Abstract
The AAA+ (ATPase Associated with various cellular Activities) machinery represents an extremely successful and widely used design plan for biological motors. Recently found crystal structures are beginning to reveal nucleotide-dependent conformational changes in the canonical hexameric rings of the AAA+ motors. However, the physical mechanism by which ATP binding on one subunit allosterically propagates across the entire ring remains to be found. Here we analyze and compare structural organization of three ring-shaped AAA+ motors, ClpX, HslU, and dynein. By constructing multimers using subunits of identical conformations, we find that individual subunits locally possess helical geometries with varying pitch, radius, chirality, and symmetry number. These results suggest that binding of an ATP to a subunit imposes conformational constraint that must be accommodated by more flexible nucleotide-free subunits to relieve mechanical strain on the ring. Local deformation of the ring contour and subsequent propagation of strains may be a general strategy that AAA+ motors adopt to generate force while achieving functional diversity.
Keywords: ClpX, HslU, dynein, motor protein, helical assembly, translocase, chaperone
Introduction
The AAA+ motor proteins are found in all kingdoms of life and carry out a broad range of cellular tasks:23, 27 With 30,000 AAA+ genes identified to date,34 they function in roles such as oscillators underlying circadian rhythms,29 inheritance in yeast following a stress response,26 control over gene expression,30 transport,9 remodeling and degradation.22, 23 The structural core defining the AAA+ superfamily has an αβα-fold that contains a conserved ATP binding pocket including Walker-A/B and sensor-I/II motifs, the catalytic glutamate, and the arginine finger.6, 12, 34 Seven AAA+ clades are known so far that differ in the insertion of additional secondary structural elements in the conserved core.6 The entire AAA+ superfamily in turn belongs to an even broader group called the “additional strand conserved E family” (ASCE), named after the catalytic glutamate (single-letter amino acid code E).6, 20
Typically six AAA+ subunits assemble into a closed ring, so the AAA+ motor can be thought of as the 6-cylinder combustion engine of the cell (cf., Fig. 2a).24 Through multiple layers of interactions, these extremely versatile motors can further form higher order assemblies, such as coupled rings as used in dynein or N-ethylmaleimide sensitive factor (NSF).6, 12, 35 A fundamental question regarding such an organization concerns structural and dynamical aspects of the motor that enable high adaptability to various tasks while using a common force-generation mechanism. For example, even though the same structural cores are used, ring-shaped AAA+ translocases use either nucleic acid or polypeptide chains as tracks.6 Also, dynein has a stack of two AAA+ rings from which two stalks extend as ‘legs’ that walk along the microtubule (cf., Fig. 6a).28 AAA+ motors should thus employ modular domains to utilize its core fuel processor component for achieving diverse functions.15
Figure 2.
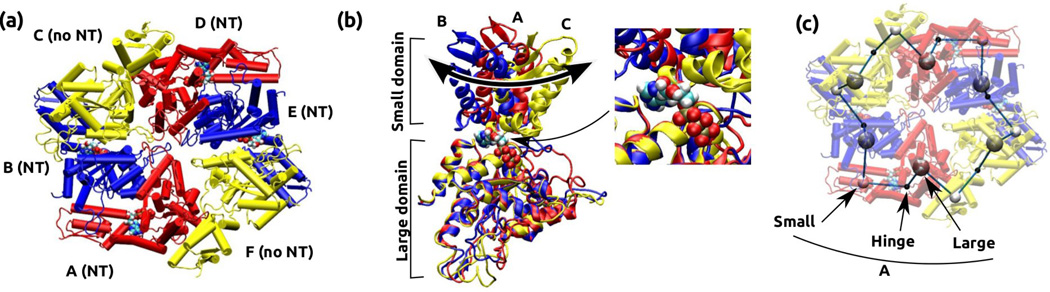
Structure of the ClpX motor with four bound nucleotides (PDB 3HWS10). (a) Overview of the hexameric ring. Chains A, B, D, and E have bound nucleotides, whereas chains C and F do not. (b) Comparison between different chains. Large domains of chain A, B, and C are optimally superposed, which reveals differences in the orientation of the small domain. Thick arrow illustrates dihedral motion of the small domain relative to the large one. Inset shows that the small domain of chain C sterically blocks the nucleotide-binding pocket. (c) The bead-on-a-chain (BOC) model of the ring contour. We used VMD14 for visualization of structures.
Figure 6.
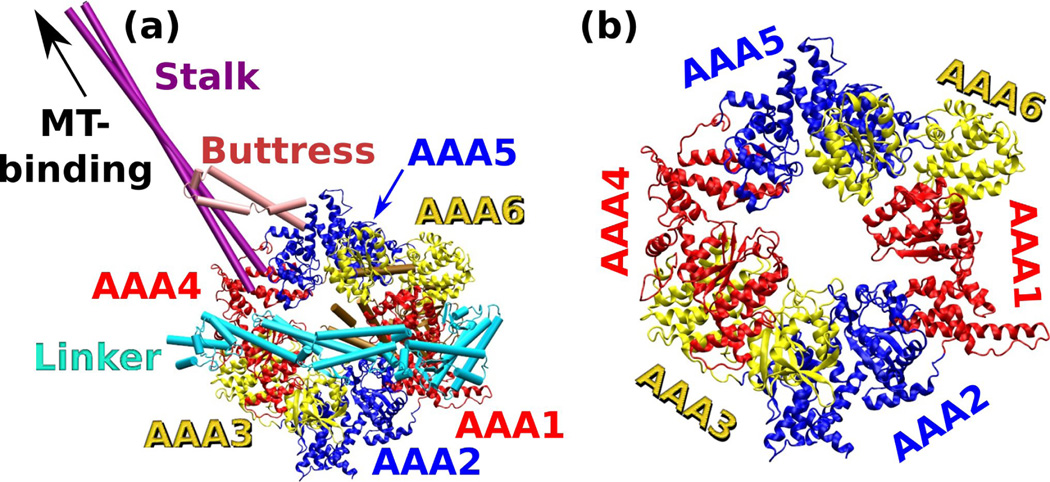
Crystal structure of cytoplasmic dynein (PDB 3VKG).18 (a) Overview. Extra α-helical domains outside of the base AAA+ ring are represented as cylinders: The N-terminal linker domain connects to AAA1. The stalk whose end binds to the microtubule (MT), inserts into the small domain of AAA4. The buttress (strut) that supports the stalk, inserts into the small domain of AAA5. (b) Magnified view of the base AAA+ ring with the extra domains not shown.
Since the ring form is a common structural motif among AAA+ motors, there must be functional advantages in having the ring geometry for adaptability or force generation, in addition to providing self-termination of the subunit assembly process. In fact, the nucleotide binding pocket is nestled between the two domains of an AAA+ subunit (called large and small domains; Fig 2b), and mutations that abolish ATP binding frequently disrupt oligomerization of AAA+ complexes, indicating the coupling between ATPase activities and the ring formation.34 Subtle conformational changes in the ATP binding site are thus likely amplified by the ring structure multiplicity, so that one can think of the AAA+ motor as a ring of 6 subunits that communicate with one another to collectively perform an overall task such as transport or unfolding.
How do subunits in an AAA+ ring communicate? Available x-ray structures show that domains forming inter-subunit contacts between neighboring subunits are strongly bound together hence they move essentially as one rigid body.11 Flexibility of the ring is conferred instead by intra-subunit hinge motion between the large and small domains, which depends on its nucleotide state (cf., Fig. 2b). Here we quantify this motion by constructing bead-on-a-chain (BOC) models that follow the contour of the ring. To investigate relative contribution of a given subunit conformation to the geometry of the ring, we construct a multimer formed by identical replicas of the subunit and study the corresponding BOC model. We use this approach to analyze crystal structures of three different AAA+ motors, ClpX, HslU (heat shock locus U), and cytoplasmic dynein. ClpX and HslU are members of the bacterial chaperone machinery, and they pull polypeptide chains through the central pore of the ring and unfold proteins tagged for destruction.23 Cytoplasmic dynein is a microtubule minus-end directed bipedal motor and is involved in processes including cell division and intracellular transport.28 Multimers constructed from these structures exhibit a wide-range of helical geometries, indicating that nucleotide-dependent strains may develop within individual subunits when the topological constraint of a hexameric ring is imposed. We also find that the large domains serve as pivots for the nucleotide-dependent changes in the contour of the ring, whereby the small domains undergo greater positional shifts. Since mechanical strain and topological constraints arise from the overall conformation of the motor, with less sensitivity with respect to the specific amino acid sequence, the AAA+ motors may be able to preserve these rather non-specific mechanisms for force generation, while substrate specificity or transduction of the generated force to other regions of the motor15 can be achieved by developing clade- or family-specific variations.
Methods
Structural manipulation and atomistic model building
The CHARMM package3, 4 was used for handling x-ray structures and coordinate manipulation. The following Protein Data Bank (PDB) structures were used: 3HWS for ClpX (resolution: 3.25 Å),10 1DO2 for HslU (resolution: 4.00 Å),2 and 3VKG for dynein (resolution: 2.81 Å).18 Loops missing in the x-ray structures that are shorter than 14 residues were constructed using the ModLoop program8 in Modeller version 9.11.7 Missing loops longer than 14 residues were either omitted or constructed using a combination of energy minimization and molecular dynamics runs of these regions. However, they were built only for convenience in handling structures and were not used for structural alignment nor analysis.
To build a multimer structure using a subunit of ClpX or HslU, Cα atoms of the large domain of the subunit (for ClpX: S62–L314, and for HslU: S2–K109 and I244–L332) were optimally superposed to those of the next subunit in the x-ray structure. In HslU, M110–A243 correspond to the I-domain (cf., Fig. 5) and was omitted in the alignment. This process was repeated up to a desired number of times to construct multimers shown in Figs. 4, 5 and 7. Since contacts between large and small domains of neighboring subunits in the original hexamer ring are nearly rigid, conformation of the multimer is determined by the relative orientation between the large and small domains within each subunit used to construct the structure.
Figure 5.
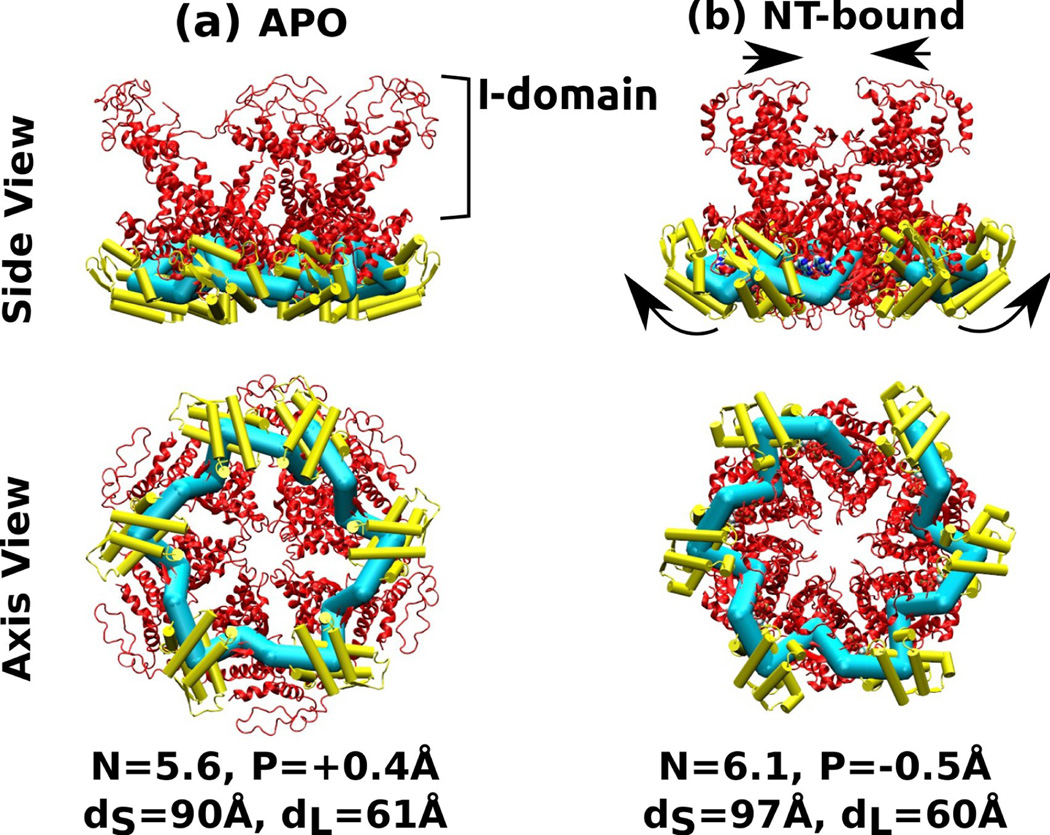
Six-mer structures constructed by connecting each of the two HslU subunits in PDB 1DO2. The I-domain is attached to the large domain, and it was not used for calculating the center of mass of the large domain. (a) Nucleotide-free, (b) AMPPNP (an ATP analog) state. Arrows in (b) indicate conformational change relative to (a) that leads to widening of the small domains and narrowing of the I-domains. Symbols, molecular representations, and color codes are the same as in Fig. 4.
Figure 4.
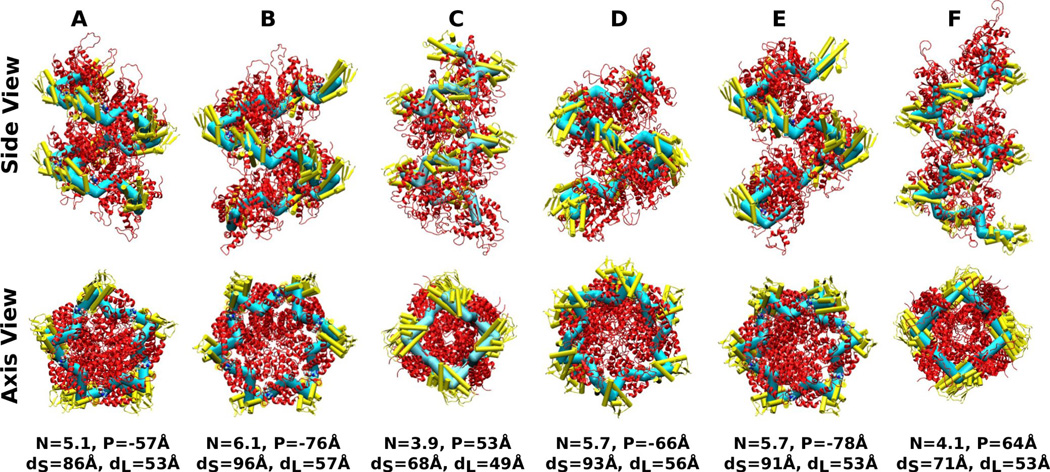
Nine-mer helices constructed using individual ClpX subunits of PDB 3HWS. Capital letters on top: chain labels in Fig. 2. N: Symmetry number (Eq. 6), P: Pitch (P > 0: right-handed, P < 0: left-handed; Eq. 7), dS,L: diameter of the cylinder spanned by center of masses of small/large domains (Eq. 8). The corresponding BOC models are represented using thick bonds to better reveal the helical contour. Alpha-helices forming the small domains are represented as cylinders and colored lighter, to distinguish from the large domains.
Figure 7.
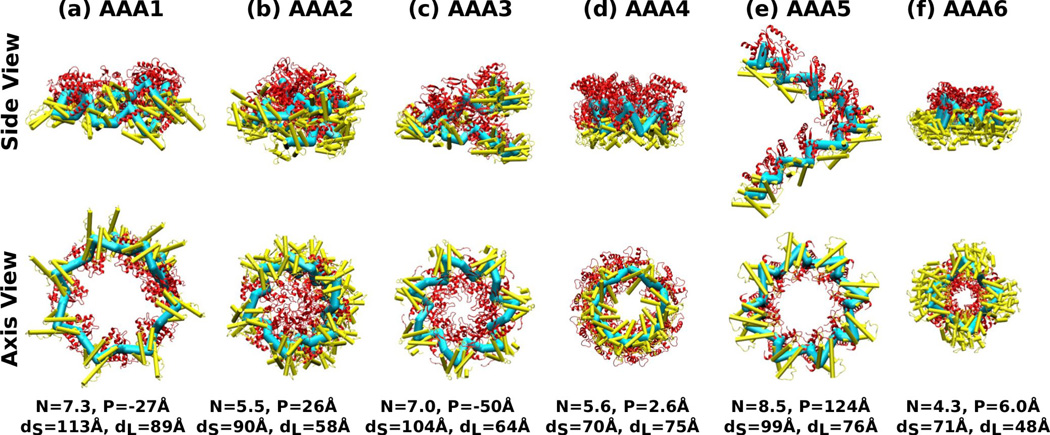
Nine-mer chains constructed using individual AAA+ domains of PDB 3VKG (dynein). Symbols, molecular representations, and color codes are the same as in Fig. 4.
Unlike ClpX or HslU that are homohexamers, dynein consists of a single polypeptide chain. We used the classification in Ref. 18 to identify large and small subdomains within dynein. Large subdomains of individual AAA+ units are: Y1936–I2101 (AAA1), P2230–E2419 (AAA2) V2634–D2817 (AAA3), P2949–Y3174 (AAA4), S3639–T3787 (AAA5), and E4115–N4240 (AAA6). Since these subdomains differ in amino acid sequence, we used the CE program25 to align their Cα atoms.
Bead-on-a-chain (BOC) model
To analyze the contour of the AAA+ ring or the constructed multimer structure, we calculated the center of mass of Cα atoms of the large, small, and hinge domains. Amino acids for the small domains are as follows. For ClpX: E320–Y413; For HslU: L335–L443; For dynein: D2104–Q2229 (AAA1), T2422–K2630 (AAA2), S2820–R2948 (AAA3) S3177–D3262 / A3597–L3638 (AAA4; F3263–S3596 is the stalk) P3790–R3806 / Y3886–L3960 (AAA5; P3807–L3885 is the buttress), and G4243–P4411 (AAA6) (stalk and buttress are shown in Fig. 6a). The hinge domain is located between the large and small domains. We assigned ‘beads’ to the three centers of masses (large-hinge-small) and ‘bonds’ were placed between these beads, which make up the BOC model representing the contour of the structure. The BOC model is represented either by explicitly using beads and bonds (Fig. 2c) or using thick bonds to better reveal the contour (Figs. 4, 5 and 7).
Contour analysis
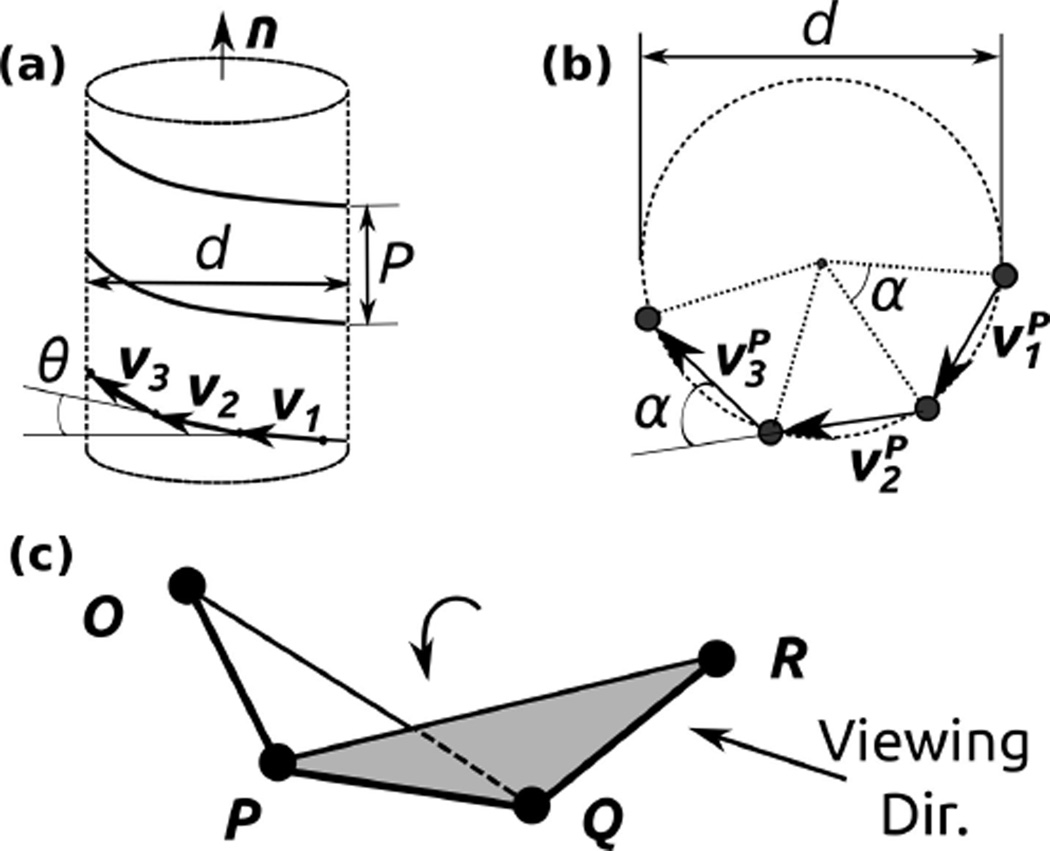
BOC models as constructed above in general have zigzag-shaped contours. To analyze their geometry, we extract a subset, only following either the large or small domains. Since identical domains are followed in the subset, the resulting contour has unique bond length, angle, and dihedral angle (Fig. 1a). We derive expressions for the parameters describing its helical contour. Since bond and dihedral angles are constant in the subset, three consecutive bonds are sufficient for determining the helical geometry. Denote the three bonds as vectors υ1, υ2, and υ3 (Fig. 1a). Let be the projection of vi on the plane perpendicular to the helix axis n (n is a unit vector). Since ,
| (1) |
| (2) |
Figure 1.
Description of the helical contour of the BOC model. (a) n: unit vector parallel to the helix axis. P: pitch (P < 0 for a left-handed helix and P > 0 for a right-handed helix). d: diameter. θ: rise angle between the i-th bond vector υi and the plane perpendicular to n. (b) Axis view. α: azimuthal angle. : projection of υi on the plane perpendicular to n. (c) Dihedral angle for three consecutive bonds , , and . The arrow labeled ‘Viewing Dir.’ provides the counterclockwise sense (positive dihedral angle up to 180°) in the rotation of the plane PQR relative to plane OPQ with as the rotation axis.
In Eqs. 1 and 2, υ1 · υ2 and υ1 · υ3 can be calculated from the coordinates of beads in the BOC model. The rise angle of the helix θ is the angle between υi and the plane perpendicular to the helix axis. Also, the angle between and is equal to the azimuthal angle α (Fig. 1b). We then get and , where b ≡ |υi|. We also have υi · n = b sin θ, which is independent of i. Subtracting Eq. 2 from Eq. 1 thus yields
| (3) |
By denoting υ12 ≡ υ1 · υ2, Eq. 1 becomes
| (4) |
Eq. 4 can be re-organized to give
| (5) |
Since the right hand side of Eq. 5 can be calculated using the coordinates of the BOC model, we can obtain α. The symmetry number N (number of subunits making one turn) is then given by
| (6) |
Knowing α, we can also get the rise angle θ using Eq. 3. The pitch of the helix is
| (7) |
The sign of P is determined by the sign of θ, which depends on the choice for the direction of n. We use the convention P > 0 for a right-handed helix. The diameter of the helix is given by:
| (8) |
For the BOC model composed of the large, hinge, and small domains, we calculated helical parameters for the two subset helices corresponding to the large and small domains. Due to the zigzag-like contour of the BOC, the diameters of the cylinders formed by large and small domains, respectively named dL and dS, are different. However, the pitch P and the symmetry number N are the same for the two cases.
Results and Discussion
ClpX
Within the hexameric ring of ClpX, the small domain in one subunit interfaces with the large one in the next subunit (Fig. 2a and c). As mentioned above, contact between the two subunits is tight, so that the large and small domains from neighboring subunits move almost as a single rigid body.10, 11 The intra-domain hinge motion can be shown by overlapping large domains in different subunits, which reveal the different orientations of the small domains (Fig. 2b). Notably, in the nucleotide-free state (chains C and F in Fig. 2), the small domain sterically blocks the ATP-binding pocket (Fig. 2b inset). This ‘blocked’ state of the nucleotide-free subunits likely accommodates conformational constraints imposed by other nucleotide-bound subunits in the ring. ATP hydrolysis and release in other subunits would thus be required to allow the blocked subunit to open and accept an ATP. In this way, nucleotide-dependent conformational changes of a subunit can affect the overall conformation of the ring, which in turn controls the subunit’s ATPase activity so that the order and phase of ATPase events across the subunits in the ring are properly laid out for processive force generation.20
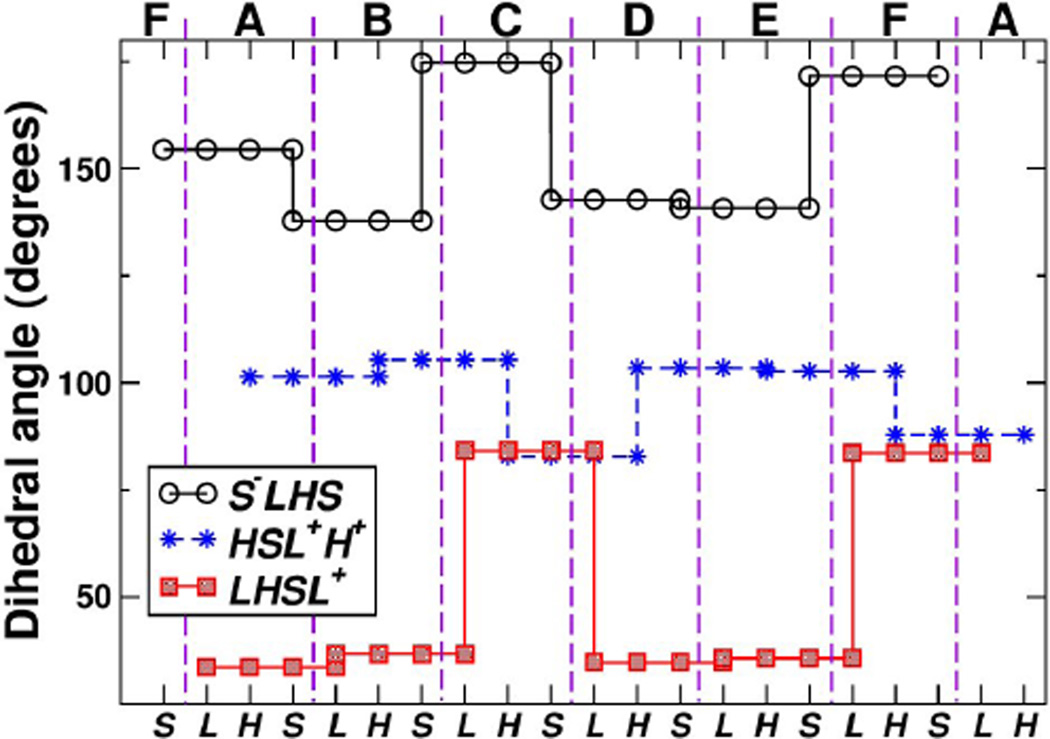
To better understand the ring’s contour, we constructed a BOC model (Fig. 2c; see Methods). Three beads named L, H, and S, are assigned respectively to the centers of mass of Cα atoms in the large, hinge, and small domains. Across the BOC ring, the bond lengths changed little: Å (mean±standard deviation), Å, and Å (superscript − or + on a bead name indicates that the bead belongs to the previous or the next subunit). Likewise, bond angles stayed almost constant (∠S−LH = 88.31° ±2.50°, ∠LHS = 108.9° ± 3.0°, ∠HSL+ = 87.91° ±1.85°). Therefore, large and small domains within each subunit, or between neighboring subunits, are not very extensible nor undergo appreciable bending motion. Instead, the conformational change arises mainly from dihedral motion (thick arrow in Fig. 2b). For four consecutive beads OPQR (they can be, for example, LHSL+), the dihedral angle ∠OPQR is defined as the angle between planes (as three points define a plane) spanned by points OPQ and PQR (Fig. 1c). It ranges between (−180°, 180°), where the positive sense follows the counterclockwise rotation of plane PQR relative to OPQ when viewed from bead Q in the direction of bead P. In Fig. 2b, the leftward rotation of the small domain from A to B is clockwise when viewed from above. This leads to a decrease of ∠S−LHS with as the rotation axis (circles in Fig. 3). The dihedral angle between B and C increases by a greater amount, consistent with the larger rightward motion from B to C in Fig. 2b. Similar changes can be seen between E and F. Changes in ∠HSL+H+ are less pronounced (stars in Fig. 3), which reflects that the hinge domain does not change its orientation greatly relative to the large and small domains of neighboring chains that form near-rigid contacts. Motion of the large domain, ∠LHSL+ with as the axis (squares in Fig. 3) shows a trend similar to that of ∠S−LHS. In all three types of dihedral angles, the largest changes are associated with chains C and F, which are nucleotide-free.
Figure 3.
Dihedral angles of four consecutive beads in the BOC model of ClpX. Top: Subunit (chain) names, delineated by vertical dashed lines (Fig. 2a). Due to the ring structure, subunits F and A reappear at both ends. Bottom: bead names (Fig. 2c). There are three types of dihedral angles, respectively formed by S−LHS, LHSL+, and HSL+H+ (legend). The same value of a dihedral angle is assigned to four consecutive beads that define it (Fig. 1c). For example, for chain C, the dihedral angle formed by S−LHS (S− is from chain B) is 175°. Changes in dihedral angles are the greatest for beads involving subunits C and F, which are nucleotide-free.
The nucleotide-dependent switching of dihedral angle has profound impact on the local twist of the ring’s contour. A previous study suggested that using only one chain in the nucleotide-bound state (A, B, D, or E in Fig. 2) leads to a lock-washer conformation instead of closing into a hexameric ring.10 Furthermore, ClpX assembled into a helix in another crystal structure.16 Thus, using a single value of dihedral angle in Fig. 3 may not be compatible with forming the toroidal geometry. To further investigate the effects of different dihedral angles on the ring’s local conformation, we constructed a multimer chain by connecting replicas of a subunit with its large domain optimally superposed to that of the next subunit in the ClpX ring (Methods). Instead of a ring, all of the resulting structures formed helices with varying pitch, symmetry, and diameter (Fig. 4). Most notably, the nucleotide-free subunits C and F formed helices with right-handed chirality (P > 0), opposite to the left-haded chirality of other chains with bound nucleotides. Furthermore, these right-handed helices had the smallest symmetry number, 4-fold, which suggest that they are the most incompatible within the hexameric ring form and thus may experience the greatest deformation.
Since having P > 0 or P < 0 across all subunits in ClpX would mean that a helix instead of a ring is formed, the right-handed chirality of nucleotide-free subunits C and F, compensate for the left-handed chirality of other nucleotide-bound subunits to maintain the topology of the ring. When nucleotide releases from one of the subunits, it in principle can take conformations of C and F. However, it is unlikely that two consecutive subunits can be in such a conformation, since it would result in nearly a 90°-bend due to the 4-fold symmetry shown in Fig. 4. It is thus expected that C and F conformations occur on oppositely positioned subunits, as in PDB 3HWS. Of note, another x-ray structure of ClpX with no nucleotide (PDB 3HTE) has similar arrangement of subunits where two in the blocked conformation are located oppositely.10 While it is possible that ClpX in the nucleotide-free state is conformationally flexible wherein the conformation of PDB 3HTE was chosen by the crystallization condition, these results demonstrate that a certain number of subunits in the ring need to be in the blocked state in order to maintain the ring structure, which is consistent with a previous solution experiment suggesting that at least two subunits remain nucleotide-free.13 Since changes in dihedral angles lead to twisting of the ring’s contour, pore loops lining the center of the ring will undergo the corresponding up-and-down motion along the ring’s axis, as suggested for HslU based on coarse-grained simulations.17 Local changes in the symmetry number will also affect the shape of the pore and orientation of the pore loops, as in Fig. 2a.
HslU
In contrast to ClpX (Fig. 2), available crystal structures of HslU are more symmetrically arranged, and structures of HslU exist with nucleotides bound to all six subunits, or with no bound nucleotide. 2,33 The less dramatic conformational changes associated with nucleotide binding may reflect that HslU can only pull a single polypeptide chain through its central pore, whereas ClpX operates on a wider range of substrates, and is even capable of pulling three strands simultaneously.10, 33 For HslU, we used PDB 1DO2 which has an alternation of nucleotide-free and nucleotide-bound subunits.2 Aligning the large domains of its subunits revealed nucleotide-dependent rotation of the small domain analogous to that of ClpX (Fig. 2b), but to a lesser extent.33 Due to the alternation of the two subunits, the BOC model of PDB 1DO2 has only two values for each of the geometric measures of the ring: , , and ; ∠S−LH = [76.7°, 78.5°], ∠LHS = [105°, 111°], and ∠HSL+ = [88.5°, 91.0°]; ∠S−LHS = [154°, 163°], ∠LHSL+ = [34.4°, 35.9°], and ∠HSL+H+ = [111°, 120°]. As for ClpX, the bond length of HslU changes little, and the dihedral angle varies more than the bond angle does. But overall the changes are less pronounced compared to ClpX.
Due to the 3-fold symmetry in the crystal’s unit cell, we constructed only two types of multimers that consist of either the nucleotide-free (APO) or nucleotide-bound subunits (Fig. 5). Helical pitches in both cases were less than 0.5 Å in magnitude, indicating that neither of the subunits generate out-of-plane strain. The two ‘rings’ differ instead in the symmetry number, with the APO state slightly less than 6-fold (N = 5.6) while the NT-bound state slightly above (N = 6.1). For a HslU ring that consists of the two types of subunits, tendency to have a higher symmetry number by the NT-bound subunits is thus compensated for by the opposite tendency of the APO subunits. It is interesting to note how this leads to changes in the size and geometry of the central pore of the ring. As Fig. 5 shows, the diameter of the circle formed by the center of masses of the large domain, dL, does not differ between the two rings. In the NT-bound multimer, on the other hand, dS increases by 7 A compared to the APO multimer, suggesting that the small domain moves outward. Thus, arrangement of large domains within the ring remains relatively fixed, while the small domains rotate and the I-domain moves inward (Fig. 5b). Similarly, in the case of ClpX, dS varies more than dL (Fig. 4). Since N = 5.6 in the APO state, in order to form a hexamer ring (N = 6), the subunits will need to open up, resulting in the increase of the pore size compared to that shown in Fig. 5a. Conversely, the requirement to decrease N from 6.1 to 6 would result in further narrowing of the central pore in Fig. 5b, which is consistent with the nucleotide-induced closure of the HslU ring observed in other crystal structures of HslU.32, 33
Dynein
X-ray structures of cytoplasmic dyneins have recently become available, with the highest resolution reaching to 2.8 Å.5,18, 19 Remarkably, the entire motor domain is made of a single polypeptide chain (Fig. 6). In addition to the canonical AAA+ domains, it also has an N-terminal linker, stalk, buttress, a 154-residue extension of the small domain of AAA5 (AAA5-extension), and a C-terminal domain (C-domain). In Fig. 6a, the AAA5-extension and the C-domain are located behind the AAA+ ring. Whereas the linker and the C-domain are attached to the termini of the AAA+ ring, the stalk, buttress, and AAA5-extension are insertions within the small domains. Since we use the large domains for alignment when constructing multimers of AAA+ domains, these insertions have little impact on our analysis. Without the insertions, the hexameric ring structure can be seen (Fig. 6b).
Belonging to an AAA+ clade different from that of the homohexameric ClpX and HslU,6 the AAA+ domains of dynein have greater sequence variations so that the number of amino acids and size of large and small domains differ within dynein. This can be seen in the corresponding BOC model. Even when extra regions such as the strut and buttress are excluded from the center of mass calculation, bond lengths vary more: Å, Å, and Å. Greater variation in indicates that the small domains have varying distances from the large domains for dynein function.5,18 Likewise, bond angles vary more: ∠S−LH = 72.31° ± 8.88°, ∠LHS = 121.8° ± 17.0°, and ∠HSL+ = 82.77° ± 11.57°, which are comparable to variations in dihedral angles: ∠S−LHS = 153.1° ± 11.8°, ∠LHSL+ = 44.74° ± 15.71°, and ∠HSL+H+ = 108.7° ± 9.55°.
Akin to ClpX, multimers built using replicas of individual AAA+ domains in dynein show great variability in contour. As shown in Fig. 7, the symmetry number N alternates above and below 6, where the maximum (8.5) and minimum (4.3) occur on AAA5 and AAA6. The multimers exhibit both left- and right-handed chirality. Diameters of small and large domains vary greatly, where the former varies more, as seen in ClpX and HslU. Among the six AAA+ domains, AAA5 and AAA6 experience the largest strain, as can be seen from their pitch and/or symmetry number. Unlike ClpX whose conformational changes are associated mostly with dihedral angles (Fig. 3), however, there is no clear correlation between the profile of dihedral angles in dynein and the multimer conformations. As mentioned above, bond length and bond angle also vary substantially among the subdomains of dynein, so that the higher strains of AAA5 and AAA6 are a combined effect of bond length, bond angle, and dihedral angle, rather than one geometric feature playing a dominant role. In PDB 3VKG, AAA1–AAA4 contain bound nucleotides, whereas AAA5 and AAA6 are nucleotide-free. Thus, although dynein may have altered individual AAA+ domains in order to control the attached moving elements such as the linker, stalk, and buttress, the basic mode of operation appears to be similar to that observed in ClpX, where the nucleotide-free domains deform the most in order to accommodate nucleotide-induced conformational changes in other domains.
Concluding Remarks
The present analysis elucidates common features in the nucleotide-dependent conformational changes among AAA+ motors, where local distortion of the ring’s contour by the nucleotide-bound subunits is countered by the nucleotide-free subunits in order to maintain the topology of the ring. The simple design wherein subunits communicate via mechanical strains generated by the topological constraint of the ring, as opposed to relying on specific amino acids, may be the basis for the widespread use of the AAA+ motors as engines of the cell.12 Furthermore, ClpP, the degradation chamber that binds to ClpX to form the protein destruction machinery, is barrel-shaped with 7-fold symmetry,23, 31 and it enhances the motility of ClpX on polypeptides.1 In addition to the local ring geometry, the boundary condition provided by the interface between ClpX and ClpP may thus play additional mechanical role for inter-subunit communication and processive motility. By contrast, AAA+ proteins in the clamp loader and initiator clades take helical forms instead of a ring, and not surprisingly, they work as single-action enzymes without processive movement on substrates.6
While the present analysis provides insight into the geometrical features of the nucleotide-dependent deformation of the ring as a whole, further studies are required to elucidate dynamical aspects of such conformational changes. For example, in the case of ClpX, single-molecule nanometry reveals that the subunits work in highly cooperative manner with a step size of 5–8 amino acids.1 In addition to cooperativity, a deeper understanding of other fundamental aspects in the AAA+ motor mechanisms remain, including the directionality of motion9, 17, 36 and the order in which individual AAA+ domains work.20, 21
Acknowledgment
This work was funded in part by the National Institute of Health grant R01GM087677 (W.H. and M.J.L.), and the National Science Foundation Career Award 0643745 (M.J.L.).
Footnotes
Conflict of interest. We declare no conflict of interest.
Contributor Information
Wonmuk Hwang, Department of Biomedical Engineering, Materials Science & Engineering Program, Texas A&M University, College Station, TX 77843, U.S.A.; School of Computational Sciences, Korea Institute for Advanced Study, Seoul 130-722, Korea
Matthew J. Lang, Departments of Chemical and Biomolecular Engineering, Molecular Physiology and Biophysics Vanderbilt University, Nashville, TN 37240, U.S.A.
References
- 1.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HslU and the ATP-dependent protease HslU-HslV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 3.Brooks BR, Brooks CL, III, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 5.Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 7.Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Protein Sci. 2008;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 9.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynn SE, Nager AR, Baker TA, Sauer RT. Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat. Struct. Mol. Biol. 2012;19:616–622. doi: 10.1038/nsmb.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Molec. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 13.Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: Allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 15.Hwang W, Lang MJ. Mechanical Design of Translocating Motor Proteins. Cell Biochem. Biophys. 2009;54:11–22. doi: 10.1007/s12013-009-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- 17.Koga N, Kameda T, Okazaki K, Takada S. Paddling mechanism for the substrate translocation by AAA+ motor revealed by multiscale molecular simulations. Proc. Natl. Acad. Sci. USA. 2009;106:18237–18242. doi: 10.1073/pnas.0904756106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8-Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 19.Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 20.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr. Op. Struct. Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 22.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 23.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 24.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shindyalov IN, Bourne PE. CE: A resource to compute and review 3-D protein structure alignments. Nucleic Acid Res. 2001;29:228–229. doi: 10.1093/nar/29.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 27.Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Op. Struct. Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 29.van Ooijen G, Millar AJ. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem. Sci. 2012;37:484–492. doi: 10.1016/j.tibs.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta Molec. Cell Res. 2011;1823:56–66. doi: 10.1016/j.bbamcr.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase–ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 34.Wendler P, Ciniawsky S, Kock M, Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta Molec. Cell Res. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Whiteheart SW, Schraw T, Matveeva EA. N-ethylmaleimide sensitive factor (NSF) structure and function. Int. Rev. Cytol. 2001;207:71–112. doi: 10.1016/s0074-7696(01)07003-6. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimoto K, Arora K, Brooks CL., III Hexameric helicase deconstructed: interplay of conformational changes and substrate coupling. Biophys. J. 2010;98:1449–1457. doi: 10.1016/j.bpj.2009.12.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]