Abstract
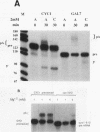
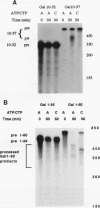
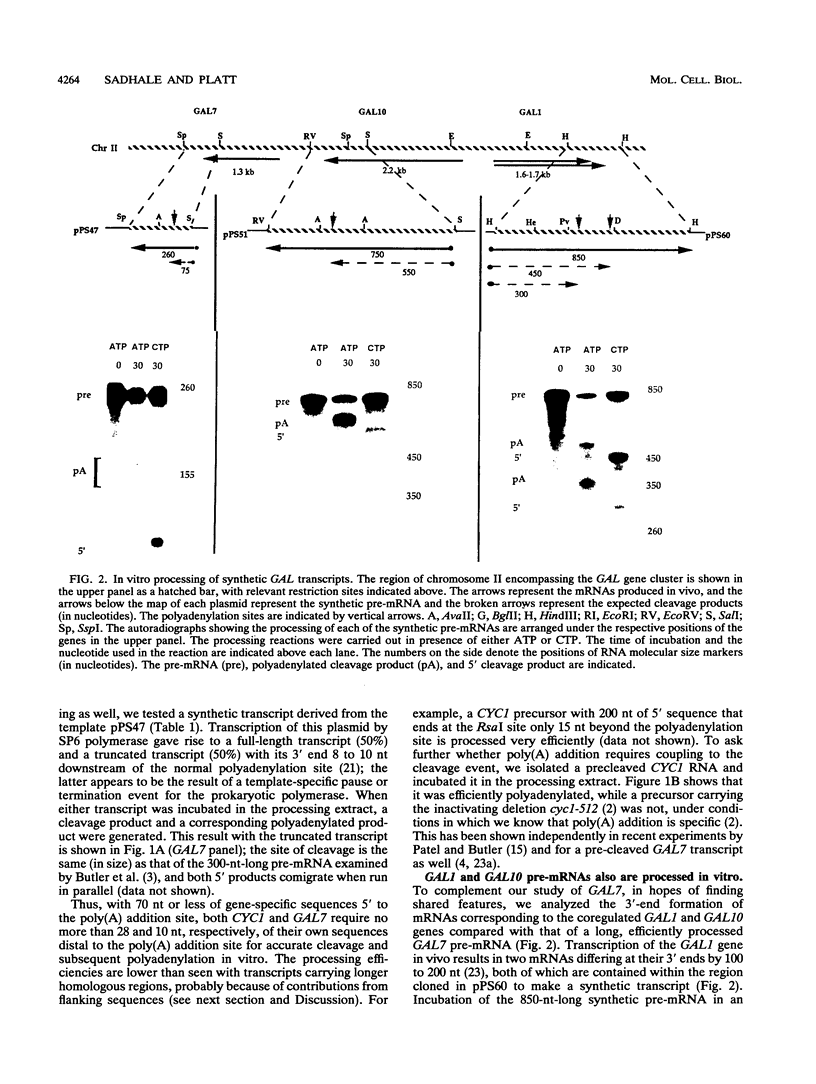
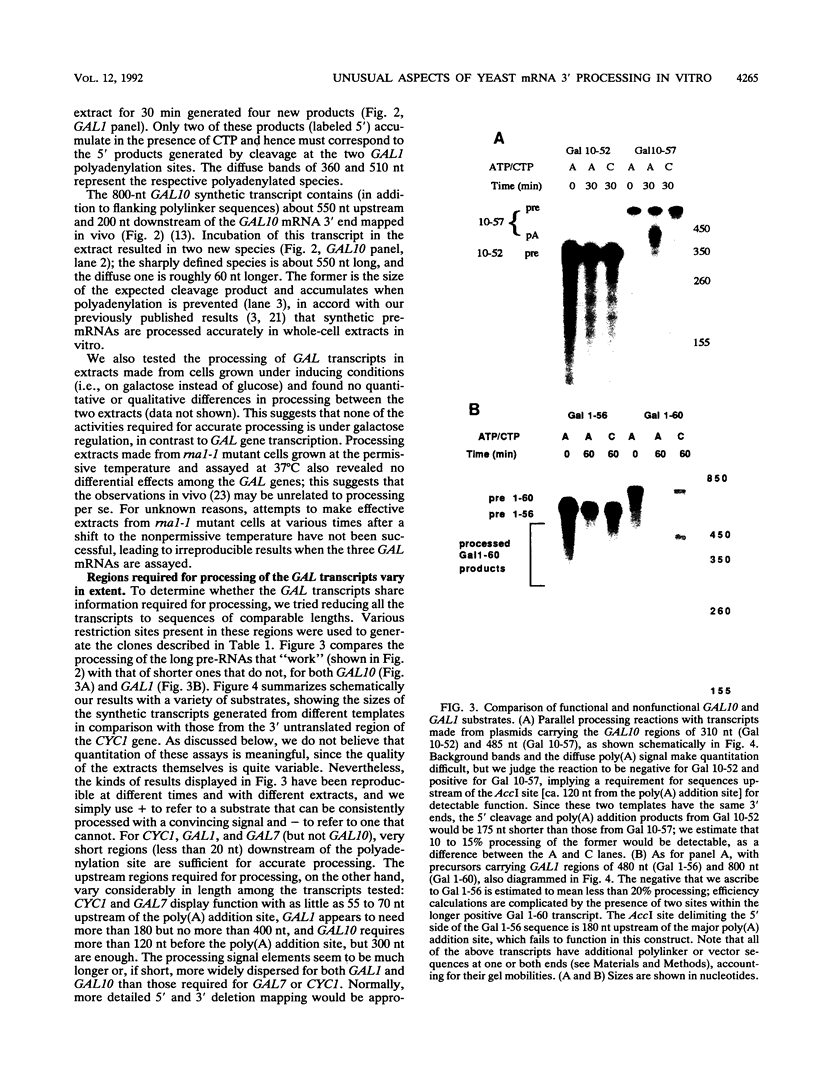
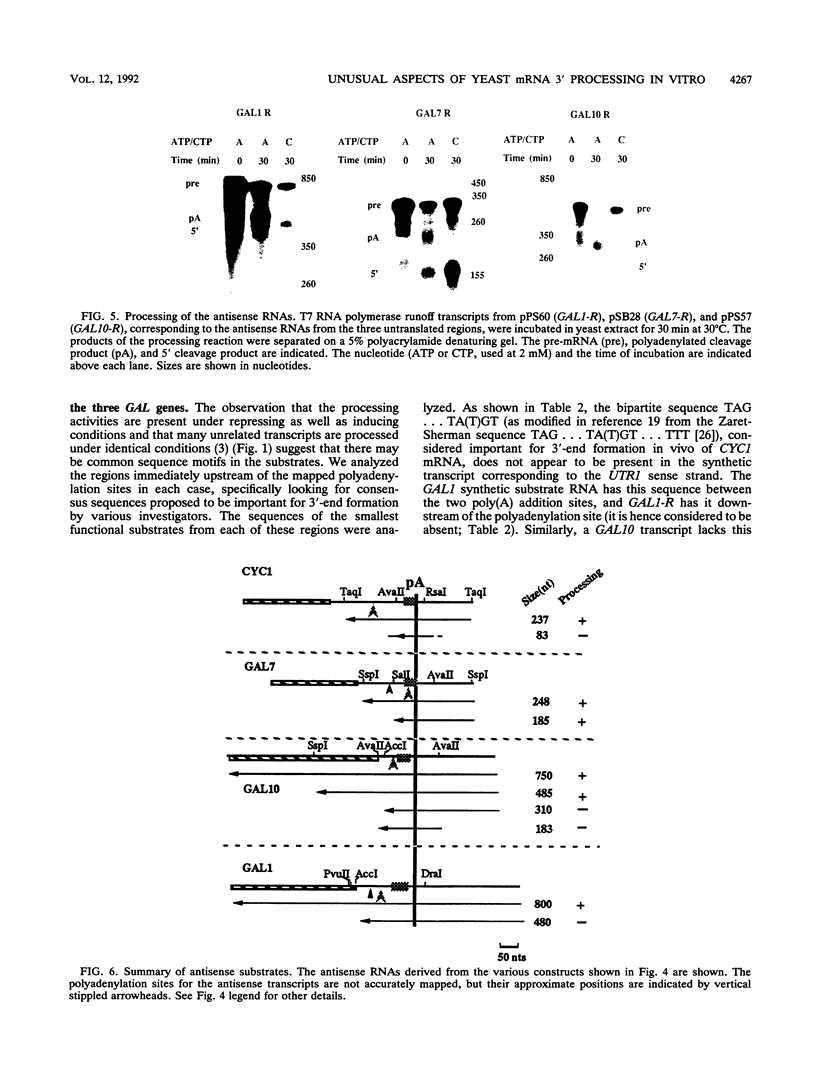
A striking feature of the 3'-end regions in polymerase II transcripts of Saccharomyces cerevisiae adjacent to their processing and polyadenylation sites is the lack of well-defined signal elements. Nonetheless, essential signals have seemed to be confined to compact regions in vivo, and we find that a short RNA with only 70 bases of GAL7 sequence upstream and 8 to 10 bases downstream of the poly(A) addition site is processed in vitro, as is an analogous CYC1 pre-RNA. Specific polyadenylation of a precleaved species further delimits the poly(A) signal and rules out obligatory coupling between cleavage and poly(A) addition. Although little proximal and even less distal sequence is required for accurate cleavage with CYC1 and GAL7, we have been unable to identify common features to which processing could be ascribed. We therefore turned to the coregulated set of genes in the galactose cluster (GAL1, GAL7, and GAL10) to assay their corresponding pre-mRNAs in vitro, in hopes of finding a common theme. By contrast to GAL7, short pre-mRNAs corresponding to GAL1 and GAL10 fail to be cleaved detectably, and only much longer transcripts are susceptible to processing. This indicates that signals, even if preserved, are more widely dispersed than the poly(A) addition site, and these results are unchanged whether extracts are from cells grown on glucose or galactose. As a further surprise, RNAs corresponding to the antisense orientation of the 3'-end regions of all three GAL genes are also effective substrates for the processing machinery in vitro. Computer analysis reveals the presence of polydisperse dyad symmetries that might account for these observations.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe A., Hiraoka Y., Fukasawa T. Signal sequence for generation of mRNA 3' end in the Saccharomyces cerevisiae GAL7 gene. EMBO J. 1990 Nov;9(11):3691–3697. doi: 10.1002/j.1460-2075.1990.tb07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S., Platt T. RNA processing generates the mature 3' end of yeast CYC1 messenger RNA in vitro. Science. 1988 Dec 2;242(4883):1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Sadhale P. P., Platt T. RNA processing in vitro produces mature 3' ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990 Jun;10(6):2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992 Aug;12(8):3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann S., Obermaier B., Vogel K., Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Sep;12(9):4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Cohen E. H. Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1515–1520. doi: 10.1128/mcb.4.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., Sadhale P., Platt T., Proudfoot N. Homologous mRNA 3' end formation in fission and budding yeast. EMBO J. 1991 Nov;10(11):3503–3511. doi: 10.1002/j.1460-2075.1991.tb04914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L. E., Seiler S. H., Whoriskey J., Moore C. L. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3'-end formation. Mol Cell Biol. 1991 Apr;11(4):2004–2012. doi: 10.1128/mcb.11.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Egli C. M., Braus G. H. Different classes of polyadenylation sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3060–3069. doi: 10.1128/mcb.11.6.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Sanfaçon H., Egli C. M., Braus G. H. Different sequence elements are required for function of the cauliflower mosaic virus polyadenylation site in Saccharomyces cerevisiae compared with in plants. Mol Cell Biol. 1992 May;12(5):2322–2330. doi: 10.1128/mcb.12.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Kellermann J., Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991 Dec 12;354(6353):496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Moore C. L., Simpson S., Clements J. B. Components required for in vitro cleavage and polyadenylation of eukaryotic mRNA. Nucleic Acids Res. 1988 Jun 24;16(12):5323–5344. doi: 10.1093/nar/16.12.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Fukasawa T. Nucleotide sequence of the transcriptional initiation region of the yeast GAL7 gene. Nucleic Acids Res. 1983 Dec 20;11(24):8555–8568. doi: 10.1093/nar/11.24.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Mutational analysis of a yeast transcriptional terminator. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4097–4101. doi: 10.1073/pnas.86.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Butler J. S. Conditional defect in mRNA 3' end processing caused by a mutation in the gene for poly(A) polymerase. Mol Cell Biol. 1992 Jul;12(7):3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Ruohola H., Baker S. M., Parker R., Platt T. Orientation-dependent function of a short CYC1 DNA fragment in directing mRNA 3' end formation in yeast. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5041–5045. doi: 10.1073/pnas.85.14.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Li W. Z., Hampsey D. M., Zaret K. S., Sherman F. Distinct cis-acting signals enhance 3' endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Sherman F. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8348–8352. doi: 10.1073/pnas.86.21.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhale P. P., Sapolsky R., Davis R. W., Butler J. S., Platt T. Polymerase chain reaction mapping of yeast GAL7 mRNA polyadenylation sites demonstrates that 3' end processing in vitro faithfully reproduces the 3' ends observed in vivo. Nucleic Acids Res. 1991 Jul 11;19(13):3683–3688. doi: 10.1093/nar/19.13.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Sapolsky R. J., Davis R. W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Yu K., Elder R. T. Some of the signals for 3'-end formation in transcription of the Saccharomyces cerevisiae Ty-D15 element are immediately downstream of the initiation site. Mol Cell Biol. 1989 Jun;9(6):2431–2444. doi: 10.1128/mcb.9.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3'-end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988 Apr 25;263(12):5780–5788. [PubMed] [Google Scholar]