Abstract
The retinal determination gene network comprises a collection of transcription factors that respond to multiple signaling inputs to direct Drosophila eye development. Previous genetic studies have shown that nemo (nmo), a gene encoding a proline-directed serine/threonine kinase, can promote retinal specification through interactions with the retinal determination gene network, although the molecular point of cross-talk was not defined. Here, we report that the Nemo kinase positively and directly regulates Eyes absent (Eya). Genetic assays show that Nmo catalytic activity enhances Eya-mediated ectopic eye formation and potentiates induction of the Eya-Sine oculis (So) transcriptional targets dachshund and lozenge. Biochemical analyses demonstrate that Nmo forms a complex with and phosphorylates Eya at two consensus mitogen-activated protein kinase (MAPK) phosphorylation sites. These same sites appear crucial for Nmo-mediated activation of Eya function in vivo. Thus, we propose that Nmo phosphorylation of Eya potentiates its transactivation function to enhance transcription of Eya-So target genes during eye specification and development.
Keywords: Eye development, Transcription factor, Cell fate, NLK
Introduction
Generation of cellular diversity in a developing organism depends on coordinated cell proliferation, differentiation, migration and morphogenesis. Dynamically controlled transcriptional programs downstream of multiple signal transduction pathways produce the specific patterns of gene expression that define unique cell types and functions. The Retinal Determination (RD) gene network, a collection of conserved transcription factors named for their essential roles in eye development in Drosophila, presents a useful model to study how input from multiple signaling pathways can modify the function of a transcriptional network to regulate specific developmental decisions.
In Drosophila, the RD network is both necessary and sufficient for eye specification. Loss of RD genes in the developing eye disk results in loss or reduction in size of the adult eye, while their misexpression in non-retinal tissues can produce ectopic eyes (Bonini et al., 1993; Czerny et al., 1999; Mardon et al., 1994; Seimiya and Gehring, 2000; Shen and Mardon, 1997). The core components of the network form a cascade of transcriptional regulation where the PAX6 homolog Eyeless (Ey) activates expression of Eyes absent (Eya) and the SIX family member Sine oculis (So), which form a bipartite transcriptional complex and drive expression of Dachshund (Dac) (Chen et al., 1997; Halder et al., 1998; Pignoni et al., 1997; Shen and Mardon, 1997). However the flow of transcriptional induction is not solely linear, as downstream members can also activate expression of upstream RD genes, thereby amplifying network output; because of these positive feedback loops, overexpression of downstream genes such as Eya or Dac can activate the entire RD circuitry to a level sufficient for driving ectopic eye formation.
The core elements of the Drosophila RD network are deployed at multiple stages of eye development. During the second instar larval stage, division of the eye-antennal imaginal disk into eye or antennal compartments occurs via downregulation of Ey in the anterior antennal region (Kenyon et al., 2003). In the third instar, Ey deploys the rest of the RD network by inducing expression of Eya, So and Dac. Their expression is maintained in the wake of the posterior-to-anterior passage of the morphogenetic furrow, a physical indentation in the epithelium that marks the transition from asynchronous proliferation to G1 arrest and differentiation (Bessa et al., 2002; Curtiss and Mlodzik, 2000; Halder et al., 1998; Pappu and Mardon, 2004; Ready et al., 1976). Cells posterior to the morphogenetic furrow develop into photoreceptor cells and nucleate formation of the ommatidia that collectively comprise the compound eye (Clandinin and Zipursky, 2002; Wolff and Ready, 1991).
Although initially identified for their role in the Drosophila eye, components of the RD gene network have multiple roles throughout development in metazoans, as evidenced by a broad spectrum of loss-of-function phenotypes. For instance, EYA1 knockout mice exhibit loss of multiple organs and defects in muscle development, while mutations in human EYA1 have been associated with branchio-oto-renal (BOR) syndrome, an autosomal dominant disorder characterized by jaw and external ear malformations, hearing loss, and renal defects (Abdelhak et al., 1997; Grifone et al., 2005; Heanue et al., 1999; Xu et al., 1999). RD network components are also expressed outside of eye tissues in Drosophila, and null mutations are generally lethal (Bonini et al., 1998; Callaerts et al., 2001; Cheyette et al., 1994).
Expression and activity of RD network members are regulated by multiple signaling pathways to produce specific developmental outcomes (reviewed by Kumar, 2009; Silver and Rebay, 2005). For example, prior to neuronal differentiation in the developing eye disk, Hedgehog (Hh) and Decapentaplegic (Dpp) signaling promote eya, so and dac expressions at the morphogenetic furrow (Pappu et al., 2003), whereas Wingless (Wg) signaling downregulates expression of eya, so and dac in the antennal disk to inhibit retinal fate (Baonza and Freeman, 2002). Although mechanisms influencing RD protein function remain less well characterized, they are likely to be equally important and to include interactions with specific binding partners and post-translational modifications. For example, distinct cofactor interactions may mediate specific roles of So during eye development (Kenyon et al., 2005), while Eya is positively regulated by MAPK phosphorylation in response to EGFR/RAS signaling during retinal determination (Hsiao et al., 2001; Rebay et al., 2000), and by Abl kinase phosphorylation during photoreceptor axon targeting (Xiong et al., 2009).
Recently, we reported that ey, eya, and dac genetically synergize with nemo (nmo) to promote eye specification (Braid and Verheyen, 2008). Drosophila nmo encodes a proline-directed serine/threonine kinase that is essential during development and is the founding member of the Nemo-like kinase (NLK) branch of the MAPK superfamily (Brott et al., 1998; Choi and Benzer, 1994; Mirkovic et al., 2002; Miyata and Nishida, 1999). NLKs are highly conserved in evolution and have multiple developmental roles in a variety of organisms, including endoderm induction in C. elegans (Meneghini et al., 1999), antero-posterior patterning and neurogenesis in zebra fish (Ishitani et al., 2010; Thorpe and Moon, 2004), and mouse hematopoiesis (Kortenjann et al., 2001). Functionally, NLKs act as regulators of downstream transcriptional effectors for multiple signaling pathways. One of the best-characterized roles for Nmo/NLK is in regulating Wnt/Wingless signaling. NLKs block activation of Wnt/Wg target genes (Zeng and Verheyen, 2004) by phosphorylating T-cell factor (TCF) and inhibiting the DNA-binding ability of the beta-catenin/TCF complexes (Ishitani et al., 1999; Ishitani et al., 2003). Nmo antagonizes BMP signaling in Drosophila where it suppresses the transcriptional activity of Mothers against Dpp (Mad) by preventing its nuclear accumulation (Zeng et al., 2007). In addition, Nmo has been implicated in planar cell polarity, programmed cell death, embryonic patterning, synaptic growth, and wing patterning, and is likely to mediate crosstalk between multiple signaling pathways in these contexts (Braid et al., 2010; Choi and Benzer, 1994; Fiehler and Wolff, 2008; Merino et al., 2009;Mirkovic et al., 2002;Mirkovic et al., 2011; Verheyen et al., 2001).
In the context of Drosophila eye development, we have previously shown that coexpression of Nmo potentiates ectopic eye formation driven by Ey, Eya and Dac transgenes in a dose-dependent manner (Braid and Verheyen, 2008). Here, we test the hypothesis that Nmo-mediated modulation of Eya-So transcriptional activity might provide a mechanistic explanation for the cooperative genetic interaction between Nmo and the RD network. We show that Nmo catalytic function is required to promote Eya-mediated retinal determination and to enhance activity of the Eya–So transcriptional complex. Mechanistically, Nmo can form a complex with and phosphorylate Eya at two MAPK consensus sites. This phosphorylation potentiates Eya activity in ectopic eye induction assays and enhances Eya–So mediated transcription of lozenge and dachshund. Together our results suggest that the Nmo kinase forms part of a novel regulatory complex that modulates Eya's transactivation function during Drosophila retinal determination.
Results
The Nemo kinase cooperates with Eya to promote eye development
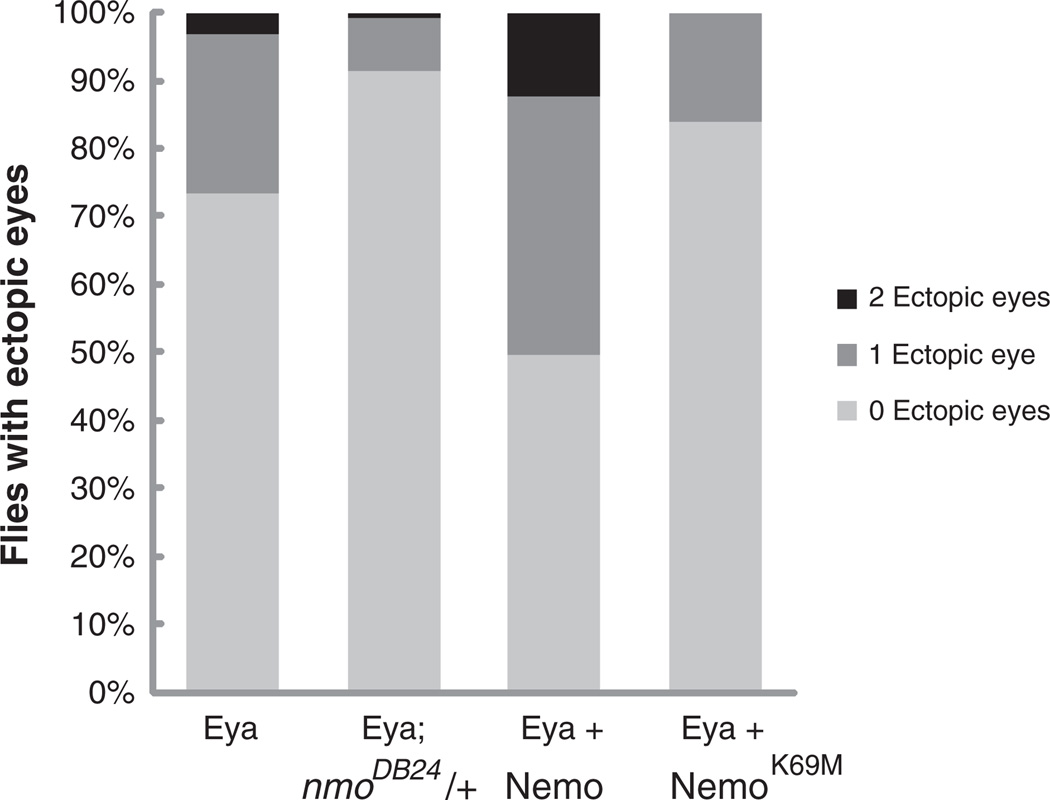
We have previously demonstrated that eya and nmo interact genetically to promote eye specification in Drosophila (Braid and Verheyen, 2008). To begin to address the underlying mechanism, we asked if the kinase function of Nmo is required for Eya activity during ectopic eye formation by comparing the effects of coexpressing wild type and kinase inactive Nmo transgenes (Fig. 1). A weak Eya transgene, whose ectopic eye induction efficiency is only ~25% (Hsiao et al., 2001), was selected to maximize the range of responsiveness to Nmo-mediated enhancement. Importantly, this background is still sensitized to nmo levels, as reduced nmo dosage decreased Eya's ectopic eye induction efficiency more than two-fold (Fig. 1). In contrast, and as previously shown with other Eya transgenes (Braid and Verheyen, 2008), coexpressing wild type Nmo increased the penetrance and the frequency of Eya-mediated ectopic eye induction (Fig. 1). Thus over 50% of adults exhibited ectopic eyes, a two-fold increase relative to Eya alone, with approximately a quarter of those animals showing ectopic eye tissue under both, rather than under just one, antennae. In this assay, we found that kinase dead Nmo failed to enhance, and slightly suppressed Eya-mediated ectopic eye formation (Fig. 1). These results extend our previously reported Eya-Nmo synergistic interaction (Braid and Verheyen, 2008), and suggest that the kinase function of Nmo is required.
Fig. 1.
Nemo's kinase activity is required for Eya-Nemo synergy during eye induction. Heterozygosity for nmo reduces the frequency of Dpp-Gal4>UAS-eya-mediated ectopic eye formation, while coexpression of UAS-Nmo increases both the frequency and penetrance of ectopic eyes. A Kinase-dead Nmo transgene (NmoK69M) fails to increase and slightly suppresses ectopic eye frequency when coexpressed with Eya. Penetrance reflects whether ectopic eye tissue was observed under one or both antennae (1 Ectopic eye, dark gray bar or 2 Ectopic eyes, black bar) and frequency refers to a binary scoring system for presence/absence of ectopic eye tissue.
Nemo potentiates Eya-So mediated induction of Lozenge and Dachshund expression
Eya has two biochemical functions, one as a transcriptional coactivator in conjunction with the DNA binding protein Sine oculis (So) and a second as a protein tyrosine phosphatase; both activities are required for full function during eye specification (Rayapureddi et al., 2003; Silver et al., 2003; Tootle et al., 2003). Taking advantage of our recent finding that cytoplasmic Eya phosphatase function contributes to photoreceptor axon targeting in the larval brain (Xiong et al., 2009), we first asked whether Nmo might interact with Eya in this context. However, neither decreasing nor increasing nmo dose modified the Eya axon guidance phenotypes, nor did Nmo knockdown or overexpression have an axonal phenotype on its own (data not shown). This suggests that Nmo may not regulate Eya phosphatase function, at least in this context. Therefore, we directed our focus to the alternate hypothesis that Nmo potentiates Eya–So transcriptional activity.
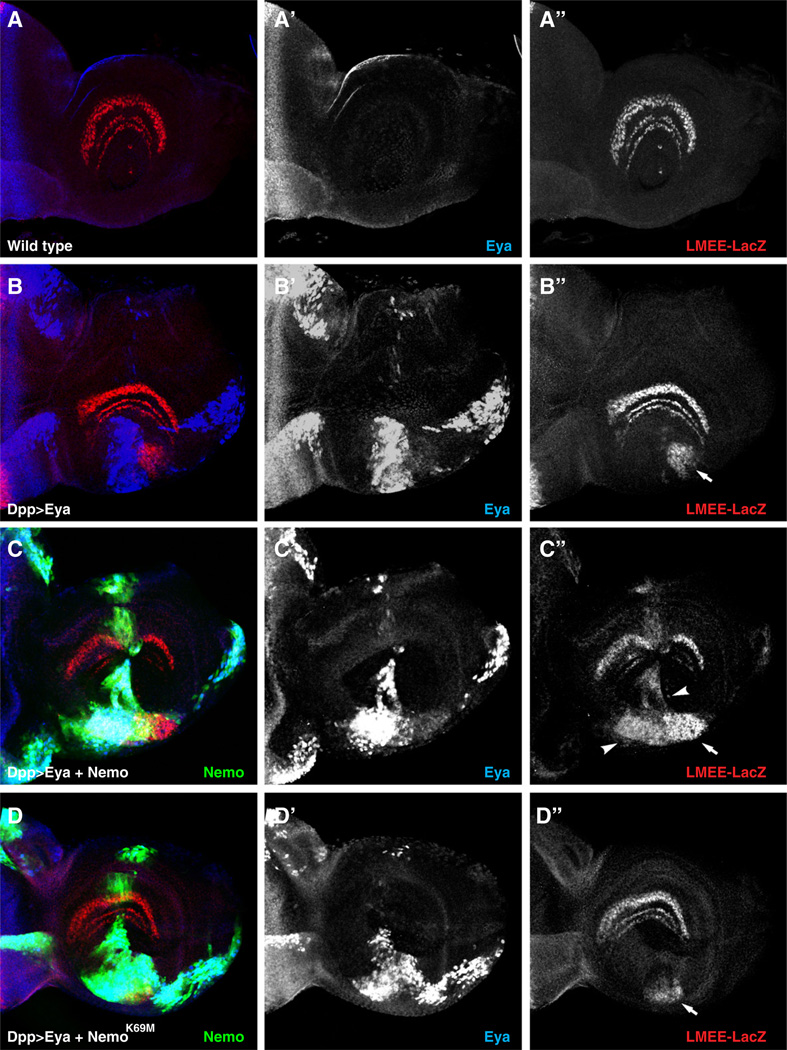
To investigate this possibility, we compared the levels of ectopic induction of two known Eya–So target genes, lozenge and dachshund (Jemc and Rebay, 2007; Pappu et al., 2005; Yan et al., 2003), in the antennal and wing imaginal disks respectively, in response to overexpression of Eya versus Eya plus Nmo. Both tissues are competent to form ectopic eye tissue, but do not normally express Eya, and have been used extensively as tractable experimental systems for probing the function and regulation of the RD network (Bonini et al., 1997; Braid and Verheyen, 2008; Jang et al., 2003; Salzer and Kumar, 2010; Shen and Mardon, 1997; Tavsanli et al., 2004; Tootle et al., 2003; Weasner et al., 2009). First, we followed induction of lozenge expression using a previously characterized Eya-So transcriptional reporter referred to as the lozenge minimal enhancer element (LMEE; Mutsuddi et al., 2005; Yan et al., 2003). Consistent with our hypothesis, in transfected cultured S2 cells, addition of Nmo resulted in a 3-fold increase in LMEE-luciferase reporter activity relative to induction with Eya-So alone (Supplemental Fig. 1). Responsiveness of this lozenge reporter was tested in flies carrying an LMEE-lacZ transgene. In wild type flies, the LMEE-lacZ reporter is expressed in three concentric circles in the antennal disk (Fig. 2A), matching the previously reported expression pattern of Lozenge in this tissue (Flores et al., 1998). Driving Eya expression with Dpp-GAL4 resulted in ectopic reporter activity in the ventral antennal disk in 24% of disks analyzed (Fig. 2B). Coexpression of Eya and Nmo increased both the frequency (~52% of disks) and the size of the tissue patch showing ectopic reporter activity (Fig. 2C). On the other hand, coexpression of Eya and a kinase inactive form of Nmo decreased the frequency of ectopic reporter expression compared to expression of Eya alone, with only ~16% of disks showing LMEE-lacZ induction (Fig. 2D). Similar frequencies of induction of ectopic Lozenge were observed in disks stained with an anti-Lozenge antibody (data not shown).
Fig. 2.
Nemo increases Eya-mediated activation of a lozenge transcriptional reporter. (A–D) Expression of the lozenge LMEE-lacZ reporter (red), Eya (blue) and Nmo-GFP (green) in third instar antennal disks oriented dorsal up, posterior left. Dpp-GAL4 was used to drive expression of Eya and Nmo. (A) LMEE-lacZ reporter expression in wild type larvae is restricted to three concentric half-circles in the center of the antennal disk. (B) 24% (n = 25) of Dpp-GAL4>UAS-Eya disks have ectopic reporter activity in the ventral antennal disk (B”, arrow). (C) Coexpression of Nmo increases both the frequency (~52%, n = 21) and the area of LMEE-lacZ induction (C”, arrowheads). (D) Coexpression of kinase inactive Nmo (NmoK69M) decreases the frequency (16%, n = 25) and area of ectopic β-gal staining.
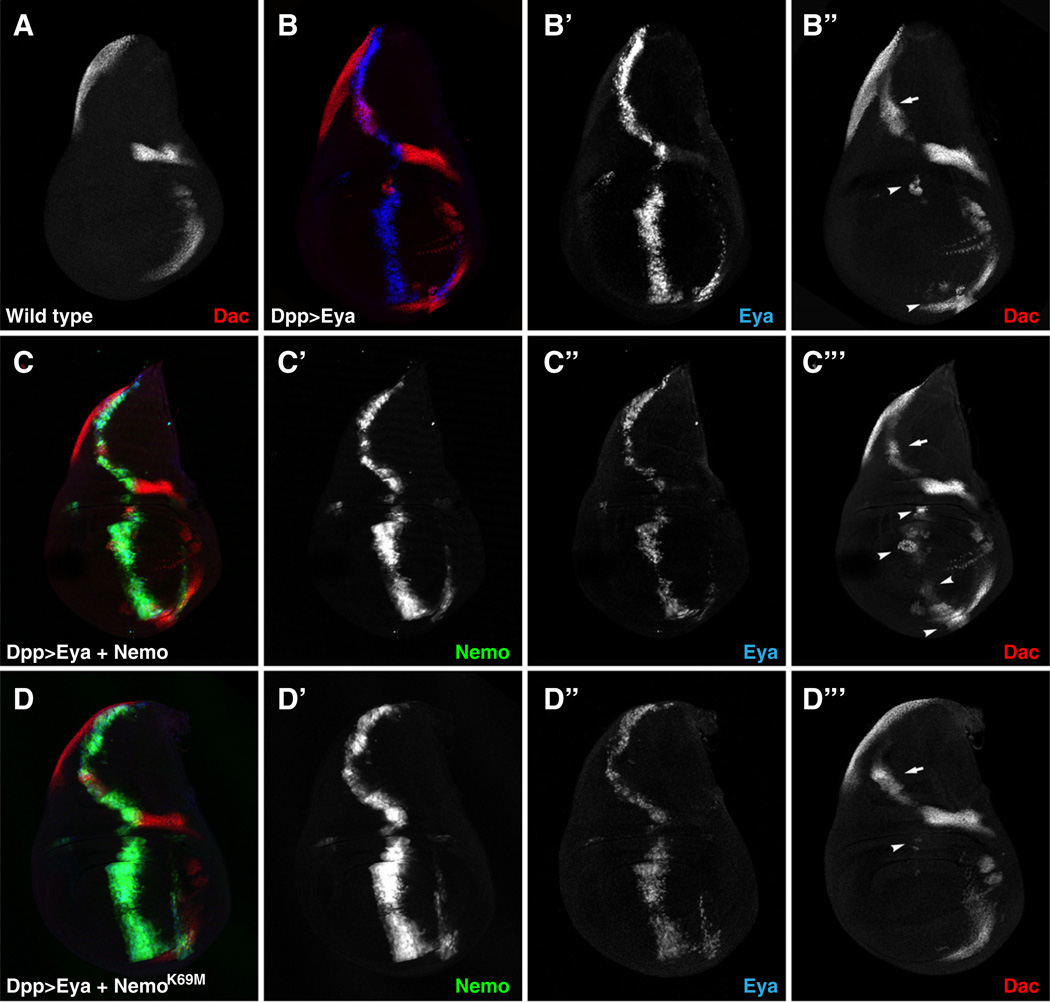
In the second set of experiments, we used Dpp-Gal4 to express Eya in combination with Nmo transgenes in the wing disk and monitored induction of Dac, which has been shown to be genetically downstream of and transcriptionally regulated by Eya-So (Chen et al., 1997; Pappu et al., 2005; Shen and Mardon, 1997). In wild type wing disks Dachshund is expressed in the dorsal posterior compartment and in two anterior regions surrounding the wing pouch (Fig. 3A; Chen et al., 1997). Expression of Eya along the Dpp stripe at the antero-posterior border of the disk induced ectopic Dac in the dorsal wing disk (Fig. 3B, arrow) and in the dorsal and ventral hinge regions (Fig. 3B, arrowheads). In agreement with our observations in ectopic eye experiments (Fig. 1), this assay is sensitized to nmo levels, as reduced nmo dosage decreased Eya-mediated ectopic Dac induction (Fig. S2). Furthermore, and consistent with the LMEE-lacZ induction assay in the antennal disk (Fig. 2), coexpression of Eya and Nmo, but not kinase dead Nmo, increased Dac induction relative to that seen with expression of Eya alone, particularly in the ventral wing pouch where endogenous nmo is not normally expressed (Figs. 3C, D, arrowheads; Chen et al., 1997). Expression of either Nmo transgene alone did not induce Dac expression (data not shown).
Fig. 3.
Nemo potentiates Eya-mediated induction of Dac expression. (A–D) Dac (red), Eya (blue) and Nmo-GFP (green) expression in third-instar larval wing disks. In this and all other figures, wing disks are oriented dorsal up, anterior left. Eya and Nmo transgenes were driven using Dpp-GAL4. (A) Endogenous Dac expression. (B) Eya induces strong ectopic Dac expression in the dorsal hinge region (B”, arrow), and weaker expression in the wing disk pouch (B”, arrowhead). (C) Coexpression of Nmo increases Dac levels, particularly in the pouch and ventral region (C”’, arrowheads). (D) Coexpression of kinase-inactive Nmo (NmoK69M) decreases Eya-mediated Dac expression in the wing disk pouch (D”’, arrowhead, compare to B”) but not in the hinge region (D”, arrow).
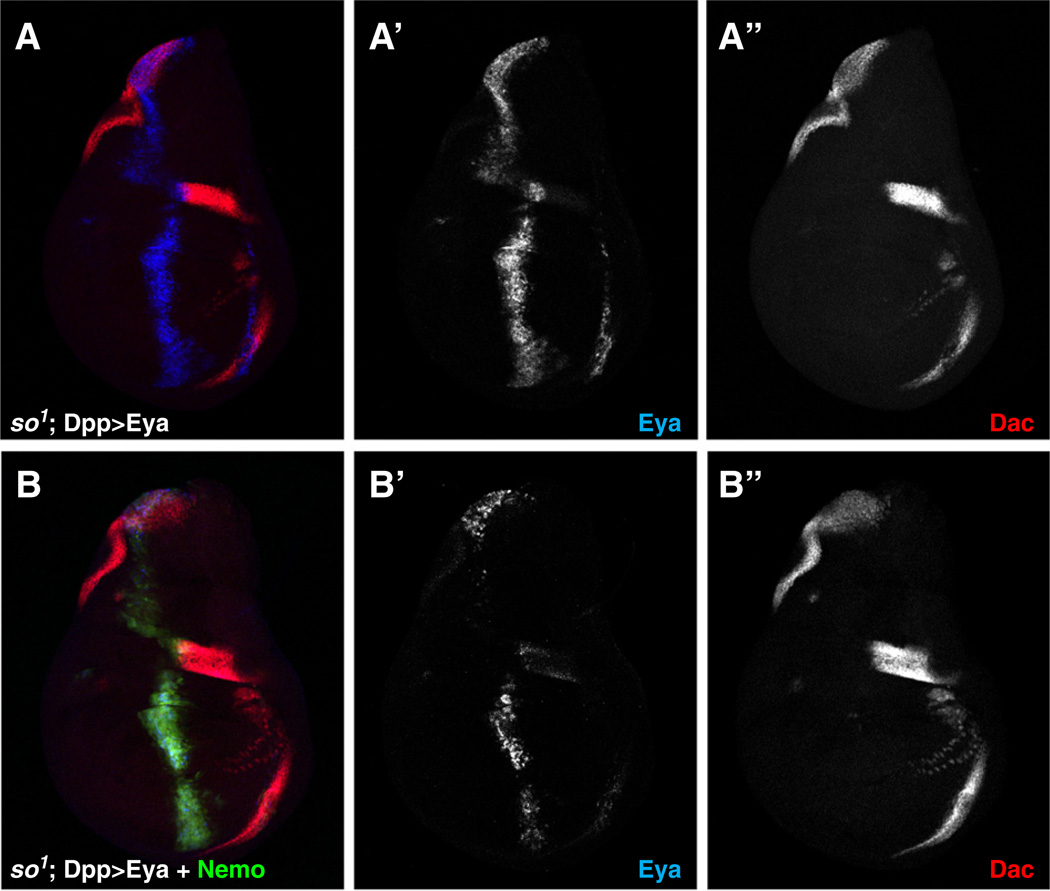
Transcriptional induction of dac presumably occurs through the normal RD network positive feedback circuitry such that ectopically expressed Eya interacts either with undetectably low levels of endogenous So in the wing or with another unknown factor to transcribe more so, raising So levels to a threshold sufficient for the Eya–So transcription factor to induce dac expression. Supporting this model, expression of Eya along the Dpp stripe at the antero-posterior border of the disk induced ectopic So expression in the wing pouch (Fig. S3), while expression of Eya in a so1 mutant background, which deletes the eye-specific enhancer in the so gene (Cheyette et al., 1994; Pignoni et al., 1997), did not, resulting in a failure to induce Dac (Fig. 4A). Similar experiments using RNAi-mediated knockdown of so gave analogous results (Supplemental Fig. S4). Coexpression of Nmo and Eya in either the so1 mutant or so RNAi background also failed to induce ectopic Dac expression (Fig. 4B, Supplemental Fig. S4), implying that the increased Dac induction seen in the Eya+Nmo background reflects increased activity of the Eya–So transcription factor.
Fig. 4.
Eya-mediated activation of Dac expression requires So. Dac (red), Eya (blue) and Nmo-GFP (green) expression in so1 third-instar larval wing disks expressing (A) Eya alone or (B) Eya+Nemo under Dpp-Gal4 control. No ectopic Dac expression is observed.
Eya is a novel substrate for Nemo phosphorylation
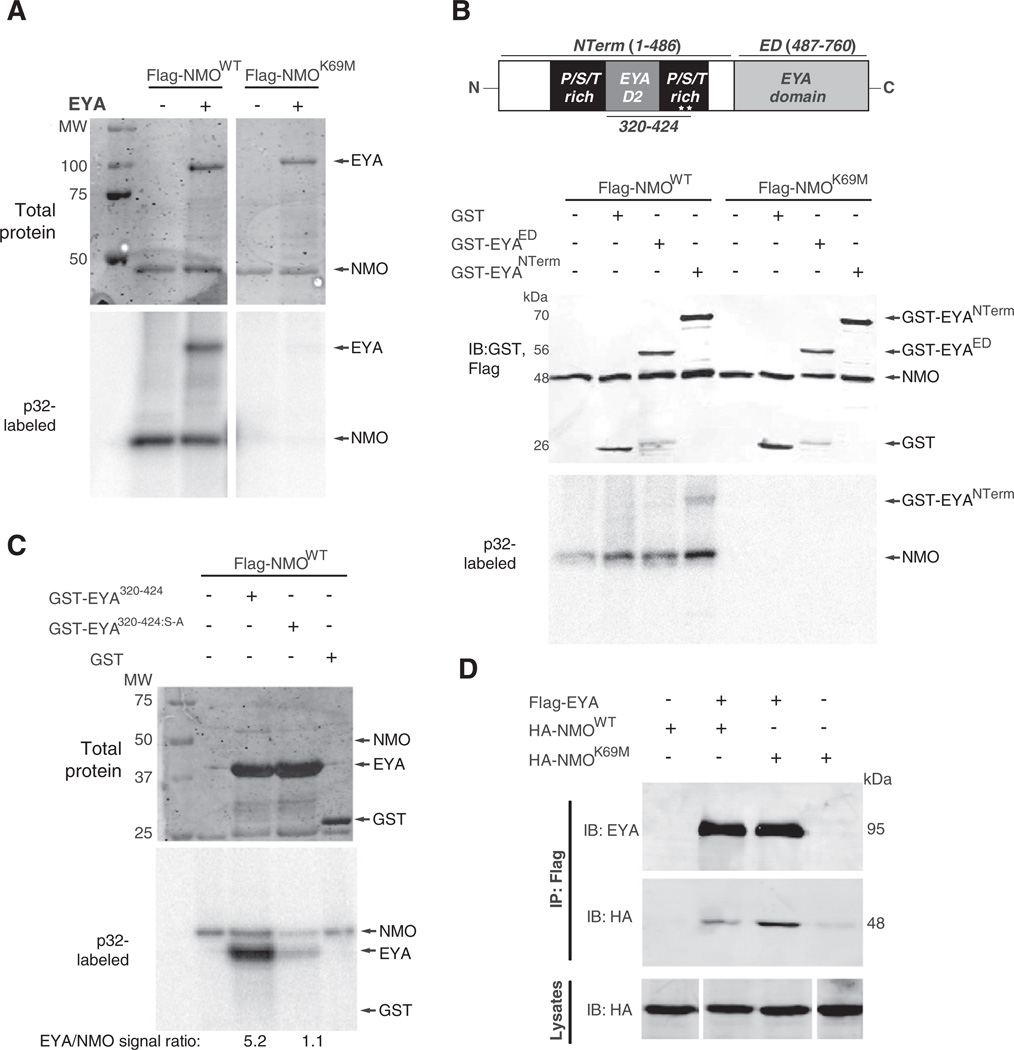
Our genetic studies consistently demonstrate that Nmo's kinase activity is necessary to promote Eya's ability to induce ectopic eyes and expression of Eya–So target genes. Since Eya activity and localization are regulated by phosphorylation (Hsiao et al., 2001; Xiong et al., 2009), we predicted that Eya may be a novel substrate for Nmo. We performed in vitro kinase assays using Nmo protein purified from HEK293T cells and recombinant full length His-tagged Eya fusion protein. Eya became phosphorylated only when incubated with the wild type Nmo protein, but not the kinase dead version; robust Nmo auto-phosphorylation was also observed, but only with the kinase active form (Fig. 5A).
Fig. 5.
Nemo phosphorylates Eya at consensus MAPK sites.(A–C). In vitro kinase assays using Flag-Nmo immunopurified from transfected HEK293T cells (see Methods for details). The upper panels show Coommassie staining for total protein, and the bottom panels show the phosphorimager exposure. (A) Full-length Eya protein is directly phosphorylated when incubated with wild type, but not the kinase inactive form of Nmo (K69M). Nmo autocatalytic activity is evident with the wild type, but not kinase dead form. (B) Schematic of the Eya protein, with the two MAPK phosphorylation consensus sites within the N-terminal P/S/T region of Eya represented by asterisks. In vitro kinase assays show that Nmo phosphorylates the N-terminal region of Eya, but not the ED. Immunoblot (IB) shows inputs for Nmo, Eya and GST. (C) In vitro kinase assays using a subfragment of the P/S/T-rich region of Eya (GST-EYA320–424). Mutating the phosphoacceptor serines in the MAPK sites significantly reduces Nmo-mediated phosphorylation. (D) Immunoblots (IB) showing HA-Nmo and Flag-Eya can be co-immunoprecipitated (IP) from transfected S2 cells. The lysates (bottom panel) were run together on the same gel, but out of order with respect to the IP gels, necessitating some cutting and splicing to align the lanes.
We next mapped the key Eya residues phosphorylated by Nmo to further clarify their enzyme-substrate relationship. We identified the N-terminal P/S/T-rich region (PST) of Eya as the primary target of Nmo-mediated phosphorylation in kinase assays using fragments of the Eya protein (Figs. 5B, C). This region carries two consensus MAPK phosphorylation sites (defined as PXS/TP), which have been previously shown to be relevant in promoting eya-mediated eye specification (Hsiao et al., 2001). Since Nemo-like kinases belong to the MAPK superfamily, we reasoned that these two sites might be good candidates for Nmo phosphorylation. We mutated both phosphoacceptor serines (S402 and S407) to alanine and tested the resulting GST-EyaS–A fusion protein as a Nmo kinase substrate (Fig. 5C). Phosphorylation of GST-EyaS–A is reduced five-fold relative to GST-Eya, but not entirely abolished, suggesting that while these sites may be primary targets of Nmo kinase activity, one or more of the seven other S/TP motifs within the Eya fusion protein might also be phosphorylated.
To test whether the Nmo-Eya kinase-substrate relationship reflects a stable molecular complex, we carried out co-immunoprecipitation experiments in S2 cultured cells transiently transfected with epitope-tagged Eya and Nmo expression constructs. Wild type and kinase dead Nmo co-immunoprecipitate with Eya (Fig. 5D), suggesting Nmo and Eya may form a molecular complex that allows Nmo to directly phosphorylate Eya and potentiate its activity.
Nemo activation of Eya during retinal specification requires two consensus MAPK sites
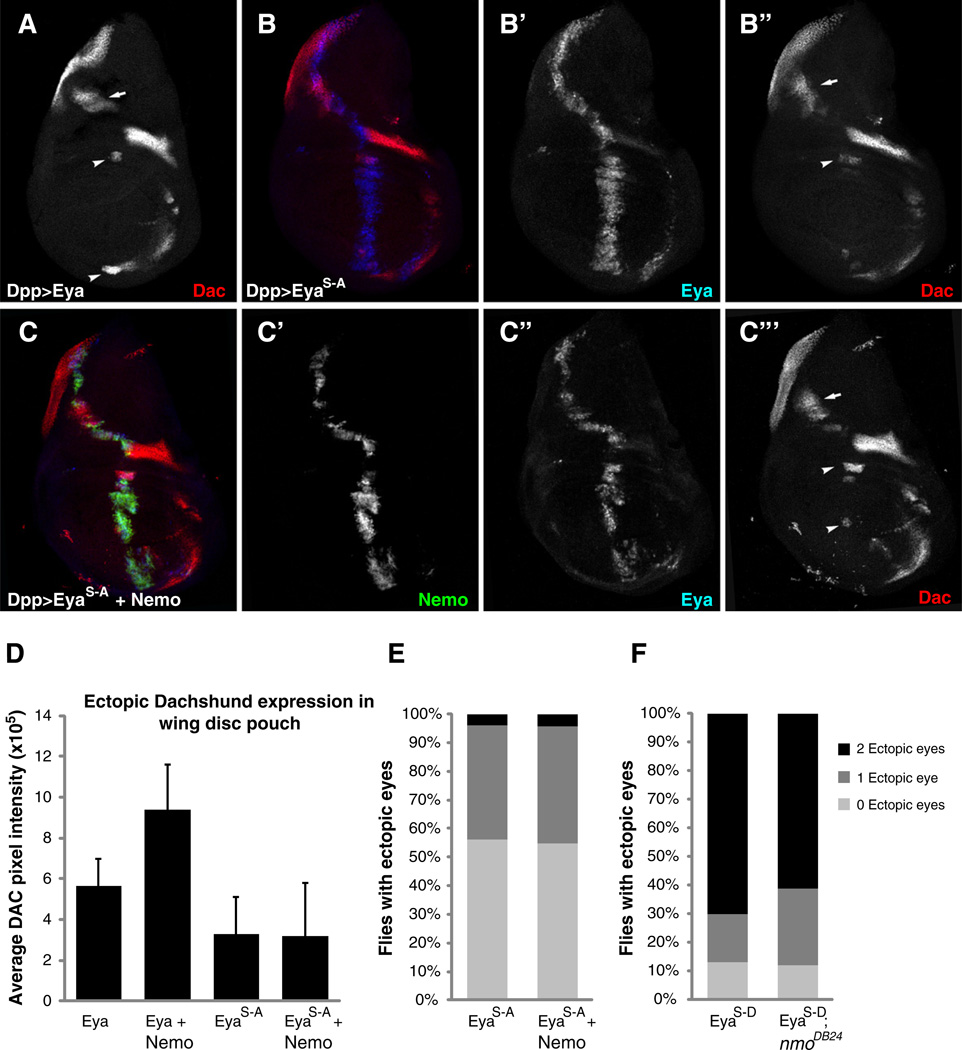
To follow up on the results from our kinase assays, we asked whether mutating the two Eya MAPK phosphorylation consensus sites would reduce the synergistic Eya-Nmo interaction in experiments measuring Dac induction in the wing. Consistent with our prior finding that UAS-EyaS–A transgenes have very low ectopic eye induction activity (Hsiao et al., 2001), most lines failed to induce ectopic Dac expression and so could not be used to test synergy with Nmo (data not shown). To circumvent this problem, EyaS–A transgenic lines providing unusually strong activity as judged by an ~40% ectopic eye induction efficiency and significant Dac induction (Figs. 6A–B, E) were used to test the hypothesis. In contrast to its synergistic interaction with wild type Eya transgenes, overexpression of Nmo did not cause a significant increase in EyaS–A mediated ectopic Dac, suggesting that the two consensus MAPK sites are crucial for the interaction (Figs. 6A–D, B” and C”’ arrowheads, compare to Fig. 3C”’). Ectopic eye induction assays showed a similar trend, such that coexpression of Nmo did not increase EyaS–A activity (Fig. 6E).
Fig. 6.
MAPK phosphorylation consensus sites are required for Nemo activation of Eya-So.(A–C) Dac (red), Eya (blue) and Nmo-GFP (green) expressions in third-instar larval wing disks. (A) Eya expression along the Dpp stripe causes ectopic Dac expression in the dorsal hinge region (arrow) and in the wing pouch (arrowhead). (B) Mutating the MAPK phosphorylation consensus sites (EyaS–A) does not alter Eya-mediated induction of Dac. (C) Nmo does not synergize with EyaS–A, with only a very slight increase in ectopic Dac expression observed in some disks (C”’, arrowheads) as compared to the robust increase seen when Nmo is coexpressed with a wild type Eya transgene (Fig. 3C”’, arrowheads). (D) Quantification of ectopic DAC along the wing disk pouch confirms the lack of significant interaction of Nmo with EyaS–A, n ≥ 16 disks. (E) Coexpression of Nmo does not alter the frequency of DppGal>UAS-EyaS–A mediated ectopic eye induction. (E) A phospho-mimetic form of Eya, EyaS–D, is refractory to loss of one copy of nmo in ectopic eye induction assays.
If Nmo potentiates Eya function by phosphorylating its two MAPK consensus sites, then constitutively activating Eya via phosphomimetic (S–D) mutations in the MAPK sites should bypass the requirement for Nmo. As predicted, heterozygosity for nmo, which markedly suppresses the ectopic eye-inducing capacity of wild type Eya transgenes (Fig. 1 and Braid and Verheyen, 2008), did not suppress ectopic eye formation by the EyaS–D transgene (Fig. 6F). Together these observations demonstrate that the two MAPK phosphorylation consensus sites are crucial for Nmo activation of Eya during retinal specification.
Discussion
Retinal determination genes are highly regulated effectors that receive input from a variety of signaling pathways to direct many aspects of Drosophila eye specification and development, including cell proliferation, differentiation and morphogenesis (reviewed by Kumar, 2009). In this study, we reveal a novel regulatory mechanism by which the proline-directed kinase Nmo phosphorylates Eya at two conserved MAPK phosphorylation consensus residues to promote activity of the Eya-So transcriptional complex during retinal specification.
While a kinase-substrate relationship often reflects a transient physical interaction, Nmo and Eya associate in a complex sufficiently stable to be detected by coimmunoprecipitation, raising the possibility that Nmo could be an intrinsic component of the Eya–So transcriptional complex assembled at target genes. Inclusion of a kinase in a transcriptional complex would provide a sensitive mechanism for rapid and dynamic modulation of transcriptional output. Mechanistically, Nmo could be recruited to DNA bound Eya–So complexes or conversely, Nmo itself could occupy specific target sites through other protein interactions and then recruit Eya–So. Consistent with such models, other MAPK superfamily members have been detected at specific target gene promoters along with their substrates (Lawrence et al., 2008; Pokholok et al., 2006). Thus chromatin immunoprecipitation studies will be a high priority for investigating possible Nmo–Eya–So co-occupancy at target genes.
Regardless of the exact biochemical mechanism, our work raises the broader question of whether interactions with Nmo augment Eya–So transcriptional activity at all target genes, or whether there are more selective, context-specific requirements for Nmo input during retinal specification and development. Overall our data support a broad, but perhaps not universal, involvement of Nmo in regulating RD network output. For example, the observation that Nmo potentiates Eya–So mediated induction of Dac expression and ectopic eye formation would be consistent with Nmo playing a global role in regulating the overall activity and output of the RD network throughout eye development. Furthermore, the ability of Nmo to potentiate induction of lozenge suggests a regulatory role for Nmo not just during early eye fate determination but later during cell specification and differentiation. lozenge encodes a RUNX family transcription factor that contributes to prepatterning in photoreceptor precursors and cell fate establishment in cone and pigment cells (Crew et al., 1997; Daga et al., 1996). The fact that lozenge expression in the developing eye is regulated by multiple transcription factors in addition to Eya–So (Behan et al., 2002; Siddall et al., 2009; Yan et al., 2003) complicates genetic analysis of the Nmo input. Thus, similar to what we reported previously for Dac (Braid and Verheyen, 2008), Lozenge protein levels do not appear reduced in nmo loss of function clones (LB and EMV unpublished observation). This suggests either Nmo does not potentiate Eya–So mediated activation of lozenge and dac in this context, or that other transcriptional inputs effectively compensate for the presumed reduction in Eya-So activity.
Considering further the issue of context specificity and combinatorial transcriptional control, we found that Nmo potentiates Eya-mediated ectopic lozenge and dachshund expression in relatively small regions of the Dpp expression domain in antennal and wing disks. Keeping in mind the caveats inherent to such overexpression experiments, these observations suggest that rather than broadly activating Eya–So-mediated transcription in all cells, Nmo regulates the function of this complex in specific cellular contexts. The ability of only certain cell populations outside of the eye imaginal disk to support retinal formation has been previously described, and in agreement with our observations from misexpressing Eya alone, the wing disk pouch is not a “hot spot” of responsiveness to RD network activity (Salzer and Kumar, 2010). However, coexpression of Nmo results in high levels of ectopic Dac expression in this region, suggesting that Nmo can activate Eya-So to drive transcription of target genes in a cellular context where this complex would normally be inactive or actively repressed. One possible explanation for this observation is that regional Nmo-mediated activation of Eya–So in wing disks correlates with high levels of endogenous Nmo, which is expressed in a ring surrounding the pouch, and along the dorsoventral boundary bisecting the wing pouch (Zeng and Verheyen, 2004). Alternatively, Nmo could drive specific activation of Eya–So by acting as a transducer of other signaling mechanisms that are active in specific regions of the wing disk.
Another context in which Nmo potentiation of Eya–So activity might be relevant is in regulating cell proliferation. Loss of eya or so results in uncontrolled cell proliferation in undifferentiated cells in the early eye epithelium, followed eventually by apoptosis (Bonini et al., 1993; Pignoni et al., 1997) while ectopic expression of Eya-So also leads to significant tissue overgrowth (Bonini et al., 1997; Pignoni et al., 1997). These results suggest that proper Eya–So activity levels are important for balancing cell proliferation and tissue growth during organ development. Our analyses show that coexpressing Nmo and Eya results in tissue overgrowth in both antennal and wing disks (Figs. 2C, 3C), suggesting that Nmo could regulate Eya–So function in cell proliferation. Furthermore, our previous observation that nmo levels can modify growth in the eye and head suggests that Nmo also plays a role in regulating cell proliferation (Braid and Verheyen, 2008).
Although the full spectrum of Eya–So transcriptional targets relevant to regulating proliferation and tissue growth remains to be identified, we have previously proposed that Eya and So control cell proliferation at least in part through direct regulation of the expression of the cell cycle regulatory gene of string (Jemc and Rebay, 2007). In the third instar eye disk, string activity contributes to synchronization of undifferentiated cells immediately anterior to the morphogenetic furrow, which is essential for subsequent cell fate specification and ommatidial assembly (Mozer and Easwarachandran, 1999; Thomas et al., 1994). Furthermore, transcriptional regulation of string in progenitor cells has been proposed as a mechanism that affects morphogenetic furrow progression (Lopes and Casares, 2010). Consistent with the possibility that Nmo potentiation of Eya–So activation of string could be important in this context, loss of nmo in eye disk clones results in a delay inmorphogenetic furrow progression, (LB and EMV, unpublished data). The specific effects of Nmo, Eya and So regulation on cell cycle progression during early eye specification remain an interesting question for future studies.
Lending further complexity to these regulatory possibilities, the two MAPK consensus residues on Eya that we report as Nmo targets were previously described as regulatory sites that modulate Eya's function in eye specification in response to RTK/RAS/MAPK signaling (Hsiao et al., 2001). Similarly to Nmo, ERK-mediated phosphorylation at these sites was proposed to positively regulate Eya activity in ectopic eye induction (Hsiao et al., 2001). Thus there may be multiple MAPK family members, perhaps even extending beyond Nmo and Erk, that depending on specific context and in response to a variety of upstream signaling inputs, function either redundantly, competitively or in non-overlapping ways to phosphorylate and regulate Eya. Whether such inputs all modulate Eya–So transcriptional output, or whether MAPK-mediated regulation may also modify other Eya functions will be an interesting direction for future investigation.
Experimental procedures
Drosophila strains
We used the following Fly stocks: dpp40C6-Gal4 (Staehling-Hampton et al., 1994), ey-Gal4, so1, (Bloomington Stock Center), nmoDB24 (Zeng and Verheyen, 2004), UAS-nmo-GFP (Fiehler and Wolff, 2008), UAS-nmoK69M-GFP (Merino et al., 2009), LMEE-lacZ (Yan et al., 2003), UAS-eyaI, UAS-eyaIIIa, UAS-eyaVb, UAS-Flag-eya, and UAS-Flag-eya1,2S–A. UAS-soRNAi, were obtained from the Vienna Drosophila RNAi Center.
For ectopic eye induction assays, dpp40C6-Gal4/TM3Sb or dpp40C6-Gal4, nmoDB24/TM3Sb; dpp40C6-Gal4/TM3Sb flies were crossed to UAS-eyaIIIa or UAS-eyaIIIa, UAS-nmo-GFP, or UAS-eyaIIIa, UAS-nmoK69M-GFP. Ectopic eyes under the antennae were scored in at least 120 progeny.
LMEE-lacZ reporter activity in antennal disks was evaluated by crossing dpp40C6-Gal4, LMEE-lacZ/TM6BTb to UAS-eyaVb or UAS-eyaVb, UAS-nmo-GFP, or UAS-eyaVb, UAS-nmoK69M-GFP/TM6B.
Induction of Dac expression was analyzed in wing disks dissected from appropriately genotyped 3rd instar progeny obtained by crossing dpp40C6-Gal4/TM6BTb to UAS-eyaVb or UAS-eyaVb, UAS-nmo-GFP or UAS-eyaVb, UAS-nmoK69M-GFP/TM6B or UAS-Flag-eya, UAS-nmo-GFP, or UAS-Flag-eya1,2S–A, UAS-nmo-GFP stocks. Requirement for So was evaluated by crossing so1; dpp40C6-Gal4/TM6BTb and dpp40C6-Gal4, UAS-nmo-GFP/TM6BTb to UAS-eyaVb or UAS-eyaVb, UAS-nmo-GFP or UAS-eyaI, UAS-soRNAi.
Immunostaining
Wing and eye-antennal imaginal disks were dissected from third instar larvae in S2 cell medium, fixed for 10 min in 4% paraformaldehyde with 0.1% Triton X-100, washed 3× in PBT (1× PBS, 0.1% Triton), and blocked for 30 min in PNT (1× PBS, 0.1% Triton, 1% normal goat serum). Disks were incubated with guinea pig α-Eya (1:1000), guinea pig α-So (1:1000), mouse α-Dac (1:20; Developmental Studies Hybridoma Bank), and rabbit α-βGal (1:1000) in PNT at 4 °C overnight as indicated, washed in PBT, incubated with donkey α-mouse-Cy3, donkey α-rabbit-Cy3, and donkey α-guinea pig-Cy5 (1:2000; Jackson ImmunoResearch) for 2 h at room temperature, washed in PBT and mounted in ProLong antifade reagent (Invitrogen) for imaging on a Zeiss 510 confocal microscope. Pictures of 0.5 µm sections were taken and stacked across the z axis using LSM Image Browser software. For quantification of Dac expression, a region of interest along the wing disk pouch was defined and analyzed using Image J software (Abramoff et al., 2004), and the integrated pixel intensity for all measured images was averaged for the analysis.
In vitro kinase assays and immunoprecipitation
Nmo protein for in vitro kinase assays was obtained by transfecting HEK293T cells with pXJ-Flag-Nmo or pXJ-Flag-NmoK69M vectors (Zeng et al., 2007) using the Effectene transfection reagent (Qiagen) and following manufacturer's instructions. Cells lysis and Nmo immunopurification were carried as previously described (Zeng et al., 2007). Recombinant full-length HIS-EYA, GST-EYA223–438, GST-EYA487–760, GST-EYA223–438, GST-SO, and GST were used as substrates. Kinase reactions were carried out as previously described (Zeng et al., 2007) and SDS-PAGE resolved samples were exposed on a Storm phosphoimager. Eya-Nmo coimmunoprecipitation was achieved by transfecting Drosophila S2 cells with pMT-Flag-EYA, pAct5-HA-Nmo, and pAct5-HA-NmoK69M and processed as previously described (Xiong et al., 2009).
Supplementary Material
Acknowledgments
We thank W. Xiong and J. Zhang for comments on the manuscript; members of the Rebay, Verheyen, Fehon and Horne-Badovinac labs for input and helpful discussions; M. DiMarco and N. Martin for confocal assistance; T. Le for help with cell culture; and R. Fehon, U. Banerjee and G. Greene for reagents. We acknowledge the Bloomington and Vienna Drosophila stock centers for flies, and the Developmental Studies Hybridoma Bank for antibodies. This research was supported by National Institutes of Health grant R01 EY12549 to I.R., an operating grant to E.M.V. and a Canada Graduate Scholarship to L.B. from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.ydbio.2012.02.030.
Contributor Information
Esther M. Verheyen, Email: everheye@sfu.ca.
Ilaria Rebay, Email: irebay@uchicago.edu.
References
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Levi-Acobas F, Cruaud C, Le Merrer M, Mathieu M, et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum. Mol. Genet. 1997;6:2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Baonza A, Freeman M. Control of Drosophila eye specification by Wingless signalling. Development. 2002;129:5313–5322. doi: 10.1242/dev.00096. [DOI] [PubMed] [Google Scholar]
- Behan KJ, Nichols CD, Cheung TL, Farlow A, Hogan BM, Batterham P, Pollock JA. Yan regulates lozenge during Drosophila eye development. Dev. Genes Evol. 2002;212:267–276. doi: 10.1007/s00427-002-0241-4. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. Multiple roles of the eyes absent gene in Drosophila. Dev. Biol. 1998;196:42–57. doi: 10.1006/dbio.1997.8845. [DOI] [PubMed] [Google Scholar]
- Braid LR, Lee W, Uetrecht AC, Swarup S, Papaianni G, Heiler A, Verheyen EM. Nemo phosphorylates Even-skipped and promotes Eve-mediated repression of odd-skipped in even parasegments during Drosophila embryogenesis. Dev. Biol. 2010;343:178–189. doi: 10.1016/j.ydbio.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Braid LR, Verheyen EM. Drosophila nemo promotes eye specification directed by the retinal determination gene network. Genetics. 2008;180:283–299. doi: 10.1534/genetics.108.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J. Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cheyette BNR, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Making connections in the fly visual system. Neuron. 2002;35:827–841. doi: 10.1016/s0896-6273(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Crew JR, Batterham P, Pollock JA. Developing compound eye in lozenge mutants of Drosophila: lozenge expression in the R7 equivalence group. Dev. Genes Evol. 1997;206:481–493. doi: 10.1007/s004270050079. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Daga A, Karlovich CA, Dumstrei K, Banerjee U. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 1996;10:1194–1205. doi: 10.1101/gad.10.10.1194. [DOI] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev. Biol. 2008;313:533–544. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GV, Daga A, Kalhor HR, Banerjee U. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development. 1998;125:3681–3687. doi: 10.1242/dev.125.18.3681. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao FC, Williams A, Davies EL, Rebay I. Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev. Cell. 2001;1:51–61. doi: 10.1016/s1534-5807(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat. Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol. Cell. Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH. Two pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 2005;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Kortenjann M, Nehls M, Smith AJ, Carsetti R, Schuler J, Kohler G, Boehm T. Abnormal bone marrow stroma in mice deficient for nemo-like kinase, Nlk. Eur. J. Immunol. 2001;31:3580–3587. doi: 10.1002/1521-4141(200112)31:12<3580::aid-immu3580>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Kumar JP. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta. 2009;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, McGlynn K, Shao C, Duan L, Naziruddin B, Levy MF, Cobb MH. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13315–13320. doi: 10.1073/pnas.0806465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CS, Casares F. hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev. Biol. 2010;339:78–88. doi: 10.1016/j.ydbio.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Merino C, Penney J, Gonzalez M, Tsurudome K, Moujahidine M, O'Connor MB, Verheyen EM, Haghighi P. Nemo kinase interacts with Mad to coordinate synaptic growth at the Drosophila neuromuscular junction. J. Cell Biol. 2009;185:713–725. doi: 10.1083/jcb.200809127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I, Charish K, Gorski SM, McKnight K, Verheyen EM. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech. Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, Mlodzik M. Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat. Struct. Mol. Biol. 2011;18:665–672. doi: 10.1038/nsmb.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Nishida E. Distantly related cousins of MAP kinase: biochemical properties and possible physiological functions. Biochem. Biophys. Res. Commun. 1999;266:291–295. doi: 10.1006/bbrc.1999.1705. [DOI] [PubMed] [Google Scholar]
- Mozer BA, Easwarachandran K. Pattern formation in the absence of cell proliferation: tissue-specific regulation of cell cycle progression by string (stg) during Drosophila eye development. Dev. Biol. 1999;213:54–69. doi: 10.1006/dbio.1999.9350. [DOI] [PubMed] [Google Scholar]
- Mutsuddi M, Chaffee B, Cassidy J, Silver SJ, Tootle TL, Rebay I. Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics. 2005;170:687–695. doi: 10.1534/genetics.104.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu KS, Chen R, Middlebrooks BW, Woo C, Heberlein U, Mardon G. Mechanism of hedgehog signaling during Drosophila eye development. Development. 2003;130:3053–3062. doi: 10.1242/dev.00534. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Mardon G. Genetic control of retinal specification and determination in Drosophila. Int. J. Dev. Biol. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T, Suh C, Voas M, Williams A, Rubin GM. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer CL, Kumar JP. Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS One. 2010;5:e8510. doi: 10.1371/journal.pone.0008510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya M, Gehring WJ. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development. 2000;127:1879–1886. doi: 10.1242/dev.127.9.1879. [DOI] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Siddall NA, Hime GR, Pollock JA, Batterham P. Ttk69-dependent repression of lozenge prevents the ectopic development of R7 cells in the Drosophila larval eye disc. BMC Dev. Biol. 2009;9:64. doi: 10.1186/1471-213X-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994;5:585–593. [PubMed] [Google Scholar]
- Tavsanli BC, Ostrin EJ, Burgess HK, Middlebrooks BW, Pham TA, Mardon G. Structure-function analysis of the Drosophila retinal determination protein Dachshund. Dev. Biol. 2004;272:231–247. doi: 10.1016/j.ydbio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Gunning DA, Cho J, Zipursky L. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Moon RT. Nemo-like kinase is an essential co-activator of Wnt signaling during early zebrafish development. Development. 2004;131:2899–2909. doi: 10.1242/dev.01171. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Verheyen EM, Mirkovic I, MacLean SJ, Langmann C, Andrews BC, MacKinnon C. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development. Mech. Dev. 2001;101:119–132. doi: 10.1016/s0925-4773(00)00574-8. [DOI] [PubMed] [Google Scholar]
- Weasner BM, Weasner B, Deyoung SM, Michaels SD, Kumar JP. Transcriptional activities of the Pax6 gene eyeless regulate tissue specificity of ectopic eye formation in Drosophila. Dev. Biol. 2009;334:492–502. doi: 10.1016/j.ydbio.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Xiong W, Dabbouseh NM, Rebay I. Interactions with the abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev. Cell. 2009;16:271–279. doi: 10.1016/j.devcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev. Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–2920. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.