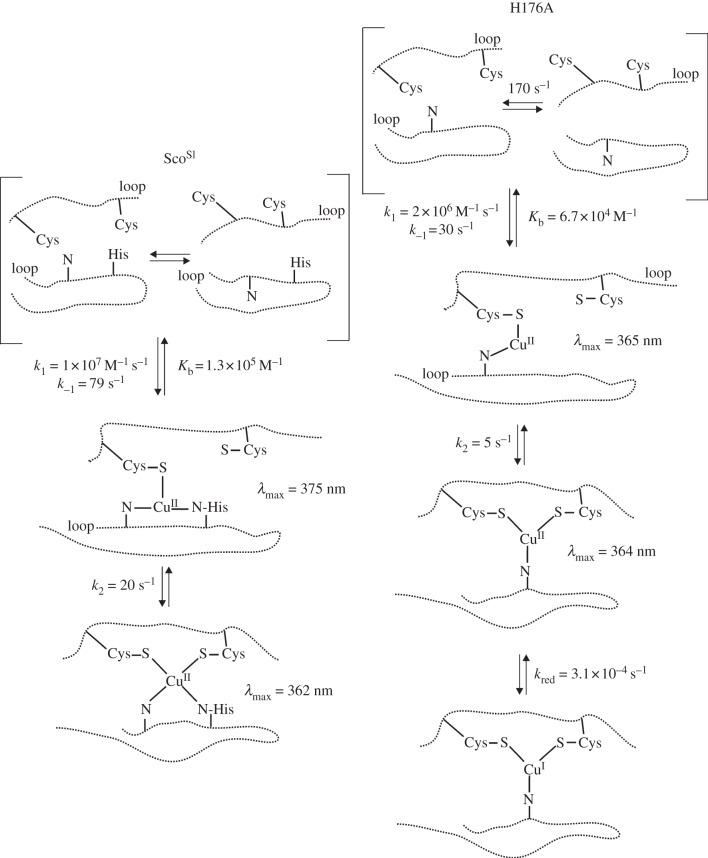
While our data establish that ScoSl acts as a Cu(II)-metallochaperone, how it acquires Cu(II) in vivo is not known. ScoSl is exported via the secretory (Sec)-pathway so is likely to obtain Cu once folded in the extracellular environment through a ligand-exchange interaction with another Cu-chaperone [51]. Under low Cu levels, it is expected that the cells' most efficient chaperone and scavenging systems operate, facilitating the efficient delivery of Cu to required locations, consistent with our in vivo data for ScoSl at low [Cu]. An emerging paradigm is that periplasmic (and possibly extracellular) cuproproteins may obtain Cu not from periplasmic or extracellular pools but from Cu that has been routed via the cytosol and delivered by P1-type ATPases [53]. Regardless of how ScoSl is co-factored in vivo, the mechanism through which Cu(II) binds to apo-ScoSl in vitro may best be understood through figure 10. The loops containing the Cu binding Cys residues and the His residue in Sco proteins (figure 1b) have been reported to be highly dynamic in the absence of Cu [22]. Therefore, the apo-ScoSl can be considered to have multiple conformations in equilibrium with one another, with some possessing a pre-formed Cu(II) binding site (figure 10). Rapid freeze EPR experiments with ScoBs have indicated prior to Cu(II)-thiolate coordination an initial capture complex exists composed of an equatorial N-Cu(II)-N complex, likely involving H135 and a N atom, possibly from a backbone amide [54]. By including this information in our model, we envisage that an initial capture complex comes close to one of the thiols and forms a site to which Cu(II) binds in a second-order process to form an intermediate in which the Cu(II) is coordinated to one thiol, His176 and a backbone amide N with a λmax = 375 nm (figure 10). This spectroscopically observable intermediate is consistent with absorption spectra reported for the single Cu(II)-thiolate coordinating mutants, C45A and C49A, in ScoBs [42] where a λmax of approximately 380 nm is observed, assigned to coordination of Cu(II) by a single Cys, His-135 and two unknown O/N ligands [42]. The protein dynamics in the apo-form is very rapid compared with Cu(II) binding to the pre-formed site and thus the spectral change we observe, dominated by binding to the thiol, is second-order. Once formed, the intermediate undergoes rearrangement in which the Cu(II)-complex is presented to the second thiol ligand leading to the final Cu(II)-ScoSl complex with a twofold increase in absorption and a λmax shift to 362 nm. This rearrangement occurs with k2 approximately 20 s−1 and a rate constant for dissociation (k−2) based on experiments with high concentrations of Cu(II) chelators of less than 10−6 s−1 (data not shown). While the binding of Cu(II) to form the intermediate is relatively weak (Kb = 105 M−1), coupling to the rearrangement that has an estimated equilibrium constant in favour of the final complex of 107 (k2/k−2) yields an overall affinity for Cu(II) greater than 1012 M−1. Although it appears that Cu(II) capture undergoes a similar mechanism to that reported for ScoBs, the rates for ScoSl are considerably quicker (k1 > three orders of magnitude, k2 > 10 times faster) indicating that ScoSl is much more efficient in Cu(II) capture and rearrangement to the final complex. Our experiments were carried out under similar conditions (temperature, pH and ionic strength) and thus structural differences inherent between proteins or the dynamics of the loops housing the Cys and His residues governing the accessibility to Cu(II) may be a reason.
Figure 10.
Mechanistic features of Cu(II) capture and binding to ScoSl and the H176A mutant.
5.6. H176 plays a significant role in Cu(II) capture
The second-order rate constant for the H176A mutant is significantly lower than wt (figure 10) and probably reflects the differences in the geometry of the intermediate site in the absence of the His residue. The spectrum of the intermediate is again dominated by thiol binding but the spectral modulation (position of λmax) given by His coordination is absent in the mutant compared with wt (figure 10). Interestingly, the His coordination appears to modulate the spectrum of the intermediate to a greater extent than the final complex in which both thiols bind to the Cu(II) (figure 10). The second-order nature of Cu(II) binding gives way to a first-order process at high [Cu(II)] (figure 9b), indicating that the rate limit for the formation of the intermediate species becomes rate limited by the rate at which the binding site can be formed in the absence of H176. This may be unremarkable given the different stereochemistries required to bring a His-N into the site as opposed to an amide-N close to the thiol. The intermediate nevertheless rearranges, albeit on a slower time scale than wt, to form the final product, in keeping with the kinetic restraints imposed by the absence of the His. The absence of H176 therefore does not impede Cu(II) capture but in its absence it becomes less efficient. H176 therefore clearly has a role in Cu(II) capture and is critical for maintaining the cupric form that is essential for the functional role of ScoSl.
6. Acknowledgement
This study was supported by the University of Essex through the award of a PhD studentship to K.L.I.M.B.
References
- 1.Chater KF. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Phil. Trans. R. Soc. B 361, 761–768 10.1098/rstb.2005.1758 (doi:10.1098/rstb.2005.1758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda K, Tomaru Y, Endoh K, Beppu T. 1997. Stimulatory effect of copper on antibiotic production and morphological differentiation in Streptomyces tanashiensis. J. Antibiot. (Tokyo) 50, 693–695 10.7164/antibiotics.50.693 (doi:10.7164/antibiotics.50.693) [DOI] [PubMed] [Google Scholar]
- 3.Keijser BJ, van Wezel GP, Canters GW, Kieser T, Vijgenboom E. 2000. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2, 565–574 [PubMed] [Google Scholar]
- 4.Fujimoto M, Yamada A, Kurosawa J, Kawata A, Beppu T, Takano H, Ueda K. 2012. Pleiotropic role of the Sco1/SenC family copper chaperone in the physiology of Streptomyces. Microbial Biotechnol. 5, 477–488 10.1111/j.1751-7915.2011.00319.x (doi:10.1111/j.1751-7915.2011.00319.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieser T, Hopwood DA. 1991. Genetic manipulation of Streptomyces: integrating vectors and gene replacement. Methods Enzymol. 204, 430–458 10.1016/0076-6879(91)04023-H (doi:10.1016/0076-6879(91)04023-H) [DOI] [PubMed] [Google Scholar]
- 6.Andreini C, Banci L, Bertini I, Rosato A. 2008. Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J. Proteome Res. 7, 209–216 10.1021/pr070480u (doi:10.1021/pr070480u) [DOI] [PubMed] [Google Scholar]
- 7.Worrall JAR, Vijgenboom E. 2010. Copper mining in Streptomyces: enzymes, natural products and development. Nat. Prod. Rep. 27, 742–756 10.1039/B804465C (doi:10.1039/B804465C) [DOI] [PubMed] [Google Scholar]
- 8.Dwarakanath S, Chaplin AK, Hough MA, Rigali S, Vijgenboom E, Worrall JAR. 2012. Response to copper stress in Streptomyces lividans extends beyond genes under direct control of a copper-sensitive operon repressor protein (CsoR). J. Biol. Chem. 287, 17 833–17 847 10.1074/jbc.M112.352740 (doi:10.1074/jbc.M112.352740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worrall JAR, Machczynski MC, Keijser BJ, di Rocco G, Ceola S, Ubbink M, Vijgenboom E, Canters GW. 2006. Spectroscopic characterization of a high-potential lipo-cupredoxin found in Streptomyces coelicolor. J. Am. Chem. Soc. 128, 14 579–14 589 10.1021/ja064112n (doi:10.1021/ja064112n) [DOI] [PubMed] [Google Scholar]
- 10.Ye Q, Imriskova-Sosova I, Hill BC, Jia Z. 2005. Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins. Biochemistry 44, 2934–2942 10.1021/bi0480537 (doi:10.1021/bi0480537) [DOI] [PubMed] [Google Scholar]
- 11.Larkin MA, et al. 2007. Clustal W and Clustal X, version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 (doi:10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 12.Glerum DM, Shtanko A, Tzagoloff A. 1996. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271, 20 531–20 535 10.1074/jbc.271.34.20531 (doi:10.1074/jbc.271.34.20531) [DOI] [PubMed] [Google Scholar]
- 13.Rentzsch A, Krummeck-Weiss G, Hofer A, Bartuschka A, Ostermann K, Rodel G. 1999. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr. Genet. 35, 103–108 10.1007/s002940050438 (doi:10.1007/s002940050438) [DOI] [PubMed] [Google Scholar]
- 14.Nittis T, George GN, Winge DR. 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276, 42 520–42 526 10.1074/jbc.M107077200 (doi:10.1074/jbc.M107077200) [DOI] [PubMed] [Google Scholar]
- 15.Mattatall NR, Jazairi J, Hill BC. 2000. Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis. J. Biol. Chem. 275, 28 802–28 809 10.1074/jbc.M002741200 (doi:10.1074/jbc.M002741200) [DOI] [PubMed] [Google Scholar]
- 16.Andruzzi L, Nakano M, Nilges MJ, Blackburn NJ. 2005. Spectroscopic studies of metal binding and metal selectivity in Bacillus subtilis BSco, a homologue of the yeast mitochondrial protein Sco1p. J. Am. Chem. Soc. 127, 16 548–16 558 10.1021/ja0529539 (doi:10.1021/ja0529539) [DOI] [PubMed] [Google Scholar]
- 17.Banci L, Bertini I, Cavallaro G, Ciofi-Baffoni S. 2011. Seeking the determinants of the elusive functions of Sco proteins. FEBS J. 278, 2244–2262 10.1111/j.1742-4658.2011.08141.x (doi:10.1111/j.1742-4658.2011.08141.x) [DOI] [PubMed] [Google Scholar]
- 18.Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ, Wang S. 2008. Mechanism of Cu(A) assembly. Nat. Chem. Biol. 4, 599–601 10.1038/nchembio.110 (doi:10.1038/nchembio.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abajian C, Rosenzweig AC. 2006. Crystal structure of yeast Sco1. J. Biol. Inorg. Chem. 11, 459–466 10.1007/s00775-006-0096-7 (doi:10.1007/s00775-006-0096-7) [DOI] [PubMed] [Google Scholar]
- 20.Williams JC, Sue C, Banting GS, Yang H, Glerum DM, Hendrickson WA, Schon EA. 2005. Crystal structure of human SCO1: implications for redox signaling by a mitochondrial cytochrome c oxidase ‘assembly’ protein. J. Biol. Chem. 280, 15 202–15 211 10.1074/jbc.M410705200 (doi:10.1074/jbc.M410705200) [DOI] [PubMed] [Google Scholar]
- 21.Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Martinelli M, Palumaa P, Wang S. 2006. A hint for the function of human Sco1 from different structures. Proc. Natl Acad. Sci. USA 103, 8595–8600 10.1073/pnas.0601375103 (doi:10.1073/pnas.0601375103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. 2003. Solution structure of Sco1: a thioredoxin-like protein involved in cytochrome c oxidase assembly. Structure 11, 1431–1443 10.1016/j.str.2003.10.004 (doi:10.1016/j.str.2003.10.004) [DOI] [PubMed] [Google Scholar]
- 23.Imriskova-Sosova I, Andrews D, Yam K, Davidson D, Yachnin B, Hill BC. 2005. Characterization of the redox and metal binding activity of BsSco, a protein implicated in the assembly of cytochrome c oxidase. Biochemistry 44, 16 949–16 956 10.1021/bi051343i (doi:10.1021/bi051343i) [DOI] [PubMed] [Google Scholar]
- 24.Badrick AC, Hamilton AJ, Bernhardt PV, Jones CE, Kappler U, Jennings MP, McEwan AG. 2007. PrrC, a Sco homologue from Rhodobacter sphaeroides, possesses thiol-disulfide oxidoreductase activity. FEBS Lett. 581, 4663–4667 10.1016/j.febslet.2007.08.058 (doi:10.1016/j.febslet.2007.08.058) [DOI] [PubMed] [Google Scholar]
- 25.Kieser TB, Buttner MJ, Chater KF, Hopwood DA. 2000. Pratcical Streptomyces genetics. Norwich, UK: John Innes Foundation [Google Scholar]
- 26.Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A. 2008. Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 78, 1065–1070 10.1007/s00253-008-1382-9 (doi:10.1007/s00253-008-1382-9) [DOI] [PubMed] [Google Scholar]
- 27.Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171, 5872–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wezel GP, White J, Hoogvliet G, Bibb MJ. 2000. Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Mol. Microbiol. Biotechnol. 2, 551–556 [PubMed] [Google Scholar]
- 29.Green GN, Gennis RB. 1983. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J. Bacteriol. 154, 1269–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller JP, Taber HW. 1989. Isolation and sequence of ctaA, a gene required for cytochrome aa3 biosynthesis and sporulation in Bacillus subtilis. J. Bacteriol. 171, 4967–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 10.1093/nar/gkg563 (doi:10.1093/nar/gkg563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitmore L, Wallace BA. 2004. DichroWeb, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 10.1093/nar/gkh371 (doi:10.1093/nar/gkh371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 10.1002/bip.20853 (doi:10.1002/bip.20853) [DOI] [PubMed] [Google Scholar]
- 34.Compton LA, Johnson WC., Jr 1986. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155, 155–167 10.1016/0003-2697(86)90241-1 (doi:10.1016/0003-2697(86)90241-1) [DOI] [PubMed] [Google Scholar]
- 35.Manavalan P, Johnson WC., Jr 1987. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167, 76–85 10.1016/0003-2697(87)90135-7 (doi:10.1016/0003-2697(87)90135-7) [DOI] [PubMed] [Google Scholar]
- 36.Sreerama N, Venyaminov SY, Woody RW. 2000. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochemistry 287, 243–251 10.1006/abio.2000.4879 (doi:10.1006/abio.2000.4879) [DOI] [PubMed] [Google Scholar]
- 37.Provencher SW, Glockner J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 10.1021/bi00504a006 (doi:10.1021/bi00504a006) [DOI] [PubMed] [Google Scholar]
- 38.Sreerama N, Woody RW. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 10.1006/abio.2000.4880 (doi:10.1006/abio.2000.4880) [DOI] [PubMed] [Google Scholar]
- 39.Lees JG, Miles AJ, Wien F, Wallace BA. 2006. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962 10.1093/bioinformatics/btl327 (doi:10.1093/bioinformatics/btl327) [DOI] [PubMed] [Google Scholar]
- 40.Holmgren A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 41.Ellman GL. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 10.1016/0003-9861(59)90090-6 (doi:10.1016/0003-9861(59)90090-6) [DOI] [PubMed] [Google Scholar]
- 42.Siluvai GS, Mayfield M, Nilges MJ, Debeer George S, Blackburn NJ. 2010. Anatomy of a red copper center: spectroscopic identification and reactivity of the copper centers of Bacillus subtilis Sco and its Cys-to-Ala variants. J. Am. Chem. Soc. 132, 5215–5226 10.1021/ja910759v (doi:10.1021/ja910759v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cawthorn TR, Poulsen BE, Davidson DE, Andrews D, Hill BC. 2009. Probing the kinetics and thermodynamics of copper(II) binding to Bacillus subtilis Sco, a protein involved in the assembly of the Cu(A) center of cytochrome c oxidase. Biochemistry 48, 4448–4454 10.1021/bi802288m (doi:10.1021/bi802288m) [DOI] [PubMed] [Google Scholar]
- 44.Calabrese L, Carbonaro M, Musci G. 1988. Chicken ceruloplasmin. Evidence in support of a trinuclear cluster involving type 2 and 3 copper centers. J. Biol. Chem. 263, 6480–6483 [PubMed] [Google Scholar]
- 45.Malone SA, Lewin A, Kilic MA, Svistunenko DA, Cooper CE, Wilson MT, Le Brun NE, Spiro S, Moore GR. 2004. Protein-template-driven formation of polynuclear iron species. J. Am. Chem. Soc. 126, 496–504 10.1021/ja036483z (doi:10.1021/ja036483z) [DOI] [PubMed] [Google Scholar]
- 46.Ludwig P, Layloff T, Adams RN. 1964. Solvent effects on hyperfine coupling constants in electron paramagnetic resonance spectra. J. Am. Chem. Soc. 86, 4568–4573 10.1021/ja01075a011 (doi:10.1021/ja01075a011) [DOI] [Google Scholar]
- 47.Horng YC, Leary SC, Cobine PA, Young FB, George GN, Shoubridge EA, Winge DR. 2005. Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280, 34 113–34 122 10.1074/jbc.M506801200 (doi:10.1074/jbc.M506801200) [DOI] [PubMed] [Google Scholar]
- 48.Lieberman RL, Arciero DM, Hooper AB, Rosenzweig AC. 2001. Crystal structure of a novel red copper protein from Nitrosomonas europaea. Biochemistry 40, 5674–5681 10.1021/bi0102611 (doi:10.1021/bi0102611) [DOI] [PubMed] [Google Scholar]
- 49.Basumallick L, Sarangi R, DeBeer George S, Elmore B, Hooper AB, Hedman B, Hodgson KO, Solomon EI. 2005. Spectroscopic and density functional studies of the red copper site in nitrosocyanin: role of the protein in determining active site geometric and electronic structure. J. Am. Chem. Soc. 127, 3531–3544 10.1021/ja044412+ (doi:10.1021/ja044412+) [DOI] [PubMed] [Google Scholar]
- 50.Siluvai GS, Nakano MM, Mayfield M, Nilges MJ, Blackburn NJ. 2009. H135A controls the redox activity of the Sco copper center. Kinetic and spectroscopic studies of the His135Ala variant of Bacillus subtilis Sco. Biochemistry 48, 12 133–12 144 10.1021/bi901480g (doi:10.1021/bi901480g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson NJ, Winge DR. 2010. Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 10.1146/annurev-biochem-030409-143539 (doi:10.1146/annurev-biochem-030409-143539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson AK, Gray J, Liu A, Hosler JP. 2012. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biochim. Biophys. Acta 1817, 955–964 10.1016/j.bbabio.2012.01.003 (doi:10.1016/j.bbabio.2012.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waldron KJ, Firbank SJ, Dainty SJ, Perez-Rama M, Tottey S, Robinson NJ. 2010. Structure and metal loading of a soluble periplasm cuproprotein. J. Biol. Chem. 285, 32 504–32 511 10.1074/jbc.M110.153080 (doi:10.1074/jbc.M110.153080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett B, Hill BC. 2011. Avoiding premature oxidation during the binding of Cu(II) to a dithiolate site in BsSCO. A rapid freeze-quench EPR study. FEBS Lett. 585, 861–864 10.1016/j.febslet.2011.02.014 (doi:10.1016/j.febslet.2011.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]