Abstract
Compared with the average transmissibility of human influenza A virus, much less attention has been paid to the potential variability in its transmissibility. We considered viral shedding as a proxy for infectiousness and explored the heterogeneity of infectiousness among patients with medically attended seasonal influenza A virus infection. The analysis revealed that viral shedding is more heterogeneous in children than in adults. The top 20% most infectious children and adults were estimated to be responsible for 89%–96% and 78%–82%, respectively, of the total infectiousness in each age group. Further investigation is required to correlate the substantial variations in viral shedding with heterogeneity in actual transmissibility.
Keywords: influenza, viral shedding, infectiousness
Superspreading, in which a small percentage of individuals in a population are responsible for a major proportion of transmissions [1], has been well documented for many infectious diseases [2]. While outbreaks of influenza with a high attack rate in a poorly ventilated environment highlighted the possibility of superspreading in influenza transmission under special settings [3, 4], the presence of superspreaders and their potential role in characterizing the transmission dynamics of influenza and how they present clinically remains poorly understood. Because of the significant global morbidity and mortality burden of influenza as a result of seasonal epidemics and occasional pandemics, it is of critical importance to clarify the individual variability of infectiousness and its clinical and epidemiological relevance, which is likely to have important public health implications for influenza management and control [5].
The amount of viral shedding has generally been regarded as a reasonable proxy measure of infectiousness [6]. Although the temporal profile of viral shedding over time in individual patients has been previously described [7–9], and age group–associated differences were observed, the existence and magnitude of interindividual variability in viral shedding within age groups among cases of naturally acquired influenza infection has yet to be reported. In this study, we attempted to quantify and analyze this heterogeneity of viral shedding among individuals with medically attended seasonal influenza A virus infection.
METHODS
Subjects
In 2008 and 2009, 2 large community-based trials studying household transmission of influenza were conducted in Hong Kong [10, 11]. Outpatients presenting within 2 days after illness onset with ≥2 signs and symptoms consistent with acute respiratory illness were recruited and tested by the QuickVue Influenza A + B test (Quidel, San Diego, CA). Subjects with a positive result of a rapid test were invited to participate in further follow-up observations involving 3 home visits over approximately 7 days. Pooled nose and throat swab specimens were collected from all subjects by study nurses regardless of the presence of respiratory symptoms and signs. Subjects kept daily symptom diaries for the duration of follow-up. Written informed consent was collected from all subjects ≥18 years of age and from parents or legal guardians of subjects <18 years old. The study was approved by the Institutional Review Board of the University of Hong Kong.
Laboratory Methods
Viral shedding was quantified by quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of the nose and throat swab specimens. The NucliSense easyMAG extraction system (bioMérieux, Boxtel, Netherlands) was used to extract nucleic acid from specimens according to the manufacturer's instructions. The Invitrogen Superscript III kit (Invitrogen) was used to prepare complementary DNA from 12 μL of extracted nucleic acid with a random primer, as described elsewhere [12].
To detect influenza A virus, 2 μL of complementary DNA was amplified in a LightCycler 2.0 system (Roche Diagnostics) containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche Diagnostics), 4.0 mmol/L MgCl2, and 0.5 mmol/L of each primer, for a total reaction volume of 20 μL. For amplification of the matrix gene of influenza A virus, the forward primer 5′-CTTCTAACCGAGGTCGAAACG-3′ and the reverse primer 5′-GGCATTTTGGACAAAKCGTCTA-3′ were used. Cycling conditions were as follows: initial denaturation at 95°C for 10 minutes, followed by 40 cycles at 95°C for 10 seconds, 60°C for 3 seconds, and 72°C for 12 seconds, with ramp rates of 20°C/seconds. At the end of the assay, PCR products were subjected to a melting-curve analysis to determine the specificity of the assay. The lower limit of detection of the RT-PCR assay was approximately 900 virus gene copies per milliliter.
Statistical Analysis
Viral shedding was plotted by time since onset of acute respiratory illness and stratified as seasonal influenza A virus subtype H1N1 (A[H1N1]) or H3N2 (A[H3N2]). Specimens with undeterminable viral shedding were treated as censored at 900 copies/mL. On the basis of previous observations of naturally acquired infections [7] and volunteer challenge studies [8], as well as previous analyses of medically attended influenza [13], we assumed that viral shedding declined approximately log-linearly over time since illness onset. We therefore fitted a random effects log-linear regression model to the viral shedding trajectories of each individual, with the random intercept representing differences between individuals at illness onset. This model permits prediction of the level of viral shedding for each individual on each day. The random effect and residual error were normally distributed around 0. For every subject, the change of the log viral shedding over time (the slope) was assumed to be identical, whereas the estimated viral shedding at illness onset (the intercept) was allowed to vary by individual.
On the assumption that infectiousness is proportional to viral shedding, variability in individual subjects’ infectiousness was quantified by the random effect of the model, and we explored heterogeneity in viral shedding by calculating the mean area under the curve. All data analysis was conducted using R, version 2.14.0 (R Foundation for Statistical Computing, Vienna, Austria), and the lmec package for linear mixed-effects models with censored responses.
RESULTS
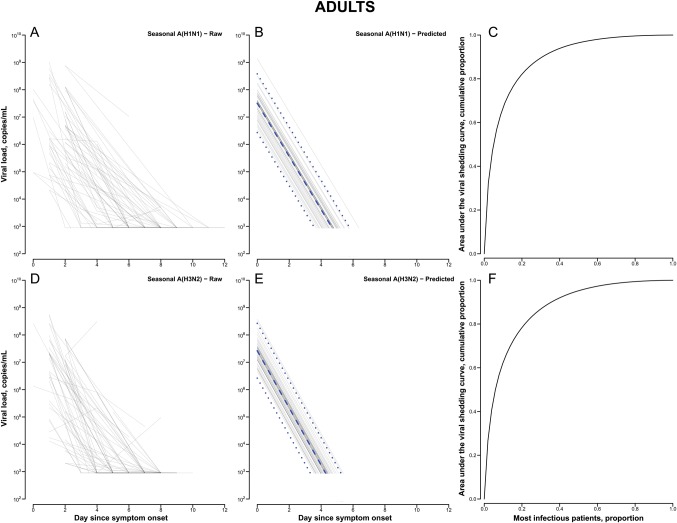
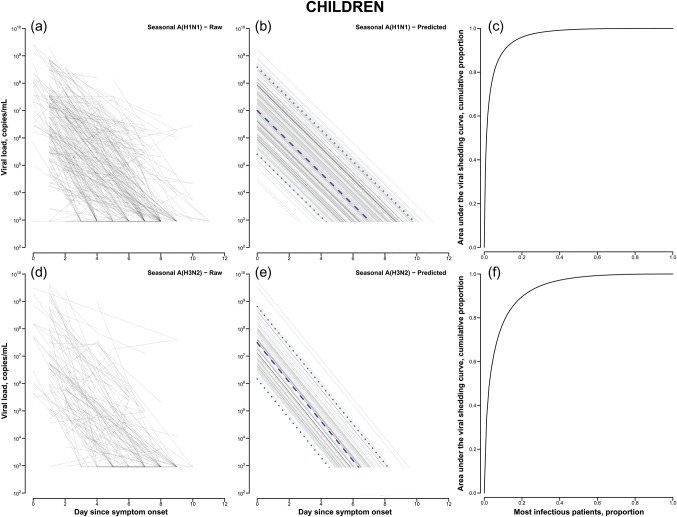
In total, 637 patients had influenza A virus detected by a rapid test between 3 January 2008 and 28 August 2009, and 204 and 130 were subsequently confirmed to have seasonal A(H1N1) and A(H3N2) infection, respectively. Among patients with seasonal A(H1N1) infection, around half (49%) were males and the mean age was 13.6 years, whereas for patients with seasonal A(H3N2) infection, 56% were males and the mean age was 21.0 years. Among these patients, the pattern of viral shedding, including the amount of virus being shed and the duration of shedding, appeared to be broadly similar between those with seasonal influenza A(H1N1) infection and those with A(H3N2) infection (Figure 1). On stratification by age, children with seasonal A(H3N2) infection appeared to shed higher quantities of virus than children with seasonal A(H1N1) infection. Compared with adults, children shed a similar amount of virus at illness onset, with a longer overall duration and slower rate of decline (Table 1).
Figure 1.
Observed and modeled seasonal influenza A virus subtypes H1N1 (A[H1N1]) and H3N2 (A[H3N2]) shedding patterns, determined by quantitative reverse transcription polymerase chain reaction analysis, and cumulative area under the viral shedding curve in adults and children. A–C, Actual (A) and predicted (B) seasonal A(H1N1) shedding patterns and cumulative viral shedding (C) in adults, by time since onset of acute respiratory illness (ARI). D–F, Actual (D) and predicted (E) seasonal A(H3N2) shedding patterns and cumulative viral shedding (F) in adults, by time since ARI onset. a–c, Actual (a) and predicted (b) seasonal A(H1N1) shedding patterns and cumulative viral shedding (c) in children, by time since ARI onset. d–f, Actual (d) and predicted (e) seasonal A(H3N2) shedding patterns and cumulative viral shedding (f) in children, by time since ARI onset. Dashed lines denote the median predicted trajectory of viral shedding, and dotted lines denote the 20th and 80th percentiles.
Table 1.
Cumulative Infectiousness Among Patients With Seasonal Influenza A Virus Infection
| Influenza A Virus Subtype | Children |
Adults |
||
|---|---|---|---|---|
| Geometric Mean (Rangea) | Top 20%b | Geometric Mean (Rangea) | Top 20%b | |
| H1N1 | 8.8 × 106 (7.9 × 105–9.9 × 107) | 0.959 | 1.9 × 107 (3.8 × 106–9.0 × 107) | 0.817 |
| H3N2 | 2.2 × 107 (3.2 × 106–1.6 × 108) | 0.891 | 1.6 × 107 (3.8 × 106–6.7 × 107) | 0.779 |
Cumulative infectiousness was calculated on the basis of the area under the curve of viral shedding.
a Data denote the range from the 20th percentile to the 80th percentile.
b Data denote the proportion of the total area under the curve for which the most infectious 20% of patients are responsible.
The results of the regression model are summarized in Supplementary Table 1, and the associated random effects are reported in Supplementary Table 2 as an estimate of the variability of the viral shedding between individuals. It consistently appeared that there was greater variability among children, compared with adults, particularly for seasonal A(H1N1) alone and for both strains combined. The cumulative infectiousness, as estimated by the total amount of viral shedding, was quantified as the area under the curve. Use of this metric revealed that the top 20% most infectious individuals were responsible for approximately 78%–82% and 89%–96% of the total infectiousness among adults and children, respectively (Table 1).
We stratified individuals into tertiles of viral shedding at illness onset and compared the prevalence of various self-reported signs or symptoms among the groups (Supplementary Table 3). None of the signs or symptoms were found to differ between the tertiles.
DISCUSSION
The patterns of viral shedding for the 2 different subtypes of seasonal influenza A virus were quite similar, with log-linear declines from illness onset to low levels typically within 4–7 days (Figure 1), as previously reported for volunteer challenge studies [8] and medically attended natural infections [9, 13]. On the assumption that shedding is a proxy of infectiousness, the higher level and longer duration of shedding in children are consistent with the notion that children are more infectious than adults. The major role of children in maintaining and propagating influenza transmission is well-known [14], but uncertainty remains over the underlying reason and, in particular, the relative importance among children of greater susceptibility to infection, more intense contact patterns, and greater infectiousness if infected. It is possible that the higher variability of viral shedding observed in children could be due to differences in exposure history against the circulating seasonal strain of influenza virus, as well as the current stage of immune development.
On the other hand, the much greater individual variability in the amount of viral shedding in children suggests that a subset of children could be substantially more infectious than others. By analyzing the total infectiousness as the area under the viral shedding curve, our results indicated that the top 20% most infectious individuals can account for around 78% and 96% of the infectiousness in adults and children, respectively. This is higher than the value previously stated by other authors, who asserted that 20% of infected individuals can be responsible for 80% of the total transmissions [5, 15]. This finding highlighted the practical importance of this heterogeneity, including its consideration in modeling studies for an improved assessment of transmissibility and the potential usefulness of any clinical indicators that may help identify these more infectious people for optimal targeting of treatment and prevention strategies. Although an infected individual's viral shedding has been hypothesized to be correlated with the severity of illness [7], this had not been demonstrated in the present study among medically attended cases when the presence or absence of symptoms was examined. Because the clinical symptomatology of a patient does not appear to be a reliable indicator of levels of virus shedding, rapid identification of the most infectious individuals in a clinical setting is difficult, and public health measures during an epidemic might need to target patients along the whole clinical spectrum in order to control transmission.
This study has a number of limitations. First, although targeting symptomatic patients in a clinical setting may represent a logical and efficient approach for recruitment, this may limit the generalizability of our results to the whole disease spectrum in the community. Additionally, it is possible that recruiting subjects on the basis of results of the QuickVue test biases the study toward patients who shed greater quantities of virus. However, our results should still complement previous data from volunteer challenge studies. Our data allowed us to look only at the pattern of viral shedding, rather than at actual transmission events, as a proxy measure for infectiousness. We assumed a direct linear relationship between viral load and infectiousness, but the exact form of the relation remains unclear. One possible area for further research would be to incorporate these viral shedding data into an analysis of the transmission of influenza virus within the household, to infer the relationship between viral shedding and infectivity. While the quantity of viral shedding is likely to be an important factor in determining the intrinsic infectiousness, secondary transmission events, or transmissibility, may also be affected by a number of other environmental and behavioral factors. Viral shedding was quantified as the number of viral gene (RNA) copies in the pooled nose and throat swab specimens, and we did not have sufficient data to investigate infectious viral shedding on the basis of the median tissue culture infective dose, which may be a closer correlate to infectiousness. Nose and throat swab specimens were pooled in the same specimen tube immediately after collection, prohibiting separate analysis of viral load in nasal swab versus throat swab specimens. With regard to the pattern of viral shedding, the single log-linear equation used in this study may be an oversimplification in capturing the actual trajectories of viral shedding after illness onset, although sensitivity analyses showed that a linear model gave a best fit to the observed pattern. Last, symptoms were self-reported, and our data allowed only for the examination of their occurrence, rather than their severity.
As a small step in further understanding the heterogeneity in transmissibility of influenza virus, our study demonstrated the substantial heterogeneity of viral shedding in children with influenza A virus infection. The findings support the previous theoretical assertion that a small proportion of individuals are responsible for most of the transmission during an epidemic. However, the degree to which these occurrences could impact transmission dynamics remains unclear.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Calvin Cheng, Jo K. M. Chan, Y. L. Ho, and Patrick Y. B. Lam, for technical support; and Rita Fung, for research support.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; contract HHSN266200700005C; ADB N01-AI-70005 [NIAID Centers for Excellence in Influenza Research and Surveillance]), the US Centers for Disease Control and Prevention (grant 1 U01 CI000439); the Research Fund for the Control of Infectious Disease, Food and Health Bureau, Government of the Hong Kong Special Administrative Region (grant HK-10-04-02); the Harvard Center for Communicable Disease Dynamics, from the National Institute of General Medical Sciences (grant U54 GM088558); and the Area of Excellence Scheme of the Hong Kong University Grants Committee (grant AoE/M-12/06). H. N. received funding support from the JST PRESTO program.
Disclaimer. The funding bodies had no role in study design, data collection and analysis, preparation of the manuscript, or the decision to publish.
Potential conflicts of interest. D. K. M. I. has received research funding from F. Hoffmann–La Roche. J. S. M. P. receives research funding from Crucell and serves as an ad hoc consultant for GlaxoSmithKline and Sanofi. B. J. C. has received research funding from MedImmune and consults for Crucell. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Stein RA. Super-spreaders in infectious diseases. Int J Infect Dis. 2011;15:e510–3. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cori A, Boelle PY, Thomas G, Leung GM, Valleron AJ. Temporal variability and social heterogeneity in disease transmission: the case of SARS in Hong Kong. PLoS Comput Biol. 2009;5:e1000471. doi: 10.1371/journal.pcbi.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moser MR, Bender TR, Margolis HS, Noble GR, Kendal AP, Ritter DG. An outbreak of influenza aboard a commercial airliner. Am J Epidemiol. 1979;110:1–6. doi: 10.1093/oxfordjournals.aje.a112781. [DOI] [PubMed] [Google Scholar]
- 4.Pestre V, Morel B, Encrenaz N, et al. Transmission by super-spreading event of pandemic A/H1N1 2009 influenza during road and train travel. Scand J Infect Dis. 2012;44:225–7. doi: 10.3109/00365548.2011.631936. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–9. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell DM. Non-pharmaceutical interventions for pandemic influenza, international measures. Emerg Infect Dis. 2006;12:81–7. doi: 10.3201/eid1201.051370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau LL, Cowling BJ, Fang VJ, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis. 2010;201:1509–16. doi: 10.1086/652241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–85. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 9.Loeb M, Singh PK, Fox J, et al. Longitudinal study of influenza molecular viral shedding in Hutterite communities. J Infect Dis. 2012;206:1078–84. doi: 10.1093/infdis/jis450. [DOI] [PubMed] [Google Scholar]
- 10.Cowling BJ, Chan KH, Fang VJ, et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med. 2009;151:437–46. doi: 10.7326/0003-4819-151-7-200910060-00142. [DOI] [PubMed] [Google Scholar]
- 11.Cowling BJ, Chan KH, Fang VJ, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010;362:2175–84. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9:628–33. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip DK, Schutten M, Fang VJ, et al. Validation of self-swab for virologic confirmation of influenza virus infections in a community setting. J Infect Dis. 2012;205:631–4. doi: 10.1093/infdis/jir803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monto AS. Epidemiology of viral respiratory infections. Am J Med. 2002;112(Suppl 6A):4S–12. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 15.Woolhouse ME, Dye C, Etard JF, et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc Natl Acad Sci U S A. 1997;94:338–42. doi: 10.1073/pnas.94.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.