Summary
Deleted in colorectal cancer (DCC) is a receptor for the axon guidance cues netrin-1 and draxin. The interactions between these guidance cues and DCC play a key role in the development of the nervous system. In the present study, we reveal the crystal structure of the N-terminal four Ig-like domains of DCC. The molecule folds into a horseshoe-like configuration. We demonstrate that this horseshoe conformation of DCC is required for guidance-cue-mediated axonal attraction. Structure-based mutations that disrupt the DCC horseshoe indeed impair its function. A comparison of the DCC horseshoe with previously described horseshoe structures has revealed striking conserved structural features and important sequence signatures. Using these signatures, a genome-wide search allows us to predict the N-terminal horseshoe arrangement in a number of other cell surface receptors, nearly all of which function in the nervous system. The N-terminal horseshoe appears to be evolutionally selected as a platform for neural receptors.
Key words: Axon guidance cue, Crystal structure, Genome-wide search, Immunoglobulin superfamily, Neural receptor
Introduction
During development, neuronal axons are guided along defined pathways by both attractive and repulsive cues in the extracellular environment. Diffusible chemoattractants attract axons to their targets, whereas repulsive guidance cues generate exclusion zones that axons avoid (Keynes and Cook, 1995; Tessier-Lavigne, 2002-2003). Different guidance cue receptors on the surface of a neuron's growth cone continually explore the environment by interacting with different guidance cues, thereby allowing the axon to correctly navigate the right trajectory among many possible routes. Remarkably, only a relatively small number of highly conserved guidance cue families and their interacting receptors are used to ensure precise wiring of the nervous system. Elaborate regulatory mechanism is apparently required to build and maintain the proper interactions of this neural network (Dickson, 2002). For example, for axon guidance during midline crossing of commissural axons, the interactions between the guidance cue netrin and its receptor DCC (deleted in colorectal cancer) mediate attractive response of the commissural axons towards the midline at the pre-crossing stage in neural development. By contrast, the interactions of the guidance cue slit and its receptor Robo (roundabout) play an important role in the repulsive response of commissural axons after the midline crossing. The attractive response during the pre-crossing stage and the repulsive response during the post-crossing stage are both strictly regulated (Yang et al., 2009). Intriguingly, netrin family members also interact with other receptors, including UNC5 (Ackerman et al., 1997; Leonardo et al., 1997), neogenin (Srinivasan et al., 2003; Wilson and Key, 2006) and DSCAM (Liu et al., 2009; Ly et al., 2008), demonstrating the complexity of the regulatory networks in the axon guidance process. Moreover, the presence of UNC5 is sufficient to convert DCC-mediated attraction to repulsion, which clearly manifests the exquisite nature of this system (Hong et al., 1999). Another developmentally important guidance cue, draxin, has recently been characterized. It shares no homology with any other known guidance cues (Islam et al., 2009). Draxin also functions through netrin receptor DCC. Therefore DCC is regarded as a convergent receptor for netrin and draxin in axon growth and guidance (Ahmed et al., 2011).
DCC was first identified in 1990 and was proposed as a putative tumor suppressor gene (Fearon et al., 1990). Expressed on spinal commissural axons, DCC was later established as a receptor for netrin-1, a neuronal guidance cue involved in determining the direction and extent of cell migration and axonal outgrowth in the developing nervous system (Chan et al., 1996; Keino-Masu et al., 1996). DCC has been proposed as a member of the ‘dependence receptor’ family (Mehlen et al., 1998). In the presence of corresponding ligands, these receptors transduce signals for cell differentiation, proliferation or migration, whereas in the absence of appropriate ligands, they may promote apoptosis. In this way, DCC might also be regarded as a tumor suppressor due to its pro-apoptotic functionality in the absence of netrin-1. In the presence of netrin-1, DCC has a dual function, first as an oncoprotein that promotes cell survival, and second as an axon guidance cue receptor (Arakawa, 2004). Using recently created mouse model, it was shown that DCC indeed functions as a tumor suppressor through its ability to trigger tumor cell apoptosis (Castets et al., 2012).
DCC is a type I transmembrane glycoprotein of the immunoglobulin superfamily (IgSF). It consists of four Ig-like domains, followed by six fibronectin type III (FNIII) domains with a single transmembrane segment and a cytoplasmic domain (Fearon et al., 1990). Netrin-1 binds to DCC's membrane-proximal FNIII domains (Geisbrecht et al., 2003; Kruger et al., 2004), whereas draxin interacts with DCC's N-terminal Ig-like region with subnanomolar affinity (Ahmed et al., 2011). NMR structures are available for each of DCC' six FNIII domains in the protein databank, by contrast, there is no structural information about the Ig domains of DCC.
We report here the crystal structure of the N-terminal four Ig-like domains. The four domains fold into a horseshoe-like configuration. Our functional data show that the integrity of DCC's N-terminal horseshoe-like configuration is required for the axonal attraction mediated by netrin-1 and draxin. Mutations that disrupt this horseshoe configuration do impair DCC function.
The horseshoe of DCC is similar to previously published structures of the N-terminal four Ig-like domains of three other neural receptors, chicken axonin-1/TAG-1 (Freigang et al., 2000), Drosophila Dscam (Meijers et al., 2007) and neurofascin (Liu et al., 2011). The conserved structural features observed in these four neural receptors have allowed us to unravel the sequence signature of the horseshoe configuration that may exist in other receptors in IgSF. A genome-wide search in human genome using the signature sequence identified has come up with 23 IgSF proteins predicted to have a horseshoe structural unit at their N-terminus. Interestingly among these 23 proteins, 22 have been demonstrated to function as neural receptors, although not necessary always exclusively. The horseshoe may therefore provide a good structural platform for these neural receptors to be engaged in homophilic as well as heterophilic interactions for neural development. The similar search has also been carried out with D. melanogaster and C. elegans and further demonstrated that the N-terminal horseshoe of these IgSF members appears evolutionally selected as a platform for the neural receptors to perform biological function.
Results
Overall structure
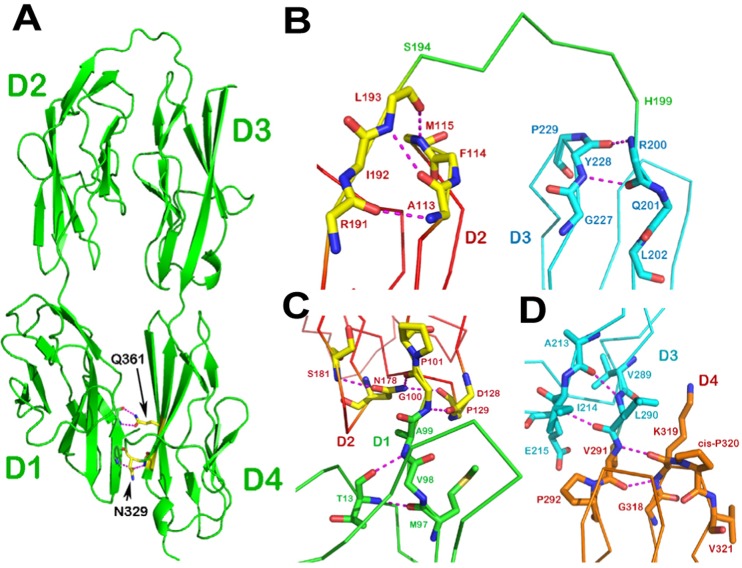
The N-terminal 383-residue of rat DCC fragment was expressed in the baculovirus system and purified to homogeneity. The crystal structure of this fragment was determined using a single-wavelength anomalous dispersion (SAD) data set collected with the PtCl4-soaked crystal at the Argonne National Laboratories. The structure consists of four Ig-like domains. The quaternary folding of the four domains results in a horseshoe-like configuration as shown in Fig. 1A, which involves the first two domains D1 and D2 bending over to contact the following two domains, D4 and D3, respectively. In this way, the structure can be viewed as having two compact modules, D2/D3 and D1/D4. The domains D3 and D4 line up relatively straight. By contrast, there is an obtuse angle (about 140°) between D1 and D2, leading to a crooked arrangement of the D1–D2 junction (Fig. 1A).
Fig. 1.
Structure of DCC horseshoe. (A) Ribbon drawing of the crystal structure of the DCC N-terminal four Ig-like domains. The molecule folds into a horseshoe configuration with a six-residue linker between domains D2 and D3. Also shown is how the conserved Asn329 and Gln361 of D4 form hydrogen bonds to the main-chain of D1 to create a specific D1/D4 interface, which defines the unique shape of the horseshoe. (B) The D2–D3 junction. At the C-terminus of D2 (in red) the last residue of D2, Leu193, participates in a pair of hydrogen bonds to Phe114 and Met115. At the N-terminus of D3 (in cyan) the first residue Arg200 is involved in a main-chain hydrogen bond with Tyr228. This clearly defines a six-residue linker (in green) from Ser194 to His199. (C) The D1–D2 junction. There is no linker present here. The last D1 residue (Ala99; in green) is still located in a part of the β sheet. The first D2 residue (Gly100; in red) is also an integrated part of D2 as it engages in a complicated hydrogen bond network. (D) The D3–D4 junction. There is no linker between these two domains either. The last D3 residue (Leu290; in cyan) is involved in a β sheet hydrogen bond network, whereas the first D4 residue (Val291; in orange) forms two main-chain hydrogen bonds with Lys319, which is on the BC loop next to the cis-Pro320. This kind of junction is commonly seen in many IgSF structures with two abutting Ig-like domains (Wang and Springer, 1998).
Homophilic interactions have been observed in crystal structures of neuronal adhesion molecules (Freigang et al., 2000; Liu et al., 2011; Meijers et al., 2007). In particular, these homophilic interactions have been proved physiologically relevant in Dscam (Meijers et al., 2007). By contrast, this is not the case for DCC. There are no similarly significant homophilic contacts in crystal lattice of DCC. Size exclusion chromatography/multi-angle scattering (SEC/MALS) analysis confirms that this 383-residue molecule is monomeric in solution (supplementary material Fig. S1). This establishes that horseshoe-shaped DCC N-terminus is not involved in homodimeric adhesion, but instead performs its biological function by interacting with guidance cues heterophilically as reported below.
The most remarkable structural features that contribute to the horseshoe configuration are the distinctive inter-domain junctions between the adjacent domains. A 6-residue linker between D2 and D3 facilitates the bend-over of D1–D2 onto D3–D4. The linker encompassing a segment of Ser194-His199 is readily noticeable (Fig. 1B). In contrast, the junctions of D1–D2 (Fig. 1C) and D3–D4 (Fig. 1D) completely lack a linker. In these two cases, the intra-domain hydrogen-bond patterns clearly indicate that the C-terminal domain immediately follows the N-terminal preceding domain without a single residue intervention. The buried surface area upon D1–D2 touching D3–D4 is 2174 Å2, among which 892 Å2 is contributed by hydrophobic residues. This implies that the horseshoe conformation is a relatively stable structure, hence the most likely functional unit.
A similar horseshoe configuration was first discovered in the X-ray structure of hemolin, an insect cell-surface receptor (Su et al., 1998). Subsequently, two neural receptors have been found to adopt a similar horseshoe configuration at their N-terminus. One is axonin-1/TAG-1, a neural adhesion molecule mediating trans-homophilic and cis-heterophilic interactions with other neural receptor (Freigang et al., 2000). Another one is Dscam, the Drosophila homolog of human DSCAM (Down syndrome cell adhesion molecule) (Schmucker et al., 2000). This 16-domain neural receptor plays a key role in neural wiring through homophilic binding involving its N-terminal horseshoe (Meijers et al., 2007) and the seventh Ig-like domain (Sawaya et al., 2008). Supplementary material Fig. S2 gives the overlay of structures of DCC, Dscam, hemolin and axonin. During the preparation of this manuscript, one more horseshoe structure has been published. This is the L1 family member neurofascin, which plays a role in axon growth and fasciculation (Liu et al., 2011). The horseshoe conformation of neurofascin was in agreement with our prediction as described below.
The finding of a horseshoe configuration shared by DCC and other neural receptors has prompted us to consider the following questions. (i) What common structural determinants constitute a horseshoe configuration? (ii) Using sequences alone, can we predict whether there are other receptors that similarly have an N-terminal horseshoe arrangement? (iii) What is the physiological relevance of the horseshoe configuration located at the receptor's N-terminus?
The horseshoe formation: the linker requirement
Considering the first two questions, the most rational explanation is that a linker exists between D2 and D3, a span that is 5–7 residues long for the four known horseshoe structures. Structural scrutiny of DCC and other IgSF members has made it possible to predict the likely existence of a linker for other neural receptors.
Similar to DCC, a number of neural receptors belong to the IgSF. Furthermore, Ig-like domains are generally located near the membrane-distal N-terminus for these type-I transmembrane proteins (Shapiro et al., 2007). Using the structure of DCC as a reference, we have compiled a structure-based local sequence alignment table, which additionally contains Dscam, axonin, neurofascin and hemolin with known structures (Fig. 2). For comparative purpose, the upper panel represents the transition from D3 to D4 without a linker, and the lower panel is its counterpart from D2 to D3 with a linker.
Fig. 2.
A comparative sequence alignment of peptide sequences containing D3–D4 transition (upper panel) and the D2–D3 transition (middle panel) shows the existence of a linker in D2–D3. The structure-based alignment was made by Dali pairwise comparison. Secondary structural elements are marked in accordance with the structure of DCC. A notable feature is the β turns between the F and G strands of the preceding domain and those between the A and B strands of the following domain. At the AB turn in particular there is a conserved Gly (shaded in orange), whereas N-terminal to the FG turn is a conserved Asn, which forms a complicated hydrogen bond network, exemplified by Asn178 shown in Fig. 1C. The end of the preceding domain can be defined by the alternate hydrophobic (shaded in gray) and hydrophilic residues at the C-terminus of the G strand. All of these make domain size at this area much less variable and the domain boundary easy to identify. In the D3–D4 transition, the interval between the conserved Cys (shaded in yellow) on the F strand of D3 and the Cys (shaded in yellow) on the B strand of D4 is 41–43 residues. By contrast, in the D2–D3 transition, this interval is 48–53 residues, which is indicative of a linker. Included at the very bottom of the alignment is the transition between D1 and D2 of UNC5 (bottom panel), which suggests a bend-over of D1/D2 with a five-residue linker.
Ig-like domains consist of a relatively fixed β-strand framework. The domain size variation stems from the “extra” β-strands at the domain's edge and the varying length of connecting loops between strands (Chothia et al., 1998; Wang and Springer, 1998). Many of Ig-like domains in cell surface receptors belong to the so-called I-set Ig-like domain, which is a truncated variable Ig-like domain (Harpaz and Chothia, 1994). The AB and FG loops of the I-set Ig-like domains of DCC, Dscam, neurofascin, axonin and hemolin are actually just a β turn with no length variation (Fig. 2). Typically the AB loop often has a Gly (Fig. 2) at the turn. Although minor, the length of the G strand itself may have some variation, as shown in Fig. 2 for D2. This makes these Ig domains to have a much less variable distance from the conserved Cys on the preceding domain's F strand to the conserved Cys on the following domain's B strand, as seen in Fig. 2 for the transition from domain 3 to domain 4 (with an interval of 41–43 residues). Therefore, the above description validates a method for predicting the existence of a linker for this I-set of Ig-like domain by counting the interval length between these two Cys, which should be approximately fewer than 45 residues. If the count exceeds 46 (like 48–53 residues from D2 to D3 in Fig. 2), the likelihood is that a linker is present.
A linker can be precisely located based on the domain boundary. A specifically conserved structural feature helps to define the end of the preceding domain and the beginning of the following domain. In Fig. 2, the gray-shaded hydrophobic residues are positioned near the end of G or G′ strand in a regular β sheet CFGA′. Based on their specific position at the middle of the β sheet, the last 5 residues of the G (or G′) strand must comply with a typical alternate hydrophobic (buried residues) and hydrophilic (exposed residues) pattern, as seen in Fig. 2. This establishes the rule of predicting the end of the Ig domain. A 5–7-residue linker segment should follow the typical 5-residue alternate hydrophobic and hydrophilic pattern at the end of domain D2. The following domain often begins one or two residues before a conserved Pro. This is because the residue N-terminal to this Pro usually forms two main-chain hydrogen bonds with a residue on the BC loop of the same domain (Wang and Springer, 1998). This can be better seen in Fig. 1D. The V291 N-terminal to the conserved P292 forms two main-chain hydrogen-bonds with K319 on BC loop. V291 defines the beginning of domain D4. In retrospect, when we started to design the constructs for structural studies on Dscam (Meijers et al., 2007) and DCC, the prediction that a linker would be present at the D2–D3 junction in both of these two receptors was indeed correct.
The horseshoe formation: the role of a conserved D1/D4 interface
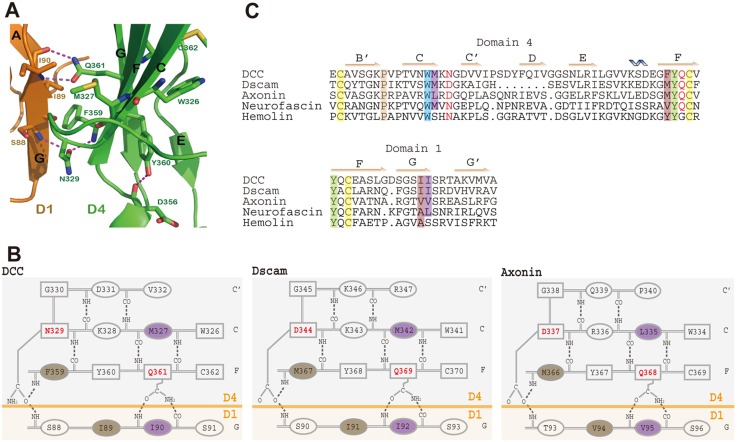
A 5–7-residue linker region in the D2–D3 junction is the essential component necessary for the bend-over of D1–D2 onto D3–D4, resulting in a horseshoe configuration. A further intriguing question is what gives rise to the unique shape of the horseshoe, common not only in DCC, Dscam and axonin, but also in hemolin?
In the horseshoe conformation there are extensive interactions between D1 and D4 in the D1/D4 module as well as between D2 and D3 in the D2/D3 module. By comparing the structure of DCC with those of Dscam and axonin, a more conserved D1/D4 interface has been discovered. In DCC D1/D4 interface, a conserved Asn329-Gly330 motif forms a β-turn from the C strand to C′ strand of the D4 domain (Fig. 3B, left panel). The amide oxygen of Asn329 makes dual hydrogen bonds to the main-chain nitrogen atom of D1's Ser88 and D4's Phe359, acting as a bridge between D1 and D4 (Fig. 3A). Furthermore, the side-chain of D4's Gln361 forms two hydrogen bonds to the main-chain atoms of Ile90 in D1. These specific hydrophilic interactions bring the side-chains of Met327 and Phe359 of D4 into hydrophobic contact with that of Ile90 and Ile89 of D1, respectively, thereby stabilizing the D1/D4 interface. Dscam and axonin have a very similar pattern for the D1/D4 interface (Fig. 3B). The structure-based sequence alignment of the above-mentioned local region for DCC and other receptors is displayed in Fig. 3C, in which recently published neurofascin is also included.
Fig. 3.
The D1/D4 interface. (A) Ribbon drawing of a local area of the D1/D4 interface of DCC. A conserved Asn329 forms two hydrogen bonds with the amide group of Phe359 in D4 (in green) and the amide group of Ser88 in D1 (in orange), respectively, to bridge the two domains together. Another conserved residue of D4 (Gln361) contributes two hydrogen bonds to the main-chain of Ile90 in D1. These specific hydrophilic interactions bring the end of the G strand of the D1 dock into a precise area of D4, allowing for hydrophobic contacts between Met327 and Phe359 of D4 to Ile90 and Ile89 of D1, respectively. This creates a defined D1/D4 interface, conserved among DCC, Dscam, and axonin, and possibly in many other neural receptors predicted to have N-terminal horseshoe arrangements. (B) Schematic of D1/D4 interfaces for DCC (left-hand panel), Dscam (middle panel), and axonin (right-hand panel) with known structures. Residues in rectangles are conserved. Asn/Asp and Gln, the key residues that contribute specific hydrogen bonds, are in a bold red font. The specific hydrogen bonds Asn/Asp and Gln mediated are drawn. The contacting hydrophobic residues are shaded with the same colors. In DCC they are the Met327-Ile90 pair (shaded in purple) and the Phe359-Ile89 pair (shaded in brown). (C) Structure-based sequence alignment of the D1/D4 interface of DCC, Dscam, axonin, neurofascin and hemolin, made by Dali pairwise comparison. The secondary structural elements are marked in accordance with the structure of DCC. The key residues Asn/Asp and Gln are in a bold red font. The conserved Cys, Tyr and Trp are shaded in yellow, light green and blue, respectively. The contacting hydrophobic pairs are shaded with the same colors, purple and brown, respectively.
There is a remarkable conservation of an Asn/Asp that is two residues following a conserved Trp, resulting in a Trp-X-X-Asn/Asp motif. This Asn/Asp is located at the corner of the CC′ turn, and stabilized by the hydrogen bond network shown in Fig. 3B. The well-structured turn offers this Asn/Asp a firm base for its side-chain to bridge together the domains D1 and D4. Moreover, the conserved Gln (Fig. 3C) is positioned between a conserved Tyr and Cys in the Tyr-Gln-Cys motif on the F strand of D4. The Tyr is located in a so-called tyrosine corner (Hemmingsen et al., 1994), as shown in Fig. 3A for Tyr360. Both the Cys and Tyr are deeply buried and stabilized by the disulfide bond and the hydrogen bond, respectively, providing the sandwiched Gln with a solid base for its exposed side-chain to interact with D1. In addition, residues engaged in the kind of hydrophobic pairing described above for DCC are also quite conserved (Fig. 3). It is also worth noting that the Gln is less conserved than the Asn/Asp. Hemolin, for instance, has Gly in the place of the conserved Gln (Fig. 3C). Interestingly, although neurofascin does have the conserved Tyr-Gln-Cys motif, it is not this particular Gln but another Gln close to it that makes two hydrogen bonds to main-chain atoms in D1 (PDB code: 3P3Y).
It is important to notice in Fig. 3C that domain D1's two consecutive hydrophobic residues (Ile in magenta and Ile in brown in DCC, etc.), both of which are involved in the interaction with D4, and are located at the end of the G strand. The bridging hydrogen bond from Asn329 and two hydrogen bonds from Gln361 all point towards the G strand of D1 (Fig. 3A,B). The combined specific hydrogen bonds and the hydrophobic interaction fix this end of D1's G strand at the specific region of D4. It is this conserved D1/D4 interface that defines the unique horseshoe shape of DCC, Dscam, axonin and hemolin. We predict that the D1/D4 interface that is observed in DCC, Dscam, axonin and hemolin should also be conserved in other receptors that contain the horseshoe structure, which was soon confirmed by the newly published structure of neurofascin (Liu et al., 2011).
A genome-wide search for horseshoe structure-containing proteins
The identification of a sequence signature for the horseshoe structure in the IgSF members described above has led us to propose that this kind of structural unit may represent a common theme in some members of IgSF. To test this hypothesis, we conducted a genome-wide search in the human genome for its existence in other IgSF receptor molecules.
The search was carried out by architectural analysis in SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de/) against Homo sapiens genome. Altogether 778 IgSF proteins were identified, which is very close to previous estimates of 765 IgSF members in the human genome (Lander et al., 2001). Among them, 105 candidates containing at least four Ig-like domains were compiled into a list for further inspection. Remarkably, 97 of these proteins contain multiple concatenated Ig-like domains at their N-terminus.
Two criteria described in the previous sections were used to examine these 97 candidates. The first step was to count the interval between the cysteine on the F strand of domain 2 and the downstream conserved cysteine on the B strand of domain 3 (Fig. 2). If the count is greater than or equal to 46, a linker is likely to exist. The second step was to inspect whether in domain 4 there is a conserved Asn/Asp located in the Trp-X-X-Asn/Asp motif on the C-C′ turn and a Gln residue in the Tyr-Gln-Cys motif on the F strand (Fig. 3). Twenty-three sequences meet these two specific criteria, and are predicted to have the horseshoe structural unit at their N-terminus (Table 1). They can be divided into several subgroups. Strikingly, 22 out of the 23 proteins that are predicted to have the N-terminal horseshoe in the human genome, function in the nervous system, according to the literature not shown here. The only exception is BCAM; from known literature we could not exclude its potential function in the nervous system.
Table 1. Predicted horseshoe-containing proteins in Homo sapiens.
| Name | Gene ID (UniProt) | Interval length, conserved motif | Extracellular domain architecture |
| DCC family | |||
| DCC | UPI00001AEDC6 | 49, Q/N | 4Ig-6FNIII |
| Neogenin | Q92859 | 48, Q/N | |
| PUNC | UPI000019908F | 50, Q/N | 4Ig-2FNIII |
| NOPE | Q8TDY8 | 52, Q/N | 4Ig-5FNIII |
| Protogenin | Q2VWP7 | 50, Q/N | |
| Contactin family | |||
| CNTN1 (gp135) | Q12860 | 53, Q/N | 6Ig-4FNIII |
| CNTN2 (TAX-1) | Q02246 | 53, Q/N | |
| CNTN3 (BIG-1) | UPI00001A7974 | 52, Q/N | |
| CNTN4 (BIG-2) | UPI00001AE9A7 | 53, Q/N | |
| CNTN5 (hNB-2) | UPI000013DBD0 | 52, Q/N | |
| CNTN6 (hNB-3) | Q9UQ52 | 53, Q/N | |
| L1 family | |||
| L1 (CD171) | UPI0000D61DF3 | 54, Q/N | 6Ig-5FNIII |
| L1-like | UPI000013CF0F | 74, Q/N | |
| NrCAM | UPI0000D61CB8 | 73, Q/N | |
| Neurofascin | UPI0000D6208B | 54, Q/N | |
| DSCAM family | |||
| DSCAM | Q59GH3 | 48, Q/N | 9Ig-4FNIII-Ig-2FNIII |
| DSCAML1 | UPI00000726E2 | 49, Q/N | |
| CDON family | |||
| CDON | UPI0000228D98 | 51, Q/N | 5Ig-3FNIII |
| BOC | A6NJ30 | 52, Q/N | 4Ig-3FNIII |
| MDGA family | |||
| MDGA1 | UPI0000D61445 | 47, N | 6Ig-FNIII-MAM |
| MDGA2 | Q7Z553 | 47, N | |
| ALCAM family | |||
| ALCAM (CD166) | Q13740 | 49, D | 5IG |
| BCAM (CD239) | P50895 | 56, D | |
A similar genome-wide search was performed for D. melanogaster and C. elegans (supplementary material Table S1). Two points have been found to be interesting. First, there are substantially more IgSF members in the human genome. Although prokaryotic homologs exist, they have probably been transferred from metazoans (Bateman et al., 1996). IgSF becomes the largest superfamily in the human genome, which can be attributed to the advanced immune system that has evolved to rapidly and effectively respond to infection (Lander et al., 2001). Secondly, there are more horseshoe structures displayed by receptors in the nervous system by ratio in the human genome compared with other two species probably as a result of evolution.
The horseshoe of DCC is required for axon guidance mediated by netrin-1 or draxin
The above genome-wide search implies that the N-terminal horseshoe might be evolutionally selected as a structural platform for a specific functional purpose. Thus an intriguing question has arisen: Does the DCC's N-terminal horseshoe structure play a role in axon guidance mediated by netrin-1 and draxin? We have used guidance cue-coated beads to test the axon growth behavior of neuronal culture.
To perform axon guidance experiments, we have designed several DCC mutants with the N-terminal horseshoe presumably disrupted, based on the structural observation discussed above. These include the four-residue (Asp-Pro-Gly-Leu) deletion at the key linker region between domains D2 and D3, as well as point mutations of N329P and Q361S that are predicted to remove the hydrogen bonds discussed above and thus impair the D1–D4 interface. The reason we chose Pro to replace Asn329 is because this residue is positioned at the β turn along with a conserved Gly (Fig. 3B), and the Pro329-Gly330 pair should form a favorable β-turn motif (Richardson, 1981). Thus, we intend to remove Asn329's function in bridging domains D1/D4, while still keep the β turn integrity within the domain D4. We aim to examine whether these mutants would affect the axon guidance function of the DCC receptor.
In order to study the axon guidance function of wild type (WT) and mutant DCC molecules, we used neuronal cultures from DCC–/– mice to eliminate the effects of endogenous DCC. Since mammalian DSCAM also has axon guidance function (Liu et al., 2009; Ly et al., 2008), neuronal cultures from the dorsal horn of spinal cords of DCC–/– mice (E15) were microinjected with small interfering RNAs (siRNAs) to mouse DSCAM1-4 to knockdown the guidance effects of endogenous DSCAM. WT DCC, DCC-N329P, DCC-Q361S and DCC with the deletion of Asp-Pro-Gly-Leu at the linker2-3 (Δlinker2-3) were injected into the neurons, respectively, with DSCAM1-4 knocked down (the efficiency of all siRNAs were verified by the manufacturer Qiagen). Heparin-coated beads were soaked with recombinant netrin-1 or draxin and placed nearby the injected cells. Western blot analysis clearly demonstrates that the beads do release netrin-1 protein into media (see Materials and Methods and supplementary material Fig. S3). After 24 hours, the axon extending directions were observed and the axons growing toward the beads were counted (Fig. 4, and Materials and Methods). Among all WT DCC-injected neurons, around 50% axons extended toward the netrin-coated beads (Fig. 5A,B, left top panel). Similarly about 40–45% axons extended toward the draxin-coated beads (Fig. 6A). By contrast, in the DCC mutant-injected neurons, remarkably less axons extended toward the beads in both netrin-coated (Fig. 5B, left top panel) and draxin-coated cases (Fig. 6A), suggesting that the DCC mutants had significant reduction of axon guidance function. As a control, beads coated with PBS or BSA did not attract axons (Fig. 5B, bottom panels). The DCC mutants also showed remarkable reduction on axon outgrowth in the case of netrin-1 (Fig. 5B, right top panel).
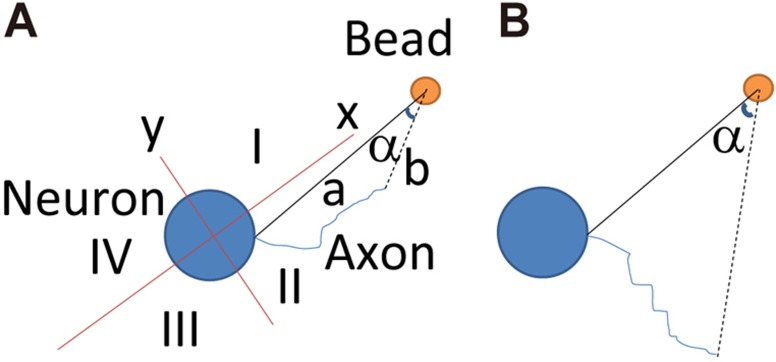
Fig. 4.
Measurement of axon guidance. From the center of the neuronal cell body to the center of the coated bead, a line was drawn as axis x. Axis y is perpendicular to x. Therefore x and y marked four quadrants: I, II, III and IV. Only the axons ending (no matter where the initiation point is) at the same side as the bead were measured (e.g. in the example diagram, quadrants I and II, but not III and IV). Then a line was drawn from the initiating point of the axon to the bead (line a). A line was drawn from the end point of the axon to the bead (line b). The angle (α) between a and b was measured. A value of α≤5 was taken as ‘towards the bead’ (A, not B, in the example diagram).
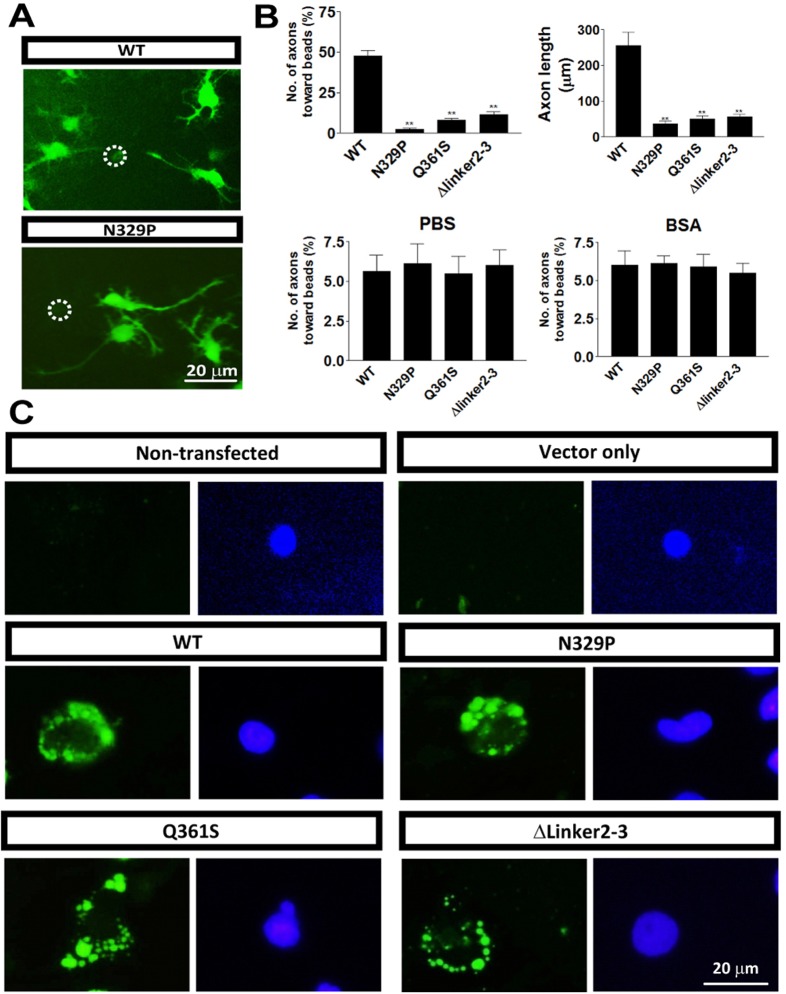
Fig. 5.
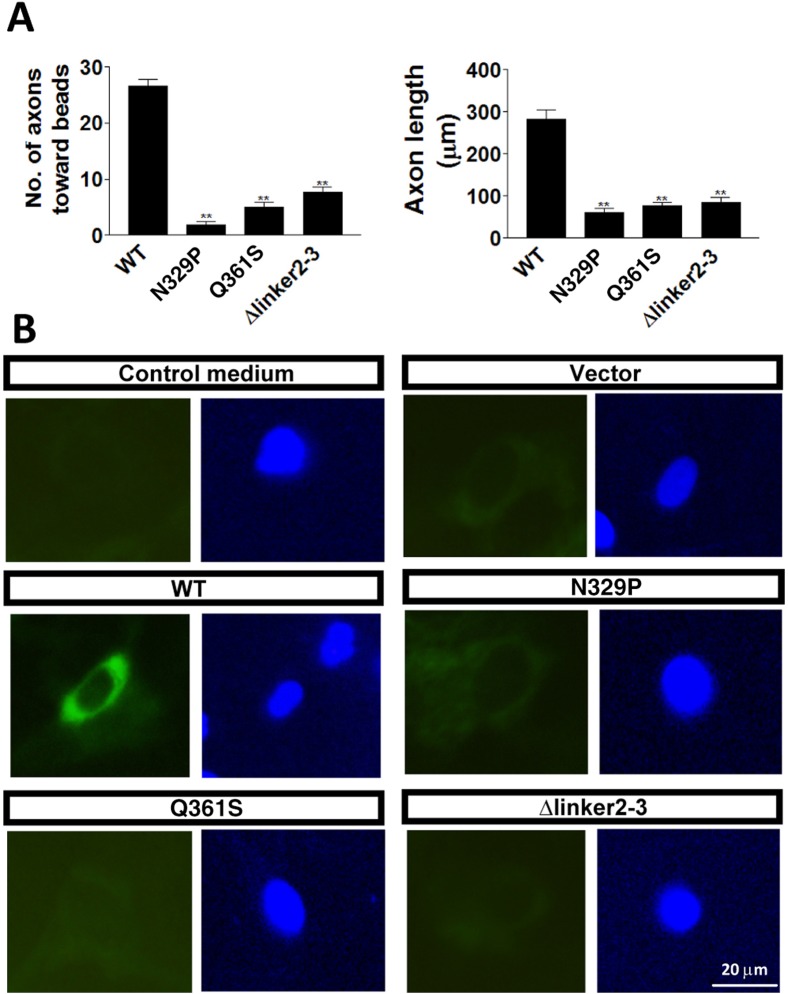
DCC mutants had reduced axon guidance induced by netrin. (A) Examples of axons growing and the netrin-coated beads (white circle). Scale bar: 20 µm. (B) Compared with WT, the numbers of axons growing towards netrin-coated beads in N329P, Q361S and Δlinker2-3 DCC mutants were significantly decreased. In addition, the length of axons was reduced in neurons microinjected with mutant DCC. Beads coated with PBS or BSA did not attract axons. Data are means±s.e.m. (n = 6). **P<0.01. (C) In COS-7 cells, netrin-binding assays showed binding ability of netrin to non-transfected cells, vector only, WT, N329P, Q361S and Δlinker2-3 DCC mutants at the cell surface. Scale bar: 20 µm.
Fig. 6.
DCC mutants had reduced axon guidance induced by draxin. (A) Compared with WT, the numbers of axons growing toward draxin-coated beads in N329P, Q361S and Δlinker2-3 DCC mutants were significantly decreased. In addition, the length of axons was reduced in neurons microinjected with mutant DCC. Data are means±s.e.m. (n = 6). **P<0.01. (B) In COS-7 cells, draxin-binding assays showed binding ability of draxin to vector only, WT, N329P, Q361S and Δlinker2-3 DCC mutants at the cell surface. The control medium was the culture medium without draxin treatment. Scale bar: 20 µm.
To exclude the possibility that the functional loss of DCC mutants was due to the loss of binding to netrin-1, netrin-binding assay was carried out following the previously published method (Kruger et al., 2004). COS-7 cells (with no endogenous DCC or DSCAM) were transfected with WT or mutant DCC constructs, respectively. Twenty-four hours later, recombinant netrin-1 was added to the culture medium to allow binding. After 24 hours of incubation, the culture medium was removed and cells were washed five times before the immunocytochemistry for netrin-1 was performed. Our data showed that the COS-7 cells transfected with vector only or cultured with control medium without netrin-1 did not have positive netrin-1 staining (Fig. 5C, top panels). By contrast, the positive netrin-1 staining appeared around the cell surface in WT and all DCC mutants (Fig. 5C, middle and bottom panels). These results have shown that although WT and DCC mutants are all expressed on cell surface and keep the same netrin-binding ability, the mutants have significantly impaired axon guidance function. Therefore, the netrin-guided function loss of these mutant DCCs is caused by the disruption of the N-terminal horseshoe of DCC.
To furthermore exclude the possibility that mutant DCC may not express as well as WT or undergo aggregation, destabilization or degradation, we performed immunoprecipitation of biotinylated cell surface DCC. The results showed that similar express levels of WT DCC, DCC-N329P, DCC-Q361S and DCC-Δlinker2-3 at 4, 8, 16 and 24 hours were observed. Our data indicate that all mutants were matured and expressed at similar levels as the WT was on the cell surface (supplementary material Fig. S4). Additionally, DCC immunostaining with DCC antibody on COS-7 cells transfected with WT, DCC-N329P, DCC-Q361S and DCC-Δlinker2-3 constructs showed that at 24 hours after transfection, all constructs were expressed (green fluorescent) at roughly similar levels at cell surface (supplementary material Fig. S4).
In the case of draxin, similar axon guidance and binding assays have been carried out. The results of axon guidance assay are very similar to that of netrin-1 (Fig. 6A). However, in contrast to netrin-1 staining experiments, the positive draxin staining appeared around the surfaces of the WT but not in all mutant DCC transfected cells (Fig. 6B). In other words, the loss of draxin-mediated axon guidance function of DCC mutants is due to the loss of the draxin binding to these DCC mutants. This agrees with the recent report showing that guidance cue draxin functions through interacting with the N-terminal region of DCC (Ahmed et al., 2011).
In conclusion, the above results have demonstrated that the N-terminal horseshoe structure is indeed required for axon guidance function of DCC mediated by guidance cues netrin-1 or draxin.
Discussion
In this work, we report a horseshoe conformation for the N-terminal four Ig-like domains of DCC. By comparing this conformation with previously published horseshoe structures we have been able to identify a sequence signature critical for this unique horseshoe conformation. Using this sequence signature to conduct a genome-wide search we have predicted the existence of an N-terminal horseshoe conformation in other neuronal receptors. From DCC, Dscam, axonin, L1 to a number of other neuronal receptors, this N-terminal horseshoe seems to be evolutionally selected, and may conceivably be of functional significance. Increasing evidences suggest the involvement of the N-terminal horseshoe in homophilic and heterophilic interactions for neural receptors to perform their functions. Drosophila Dscam provides the first convincing example of the N-terminal horseshoe involved in functionally critical homophilic interactions (Meijers et al., 2007). The functional heterophilic interactions of a horseshoe have been elegantly described in the recent structure of PTPRGCA/CNTN4, the ligand-binding carbonic anhydrase-like domain (CA) of the mouse protein tyrosine phosphatase G (PTPRG) in complex with the N-terminal four Ig-like domains of a member of the contactin family, CNTN4 (Bouyain and Watkins, 2010).
The DCC story is an intricate one. Our work shows no evidence of the functional homophilic engagement of its horseshoe. On the contrary, the scenario is that the cell surface receptor DCC heterophilically interacts with two guidance cues, the soluble matrix proteins, netrin-1 and draxin. Whereas draxin appears to bind to the N-terminal Ig-like domains of the DCC molecule, netrin-1 does not directly bind DCC horseshoe. Instead, netrin reaches DCC's membrane-proximal fibronectin type III domains FNIII-4 and/or FNIII-5. Yet, our experiments suggest that DCC's N-terminal horseshoe is functionally required in axon guidance mediated by both draxin and netrin-1. What is the specific mechanism the horseshoe of DCC utilizes to exert its observed netrin-mediated role? One speculation might be that the extracellular part of DCC receptor may fold into a relatively compact unit such that the integrity of the horseshoe, and hence the integrity of the ectodomains of the receptor might be required for the function. Drosophila Dscam, for example, has its N-terminal 8-domain fold into a S-shaped configuration with its N-terminal horseshoe closely touching domain 8 C-terminal to the horseshoe. This integrated 8-domain structure is known to be a functional unit (Sawaya et al., 2008). The large ectodomains of αβ subunits of integrins in their resting state also assume a compact conformation with the headpiece intimately contacting the legs near plasma membrane (Luo et al., 2007). It would be interesting to further explore the manner, by which DCC horseshoe structurally affects the netrin-binding FN region C-terminal to it.
In the case of draxin, we observed an axon attraction effect, but not a repulsion effect as reported (Ahmed et al., 2011). A generally accepted concept is that guidance cues are multifunctional. A single cue can either attract or repel axon. On the other hand, growth cone responses to guidance cues are quite plastic depending on intrinsic and extrinsic factors (Dickson, 2002). Since we employ a different system for our functional work than that implemented by others, this may lead to the complication. What we want to emphasize is the functional importance of having a horseshoe at the N-terminal region of the DCC molecule. We have clearly demonstrated the relationship between draxin binding to the N-terminal horseshoe and its functional outcome. The binding of draxin and the ability to perform its function are both impaired by disrupting the horseshoe structure.
Materials and Methods
Protein expression and purification
Protein was expressed in a baculovirus system. The cDNA of rat DCC Ig1-4 was cloned into pMelBac-B (Invitrogen) with an N-terminal His6-tag in the N-terminus. The vector pMelBacB has a honeybee melittin secretion signal for secreted expression. The recombinant plasmid containing the target gene was co-transfected with Bac-N-Blue DNA to Spodoptera frugiperda 9 (SF9) cells (Invitrogen). Recombinant virus was amplified in SF9 cells to prepare high-titer viral stocks.
For expression, High five cells with a density of ∼1.5×106 cells/mL were infected with the recombinant baculovirus at a multiplicity of infection between 5 and 10. At 2–3 days post infection, cells were spun down and the medium was collected for purification. The medium was filtered by a 0.45 µm CA membrane (Corning) and concentrated to 100 ml with a Stirred Cell 8400 (Millipore). The small volume of concentrated medium was dialyzed with 20 mM Tris pH 7.4, 250 mM NaCl and loaded onto a Ni-NTA column (Qiagen) equilibrated with the same buffer. The column was then washed by 10 column volumes of equilibration buffer containing 20 mM imidazole to remove impurity, and the target protein was then eluted by equilibration buffer containing 200 mM imidazole. The eluted target protein was concentrated to 0.5 ml with an Amicon Ultra centrifugal filter (Millipore) and further purified via a Superdex 200 gel filtration column (GE Healthcare) with 20 mM HEPES pH 7.5, 100 mM NaCl. The fractions containing target protein were pooled and concentrated to 15 mg/ml for crystallization.
Crystallization and structure determination
Crystals were grown at room temperature by the hanging-drop vapor diffusion method. Equal volumes of protein solution (15 mg/ml in 20 mM HEPES pH 7.5, 100 mM NaCl) and reservoir solution (100 mM Bis-tris pH 5.5, 1.0 M ammonium sulfate) were mixed and crystals appeared in 1–2 days. 3.0 M sodium malonate was used as a cryoprotectant to freeze the crystals. For preparation of heavy-atom derivative, crystals were soaked in 1 mM K2PtCl4 in cryoprotectant for 3 hours. Diffraction data were collected at beamline 24-ID-C at the Argonne National Laboratories. 3.1 Å data set collected at the peak wavelength 1.0720 Å was used for structure determination using the SAD method. The platinum positions were determined by SHELEX (Sheldrick, 2008), while the subsequent refinement of Pt positions, phasing, density modification, and automated model building were done by autoSHARP (Vonrhein et al., 2007). The entire model was then built manually by Coot (Emsley and Cowtan, 2004).
2.4 Å native data was obtained by dehydration of the crystal. Crystals were transferred to 50 µl 1.5 M ammonium sulfate and the solution was exposed to air until small salt crystals appeared around the edge. Crystals were then fished immediately and frozen in liquid nitrogen. The diffraction data was collected using wavelength 0.9794 Å at 19-ID at the Argonne National Laboratories. This 2.4 Å data set was used for structure refinement by CNS (Brünger et al., 1998), and further by PDB_Redo. The model was validated with MolProbity (Davis et al., 2007). Residues in the favored, allowed, and outlier region on Ramachandran plot are 95.6%, 4.4%, and 0%, respectively. The data collection and refinement statistics are listed in Table 2. The coordinates of the DCC structure have been deposited in the Protein Data Bank with the code 3LAF.
Table 2. Data collection and refinement statistics.
| Pt-derivative | Native | |
| Data collection | ||
| Space group | P6(5) | P6(5) |
| Cell dimensions | ||
| a, b, c (Å) | 109.13, 109.13, 130.03 | 109.38, 109.38, 129.44 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å)* | 50-3.10 (3.21-3.10) | 50-2.40 (2.46-2.40) |
| Rsym (%)* | 12.8 (61.4) | 6.0 (61.6) |
| I/σI* | 37.0 (4.3) | 33.2 (3.3) |
| Completeness (%)* | 100 (100) | 100 (100) |
| Redundancy* | 22.5 (21.9) | 11.4 (11.3) |
| Refinement | ||
| Resolution (Å) | 50-2.40 | |
| No. reflections | 32,630 | |
| Rwork/Rfree | 20.9/24.1 | |
| No. atoms | ||
| Protein | 2944 | |
| Ligand/ion | 166 | |
| Water | 218 | |
| B-factors | ||
| Protein | 31.2 | |
| Ligand/ion | 84.1 | |
| Water | 32.8 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.760 |
One crystal was used for each data set.
<?ENTCHAR ast?>: Values in parentheses are for the highest-resolution shell.
Molecular weight measurement
Size exclusion chromatography/multi-angle light scattering (SEC/MALS; Wyatt Technology Inc., Santa Barbara, CA. USA) analysis was used to measure the molecular weight of the DCC Ig1-4 fragment in solution. Protein (11.6 mg/ml, 200 µl) was loaded on a Superdex 200 column equilibrated with 20 mM HEPES pH 7.5, 100 mM sodium chloride. The molecular weight was measured according to the user manual.
Cell culture
Mouse primary neurons were cultured from the dorsal horn of the spinal cord of E15 embryo of WT or DCC−/− mice (Rigato et al., 2011), following the regulations of Peking University Animal Care and Use Committee. In brief, fresh mouse hippocampal tissues were dissociated with 0.25% trypsin (Invitrogen, Carlsbad, CA), which was then inactivated by 10% decomplemented fetal bovine serum (FBS, HyClone, Logan, UT). The mixture was triturated through pipette to make a homogenous mixture. After filtering the mixture through 70 µm sterilized filters, the flow-through was centrifuged. The pellet was then washed once by PBS and once by DMEM in Earle's balanced salt solution containing 0.225% sodium bicarbonate, 1 mM sodium pyruvate, 2 mM L-glutamine, 0.1% dextrose, 1×antibiotic Pen-Strep (all from Invitrogen, Carlsbad, CA) with 5% FBS. Cells were then plated on poly-l-lysine (Sigma, St. Louis, MO) coated plates or glass coverslips at the density of 3×106 cells/ml. Neurons were incubated at 37°C in DMEM without phenol red with 5% FBS and with 5% circulating CO2. Cytarabine was added to culture media 24 hours after plating at 10 µM to inhibit cell growth. Medium was changed every 48 hours. Cells were treated for experiments at 2 days in culture. COS-7 cells were maintained in DMEM medium.
Microinjection
Thin-walled Borosilicate glass capillaries (outer diameter = 1.0 mm, inner diameter = 0.5 mm) with microfilament (MTW100F-4, World Precision Instrument, Sarasota, FL) were pulled with a Flaming/Brown Micropipette Puller (P-97, Sutter, Novato, CA) to obtain injection needles with a tip diameter of ∼0.5 µm. Microinjections were performed in the cytosol of each cell using the Eppendorf Microinjector FemtoJet and Eppendorf Micromanipulator (Eppendorf, Hamburg, Germany). Neurons were injected with 25 fl/shot at an injection pressure of 100 hPa, a compensation pressure of 50 hPa, and an injection time of 0.1 seconds. The solutions were injected at the indicated concentrations with 100 µg/ml dextran Texas Red (DTR, MW: 3000, Molecular Probes, Eugene, OR) as a fluorescent marker to recognize the injected cells. Thirty neurons were injected in each treatment. Approximately 90% neurons survive the injections for at least 16 days (Zhang et al., 2000).
Axon guidance assay
At 2 DIV, the mouse neurons were injected with siRNAs to DSCAM1-4 and WT DCC/DCC-N329P/DCC-Q361S/DCC-Δlinker2-3 with DTR. The heparin beads coated with 100 µg/ml recombinant mouse netrin-1 or recombinant human draxin were placed at one corner of the culture dish. After 24 hours' incubation, the direction of axon growth of the injected cells (red fluorescence) towards the beads was observed. Nikon Super Resolution N-SIM Microscope (Nikon, Japan) was used for analyzing data. Each image was collected using a 40×oil immersion objective. The average fluorescence intensities were measured at various distances from the soma area using NIS-Ements AR software (Nikon, Japan). To quantify the data, 15 frames were averaged with subtraction of background fluorescence intensity. Pictures were taken from the fields of differently treated neurons. The length of axon was measured in the picture with the software. The direction of the axon was determined by a method derived from Yam et al. (Yam et al., 2009), and was demonstrated in Fig. 4.
DCC–netrin- or draxin-binding assay
COS-7 cells were transfected with WT DCC or DCC mutants (DCC-N329P, DCC-Q361S, DCC-Δlinker2-3) by Lipofectamine 2000 at 50% confluence. After 24 hours, netrin (Enzo, NO. 522-100-C010, 10 µg/ml) or draxin (R&D, NO. 6148-DR/CF, 10 µg/ml) was added to the culture medium. After 24 hours' incubation, the medium was removed and cells were washed 5 times by PBS and fixed for immunostaining with netrin-1 (Abcam, NO. ab39370) or draxin (R&D, NO. AF6148) antibody.
Immunocytochemistry
Cells were fixed with in fresh 4% paraformaldehyde, 4% sucrose in PBS for 20 minutes at room temperature and permeabilized in PBS-Triton at 4°C, blocked 10% donkey serum at room temperature, followed by incubation with anti-netrin-1 (1:500) or anti-draxin (1:1000) at 4°C for 24 hours. Cy2-conjugated donkey anti-rabbit antibody was applied as the secondary antibody. The nuclei were then staining by Hoechst 33258 (1 µg/ml, Sigma, St. Louis, MI) for 15 minutes in dark. The coverslips were mounted with Immunon™ mounting medium (Shandon, Pittsburgh, PA) onto glass slides the results were analyzed by using fluorescence microscope (Olympus BH2-RFCA, Olympus, Tokyo, Japan) with digital camera (Olympus DP70 Digital Microscope Camera, Olympus, Tokyo, Japan).
Biotinylation and immunoprecipitation
COS-7 cells were transfected with vectors containing WT-DCC, DCC-N329P, DCC-Q361S and DCC-Δlinker2-3 by Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 4, 8, 16 and 24 hours after transfection, the cells were rinsed with PBS on ice 3 times. Sulfo-NHS-LC-LC biotin (Pierce, IL) at 0.5 mg/ml was added to each well, and shaken for 30 min at 4°C. The cells were then washed with PBS, incubated in 50 mM glycine for 15 min at 4°C, washed again using PBS, and then collected with non-enzymatic dissociation solution (Sigma, St. Louis, MO) at 37°C.
The collected cells were subjected to subcellular fractionation using centrifugation first, resuspended with hypotonic lysis buffer (2 mM Tris-HCl, pH 7.4, 0.02% Triton-X100, 0.1% MgCl2), and then homogenized with Dounce homogenizer. The nuclei were removed by centrifugation twice at 400 g for 10 min at 4°C. The heavy membrane fraction was removed by centrifugation twice at 10,000 g for 10 min at 4°C, whereas the light membrane fraction was separated at 55,000 g for 90 min at 4°C. After resuspending in solubilization buffer (20 mM sodium phosphate, pH7.5, 500 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.02% sodium azide), the supernatant from light membrane fraction was incubated in NeutrAvidin-conjugated agarose (Pierce, IL) for 1 hour at 4°C. Then the pellet was centrifuged and washed 3 times. The samples were boiled in SDS loading buffer and put onto SDS-PAGE gel. The DCC were detected with DCC antibody (Abcam, MA).
Supplementary Material
Acknowledgments
We thank Yi Rao for stimulating discussion and the critic reading of this manuscript. We also thank Lorenzo Finci for critical reading of the manuscript.
Footnotes
Funding
This work was supported by the National Institutes of Health (NIH) [grant numbers HL48675 to J.-h.W. and CA107193 to J.W.]. The work was also supported by the National Program of Basic Research sponsored by the Ministry of Science and Technology of China [grant number 2009CB941301]; a National Science Foundation of China (NSFC) Major Research Grant [grant number 91132718]; a Peking University President Research Grant to Y.Z. and the Ministry of Education of China (to J.-h.W. and Y.Z.). In addition, this work is partly based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by the National Center for Research Resources at the NIH [award number RR-15301]. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences [contract number DE-AC02-06CH11357]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.111278/-/DC1
References
- Ackerman S. L., Kozak L. P., Przyborski S. A., Rund L. A., Boyer B. B., Knowles B. B. (1997). The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature 386, 838–842 10.1038/386838a0 [DOI] [PubMed] [Google Scholar]
- Ahmed G., Shinmyo Y., Ohta K., Islam S. M., Hossain M., Naser I. B., Riyadh M. A., Su Y., Zhang S., Tessier–Lavigne M.et al. (2011). Draxin inhibits axonal outgrowth through the netrin receptor DCC. J. Neurosci. 31, 14018–14023 10.1523/JNEUROSCI.0943-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H. (2004). Netrin-1 and its receptors in tumorigenesis. Nat. Rev. Cancer 4, 978–987 10.1038/nrc1504 [DOI] [PubMed] [Google Scholar]
- Bateman A., Eddy S. R., Chothia C. (1996). Members of the immunoglobulin superfamily in bacteria. Protein Sci. 5, 1939–1941 10.1002/pro.5560050923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyain S., Watkins D. J. (2010). The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc. Natl. Acad. Sci. USA 107, 2443–2448 10.1073/pnas.0911235107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse–Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S.et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- Castets M., Broutier L., Molin Y., Brevet M., Chazot G., Gadot N., Paquet A., Mazelin L., Jarrosson–Wuilleme L., Scoazec J. Y.et al. (2012). DCC constrains tumor progression via its dependence receptor activity. Nature 482, 534–537 10.1038/nature10708 [DOI] [PubMed] [Google Scholar]
- Chan S. S., Zheng H., Su M. W., Wilk R., Killeen M. T., Hedgecock E. M., Culotti J. G. (1996). UNC-40, a C. elegans homolog of DCC (Deleted in Colorectal Cancer), is required in motile cells responding to UNC-6 netrin cues. Cell 87, 187–195 10.1016/S0092-8674(00)81337-9 [DOI] [PubMed] [Google Scholar]
- Chothia C., Gelfand I., Kister A. (1998). Structural determinants in the sequences of immunoglobulin variable domain. J. Mol. Biol. 278, 457–479 10.1006/jmbi.1998.1653 [DOI] [PubMed] [Google Scholar]
- Davis I. W., Leaver–Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S.et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964 10.1126/science.1072165 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fearon E. R., Cho K. R., Nigro J. M., Kern S. E., Simons J. W., Ruppert J. M., Hamilton S. R., Preisinger A. C., Thomas G., Kinzler K. W.et al. (1990). Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 247, 49–56 10.1126/science.2294591 [DOI] [PubMed] [Google Scholar]
- Freigang J., Proba K., Leder L., Diederichs K., Sonderegger P., Welte W. (2000). The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell 101, 425–433 10.1016/S0092-8674(00)80852-1 [DOI] [PubMed] [Google Scholar]
- Geisbrecht B. V., Dowd K. A., Barfield R. W., Longo P. A., Leahy D. J. (2003). Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin. J. Biol. Chem. 278, 32561–32568 10.1074/jbc.M302943200 [DOI] [PubMed] [Google Scholar]
- Harpaz Y., Chothia C. (1994). Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238, 528–539 10.1006/jmbi.1994.1312 [DOI] [PubMed] [Google Scholar]
- Hemmingsen J. M., Gernert K. M., Richardson J. S., Richardson D. C. (1994). The tyrosine corner: a feature of most Greek key beta-barrel proteins. Protein Sci. 3, 1927–1937 10.1002/pro.5560031104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K., Hinck L., Nishiyama M., Poo M. M., Tessier–Lavigne M., Stein E. (1999). A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell 97, 927–941 10.1016/S0092-8674(00)80804-1 [DOI] [PubMed] [Google Scholar]
- Islam S. M., Shinmyo Y., Okafuji T., Su Y., Naser I. B., Ahmed G., Zhang S., Chen S., Ohta K., Kiyonari H.et al. (2009). Draxin, a repulsive guidance protein for spinal cord and forebrain commissures. Science 323, 388–393 10.1126/science.1165187 [DOI] [PubMed] [Google Scholar]
- Keino–Masu K., Masu M., Hinck L., Leonardo E. D., Chan S. S., Culotti J. G., Tessier–Lavigne M. (1996). Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell 87, 175–185 10.1016/S0092-8674(00)81336-7 [DOI] [PubMed] [Google Scholar]
- Keynes R., Cook G. M. (1995). Axon guidance molecules. Cell 83, 161–169 10.1016/0092-8674(95)90157-4 [DOI] [PubMed] [Google Scholar]
- Kruger R. P., Lee J., Li W., Guan K. L. (2004). Mapping netrin receptor binding reveals domains of Unc5 regulating its tyrosine phosphorylation. J. Neurosci. 24, 10826–10834 10.1523/JNEUROSCI.3715-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al. ; International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Leonardo E. D., Hinck L., Masu M., Keino–Masu K., Ackerman S. L., Tessier–Lavigne M. (1997). Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature 386, 833–838 10.1038/386833a0 [DOI] [PubMed] [Google Scholar]
- Liu G., Li W., Wang L., Kar A., Guan K. L., Rao Y., Wu J. Y. (2009). DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc. Natl. Acad. Sci. USA 106, 2951–2956 10.1073/pnas.0811083106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Focia P. J., He X. (2011). Homophilic adhesion mechanism of neurofascin, a member of the L1 family of neural cell adhesion molecules. J. Biol. Chem. 286, 797–805 10.1074/jbc.M110.180281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B. H., Carman C. V., Springer T. A. (2007). Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A., Nikolaev A., Suresh G., Zheng Y., Tessier–Lavigne M., Stein E. (2008). DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell 133, 1241–1254 10.1016/j.cell.2008.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P., Rabizadeh S., Snipas S. J., Assa–Munt N., Salvesen G. S., Bredesen D. E. (1998). The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature 395, 801–804 10.1038/27441 [DOI] [PubMed] [Google Scholar]
- Meijers R., Puettmann–Holgado R., Skiniotis G., Liu J. H., Walz T., Wang J. H., Schmucker D. (2007). Structural basis of Dscam isoform specificity. Nature 449, 487–491 10.1038/nature06147 [DOI] [PubMed] [Google Scholar]
- Richardson J. S. (1981). The anatomy and taxonomy of protein structure. Adv. Protein Chem. 34, 167–339 10.1016/S0065-3233(08)60520-3 [DOI] [PubMed] [Google Scholar]
- Rigato C., Buckinx R., Le–Corronc H., Rigo J. M., Legendre P. (2011). Pattern of invasion of the embryonic mouse spinal cord by microglial cells at the time of the onset of functional neuronal networks. Glia 59, 675–695 10.1002/glia.21140 [DOI] [PubMed] [Google Scholar]
- Sawaya M. R., Wojtowicz W. M., Andre I., Qian B., Wu W., Baker D., Eisenberg D., Zipursky S. L. (2008). A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell 134, 1007–1018 10.1016/j.cell.2008.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D., Clemens J. C., Shu H., Worby C. A., Xiao J., Muda M., Dixon J. E., Zipursky S. L. (2000). Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 10.1016/S0092-8674(00)80878-8 [DOI] [PubMed] [Google Scholar]
- Shapiro L., Love J., Colman D. R. (2007). Adhesion molecules in the nervous system: structural insights into function and diversity. Annu. Rev. Neurosci. 30, 451–474 10.1146/annurev.neuro.29.051605.113034 [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. (2008). A short history of SHELX. Acta Crystallogr. A 64, 112–122 10.1107/S0108767307043930 [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Strickland P., Valdes A., Shin G. C., Hinck L. (2003). Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev. Cell 4, 371–382 10.1016/S1534-5807(03)00054-6 [DOI] [PubMed] [Google Scholar]
- Su X. D., Gastinel L. N., Vaughn D. E., Faye I., Poon P., Bjorkman P. J. (1998). Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion. Science 281, 991–995 10.1126/science.281.5379.991 [DOI] [PubMed] [Google Scholar]
- Tessier–Lavigne M. (2002–2003). Wiring the brain: the logic and molecular mechanisms of axon guidance and regeneration. Harvey Lect. 98, 103–143 [PubMed] [Google Scholar]
- Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007). Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- Wang J., Springer T. A. (1998). Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev. 163, 197–215 10.1111/j.1600-065X.1998.tb01198.x [DOI] [PubMed] [Google Scholar]
- Wilson N. H., Key B. (2006). Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev. Biol. 296, 485–498 10.1016/j.ydbio.2006.06.018 [DOI] [PubMed] [Google Scholar]
- Yam P. T., Langlois S. D., Morin S., Charron F. (2009). Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 62, 349–362 10.1016/j.neuron.2009.03.022 [DOI] [PubMed] [Google Scholar]
- Yang L., Garbe D. S., Bashaw G. J. (2009). A frazzled/DCC-dependent transcriptional switch regulates midline axon guidance. Science 324, 944–947 10.1126/science.1171320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Goodyer C., LeBlanc A. (2000). Selective and protracted apoptosis in human primary neurons microinjected with active caspase-3, -6, -7, and -8. J. Neurosci. 20, 8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.