Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) is a disease of progressive lung fibrosis with a high mortality rate. In organ repair and remodeling, epigenetic events are important. MicroRNAs (miRNAs) regulate gene expression post-transcriptionally and can target epigenetic molecules important in DNA methylation. The miR-17∼92 miRNA cluster is critical for lung development and lung epithelial cell homeostasis and is predicted to target fibrotic genes and DNA methyltransferase (DNMT)-1 expression.
Objectives: We investigated the miR-17∼92 cluster expression and its role in regulating DNA methylation events in IPF lung tissue.
Methods: Expression and DNA methylation patterns of miR-17∼92 were determined in human IPF lung tissue and fibroblasts and fibrotic mouse lung tissue. The relationship between the miR-17∼92 cluster and DNMT-1 expression was examined in vitro. Using a murine model of pulmonary fibrosis, we examined the therapeutic potential of the demethylating agent, 5′-aza-2′-deoxycytidine.
Measurements and Main Results: Compared with control samples, miR-17∼92 expression was reduced in lung biopsies and lung fibroblasts from patients with IPF, whereas DNMT-1 expression and methylation of the miR-17∼92 promoter was increased. Several miRNAs from the miR-17∼92 cluster targeted DNMT-1 expression resulting in a negative feedback loop. Similarly, miR-17∼92 expression was reduced in the lungs of bleomycin-treated mice. Treatment with 5′-aza-2′-deoxycytidine in a murine bleomycin-induced pulmonary fibrosis model reduced fibrotic gene and DNMT-1 expression, enhanced miR-17∼92 cluster expression, and attenuated pulmonary fibrosis.
Conclusions: This study provides insight into the pathobiology of IPF and identifies a novel epigenetic feedback loop between miR-17∼92 and DNMT-1 in lung fibrosis.
Keywords: microRNA, miR-17∼92, pulmonary fibrosis, DNA methylation, DNMT-1
At a Glance Commentary
Scientific Knowledge on the Subject
Through the use of genome-wide studies, investigators have attempted to uncover the molecular changes responsible for the development of idiopathic pulmonary fibrosis (IPF). However, these studies have been met with little success. Recently, microRNAs (miRNA) have emerged as regulators of numerous genes and unique targets in IPF. The mechanisms by which miRNAs are altered in IPF remain unknown.
What This Study Adds to the Field
We identified an association between aberrant DNA methylation and miRNA expression in IPF. We report that miRNA expression in IPF is susceptible to epigenetic regulation. Based on this observation, our study suggests the potential for targeting DNA methylation events as a therapeutic approach for IPF.
Idiopathic pulmonary fibrosis (IPF) represents the most aggressive form of interstitial lung disease with a median survival of 3–5 years (1). Failure to resolve epithelial cell injury in the lung is critical to the pathogenesis of IPF (2–4). In addition, epithelial-mesenchymal transition (EMT) (5), fibroblast proliferation and activation (6), and recruitment of inflammatory cells (7, 8) all contribute to extracellular matrix accumulation in the lung (7). The current study focused on identifying the molecular mechanisms underlying the pathogenesis of IPF.
Because changes in fibrotic gene expression (2, 9–11) and few genetic mutations have been identified in IPF (12, 13), we focused on microRNA (miRNA, miR) expression and epigenetic regulators in lung epithelial cells and fibroblasts. MiRNAs can either block translation or degrade target mRNAs (14, 15). Notably, a single miRNA can regulate upward of 30 genes. MiRNAs can be encoded in intronic or exonic DNA regions and encoded in their own open reading frame and controlled by DNA promoter elements, such as DNA methylation by DNA methyltransferases (DNMTs) of CpG islands (15, 16). Of the three DNMTs expressed in humans, DNMT-1 is associated with epigenetic repair of injured tissue, whereas DNMT-3a and -3b are critical for and de novo maintenance of DNA methylation (17, 18).
Based on previously observed changes in miRNA expression in IPF lung tissue (9), we focused on the miR-17∼92 cluster, which encodes six miRNAs within a single open reading frame (19, 20). In solid tumors, miR-17∼92 expression is elevated and targets antiangiogenic and fibrotic genes (19, 21–25), many of which are altered in IPF (2, 9, 10). This cluster is also important in lung epithelial cell development (26) with high expression during embryonic development, and then declines into adulthood (15). Importantly, mice lacking the miR-17∼92 cluster, but not the duplicate paralog copies (miR-106a∼363 and miR-106b∼25), have few lung epithelial cells and die from asphyxia at birth (26). Conversely, lung-specific miR-17∼92 overexpression in mice results in a highly proliferative undifferentiated lung epithelium (27). Thus, this miRNA cluster seems to maintain lung epithelial cell homeostasis, an important factor in effective lung repair.
Here, we demonstrate that epigenetic silencing of miR-17∼92 occurred in lung tissue and fibroblast cell lines from patients with IPF because of enhanced DNA methylation. Diminished miR-17∼92 expression inversely correlated to DNMT-1 expression. Introduction of the miR-17∼92 cluster in IPF lung fibroblasts reduced fibrotic gene and DNMT-1 expression, normalized cellular phenotype, and reduced DNA methylation of the cluster. We further investigated if this regulation was conserved in mice. In a murine model of pulmonary fibrosis, enhancing miR-17∼92 expression using a demethylating agent in vivo reduced fibrotic gene and DNMT-1 expression suggesting augmented in vivo lung repair. These data reveal a potential new therapeutic approach for IPF and intimate interplay among miR-17∼92, DNMT-1 activity, and lung fibrosis.
Methods
Primary Tissue Samples
Deidentified tissue samples from patients with IPF and chronic obstructive pulmonary disease (COPD) were acquired through the Lung Tissue Research Consortium. IPF and COPD samples were segregated into three FVC groups and two FVC groups, respectively. Patient clinical information and control samples were previously described (9).
Cell Culture and Treatments
Normal and IPF lung fibroblasts (American Type Culture Collection, Rockville, MD) were cultured per instructions. Cells (2–3 × 106) were treated with 0.5–2 μM of 5′-aza-2′-deoxycytidine for 24–72 hours. Nonspecific toxicity was measured. Fibroblasts (1 × 106) were transfected using siPORTNeoFX transfection reagent for 24 hours.
Bleomycin-induced Pulmonary Fibrosis and 5′-Aza-2′-Deoxycytidine Treatment in Mice
C57/BL6 mice (Jackson Laboratory, Bar Harbor, ME) were injected intraperitoneally with 0.035 U bleomycin/kg or vehicle (phosphate-buffered saline) (28). After 2 weeks, mice were injected intraperitoneally twice weekly with endotoxin-free 0.156 mg/kg 5′-aza-2′-deoxycytidine or dimethyl sulfoxide on alternating days in respect to bleomycin injections. Tissue and primary cells were isolated (28, 29).
Quantitative Real-Time Polymerase Chain Reaction Analyses
RNA was isolated and subjected to polymerase chain reaction analysis for miRNA and mRNA expression (9, 30). For mRNA analysis, the endogenous controls glyceraldehyde phosphate dehydrogenase and adenylyl cyclase–associated protein-1 (CAP-1) were used for normalization. For miRNA analysis, several endogenous controls including miR-191, small nucleolar (sno) RNA, RNU38B, RNU48, snoRNA 202, and snoRNA 220 were evaluated for each experiment. The endogenous control with the most consistent Ct value and little variation was used for normalization as indicated.
Examination of DNA Methylation Patterns
DNA was isolated using Perfect Pure DNA Cultured Cell Kit (5 Prime Inc., Gaithersburg, MD) or QIAamp DNA Mini Kit (Qiagen, Valencia, CA) from cells or tissue, respectively. DNA methylation was analyzed using the Methyl-Profiler DNA Methylation qPCR Primer Assays (SABiosciences/Qiagen, Frederick, MD) according to instructions.
miRNA Colocalization Studies
Antibodies recognizing AE 1/3, CD45, and CD31 were used to colocalize with miR-19b and -20a expression by in situ hybridization within fixed lung tissue (31, 32). In situ hybridization was performed as previously described (31) using 5′-digoxigenin–labeled LNA probes (1–2 pmol/μl) for either miR-19b or -20a.
Luciferase Reporter Experiments
The 3′ untranslated region (UTR) of DNMT-1 cloned into pGL-3 Luciferase reporter vector was provided by Dr. Muller Fabbri (The Ohio State University) (33) and the 3′ UTR of Col13a in the pEZX-MT05 reporter vector was purchased from GeneCopoeia (Rockville, MD). The miRNA recognition sites were mutated as indicated in the online supplement. Firefly and Renilla luciferase activities were measured consecutively in HEK 293 cells transfected with reporter vectors (33).
Statistical Analysis
All data are expressed as the mean ± SD. One-way analysis of variance was performed with SPSS16 (SPSS Inc., Chicago, IL) and JMP/SAS v9.1 software (SAS Institute, Inc., Cary, NC). Holm method was used to adjust for multiplicity and control the overall type I error rate at α = 0.05. To test for outliers, normality test was performed and none were identified. Statistical significance was defined as P less than 0.05.
Results
miR-17∼92 Expression Is Decreased in Lung Tissue from Patients with IPF
Given the predictive value of lung function in IPF patient outcomes (1), we stratified IPF lung tissue into severity groups based on FVC: group 1, FVC less than 50% (severe); group 2, FVC 50–80% (moderate); and group 3, FVC greater than 80% (mild). IPF lung tissue samples demonstrated reduced expression of pre-miRNAs (data not shown) and mature miRNAs encoded by the miR-17∼92 cluster compared with control lung tissue samples including pathologically normal lung tissue adjacent to lung cancer (Figure 1A). Based on miRNA target prediction software programs, the miR-17∼92 cluster was predicted to target several fibrotic genes including collagen, type I, α1 (Col1a1), transforming growth factor (TGF)-β, and metalloproteinases (Table 1). We validated that expression of the above mRNA targets of the miR-17∼92 cluster was increased in lung tissue from patients with IPF (see Figure E1A in the online supplement).
Figure 1.
Decreased expression of the miR-17∼92 cluster in human idiopathic pulmonary fibrosis. (A) Expression of each microRNA (miRNA) from the miR-17∼92 cluster was determined by quantitative real-time polymerase chain reaction from control (n = 10), >80% FVC (n = 7), 50–80% FVC (n = 8), and <50% FVC (n = 9) lung tissue samples. Data were normalized to miR-191. Relative copy number (RCN, 2−dCT × 100) was determined. Data are expressed as the average RCN ± SD, *P < 0.018 compared with control tissue. Comparison of the mild disease (80% FVC) with control tissue for miR-19b and -92, P = 0.1325 and 0.1320, respectively. (B) In situ hybridization was performed using LNA-modified DNA probes for let-7c (positive control) and miR-19b and -20a at a magnification of ×400. Scrambled probes were used as a negative control. The arrows denote positively stained cells (dark blue/purple). Shown are representative images (n = 3 per group).
TABLE 1.
miRNAs PREDICTED TO REGULATE GENE EXPRESSION INVOLVED IN IDIOPATHIC PULMONARY FIBROSIS
| Genes | Predicted miRNAs |
||||
| COL13A1 | miR-19a,b | miR-18a,b | miR-29a,b | miR-377 | miR-544 |
| COL1A1 | miR-92a,b | miR-19a,b | miR-29a,b,c | miR-224 | miR-196a |
| CTGF | miR-18a,b | miR-19a,b | miR-559 | miR-600 | miR-377 |
| VEGF | miR-17 | miR-20a | miR-106a,b | miR-93 | miR-363 |
| TGF-β | miR-17 | miR-19a,b | miR-20a | miR-29a,b | miR-34b,c |
| TSP-1 | miR-17 | miR-18a,b | miR-19a,b | miR-20a | miR-34a.c |
| MMP-1 | miR-20a | miR-19a,b | miR-29a,c | miR-190b | miR-22 |
| MMP-9 | miR-19a,b | miR-92a,b | miR-581 | miR-34a,c | miR-223 |
| MMP-7 | miR-17 | miR-18a,b | miR-34b,c | miR-106b | miR-122 |
Definition of abbreviations: COL = collagen; CTGF = connective tissue growth factor; miRNA = microRNA; MMP = metalloproteinases; TGF-β = transforming growth factor-β; TSP-1 = thrombospondin-1; VEGF = vascular endothelial growth factor.
To determine if similar changes occurred in lung tissue from patients with other chronic lung diseases, we examined miR-17∼92 cluster expression in lung tissue from patients with COPD. In contrast to IPF lung tissue, we observed significant elevation of the miRNAs in lung tissue from patients with COPD compared with control tissue samples (see Figure E1B), suggesting that reduced lung miR-17∼92 cluster expression was not a nonspecific feature of chronic lung disease.
Detection of miR-19b and -20a by In Situ Hybridization
Members of the miR-17∼92 cluster, including miR-19b and -20a, were assessed in normal lung tissue by in situ hybridization (Figure 1B). Interestingly, miR-19b expression colocalized to lung epithelial cells expressing AE1/3 cytokeratin (see Figure E2). In IPF lung tissue, miR-19b and -20a were nearly undetectable in lung tissue from patients with moderate (50–80% FVC) and severe (<50% FVC) lung disease sections staining positive for α smooth muscle actin (α-SMA) and CD45 (see Figures E3 and E7). As expected, there was no nonspecific staining of miRNAs in tissue stained with scrambled probes. A probe recognizing let-7c miRNA was used as a positive control and was similarly expressed among all tissue samples. Importantly, miRNAs from the miR-17∼92 cluster, including miR-19b and -20a, were expressed in freshly isolated lung epithelial and fibroblast cells from murine lung tissue (see Figure E4A).
Introduction of the miR-17∼92 Cluster in IPF-derived Lung Fibroblasts Results in Molecular and Phenotypic Changes
In IPF, fibroblast activation leads to extracellular matrix deposition (6, 34). Therefore, we analyzed miR-17∼92 expression in human normal and IPF lung fibroblast cell lines. Similar to IPF lung tissue, miR-17∼92 expression was decreased in IPF fibroblast cell lines compared with normal lung fibroblast cell lines (see Figure E4B).
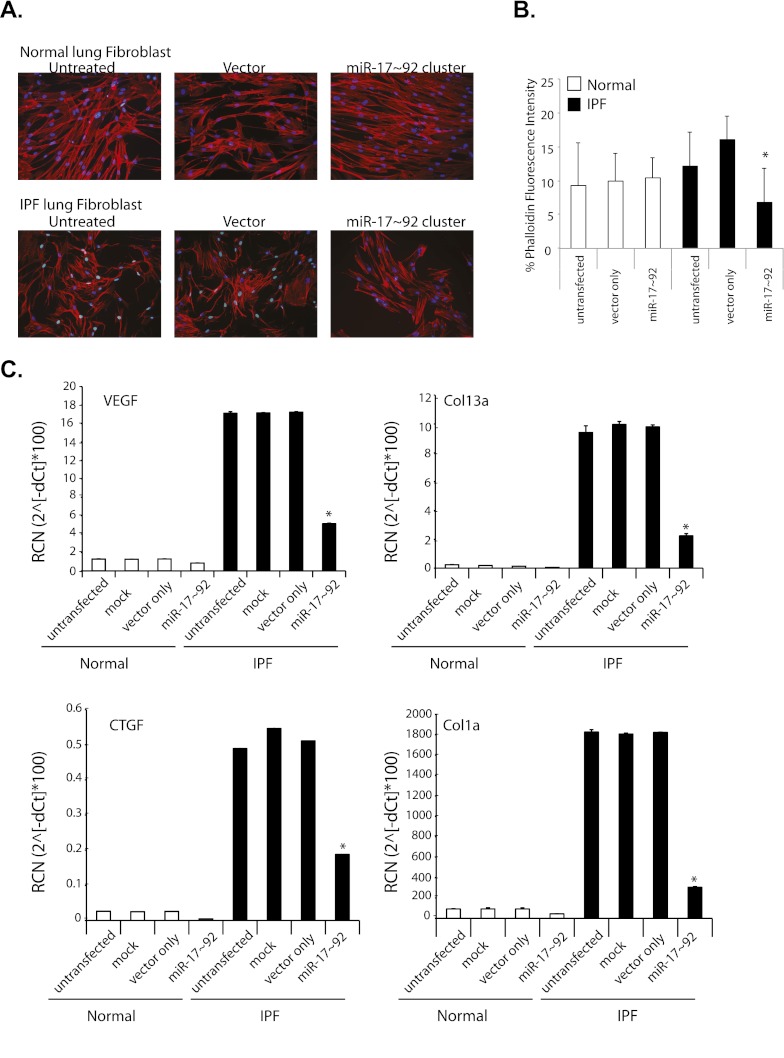
Gene profiles in IPF lung tissue are consistent with EMT and wound repair (2, 5, 9–11). Wound repair involves fibroblast migration to areas of damage (35) and cells undergoing EMT gain filopodia as they assume fibroblast phenotypes (5, 35). After transfecting miR-17∼92 in IPF and normal fibroblast cell lines, cell morphology, target gene, and miR-17∼92 cluster expression were assessed (Figure 2; see Figure E4B). Cytoskeleton morphology as indicated by numerous filopodia and increased actin staining was different in untransfected and vector-transfected IPF lung fibroblasts compared with the normal lung fibroblast samples (Figures 2A and 2B). Introduction of the miR-17∼92 cluster into IPF cells reduced the actin staining to levels similar to the normal fibroblast (Figure 2B). Interestingly, overexpression of the miR-17∼92 cluster did not alter the normal lung fibroblasts (Figure 2B).
Figure 2.
Introduction of miR-17∼92 in idiopathic pulmonary fibrosis (IPF) lung fibroblasts induces phenotypic and molecular changes. Normal and IPF lung fibroblasts were transfected with an expression vector containing the miR-17∼92 cluster. Empty vector-transfected cells (vector) also served as a negative control. (A) After 24 hours, cells were stained with phalloidin and DAPI 0.5 μg/ml then photographed using an Olympus inverted fluorescent at ×10 magnification. Shown are representative images from three independent experiments. (B) To quantity actin fibers, the red fluorescence of phalloidin was quantitated and indicated as a percentage compared with cell number as enumerated by DAPI-positive nuclei. Shown is the average intensity ± SD. Significant decrease was apparent in miR-17∼92 transfected cells compared with vector-only transfected IPF cells, *P = 0.0327; differences in phalloidin staining for IPF vector-transfected IPF cells compared with untransfected (P = 0.2549) or vector-transfected (P = 0.3534) normal lung fibroblasts. (C) RNA was isolated and quantitative real-time polymerase chain reaction was performed for the indicated genes. Data were normalized using glyceraldehyde phosphate dehydrogenase as a housekeeping control. Data are expressed as the average relative copy number (RCN) ± SD from three experiments. Comparison was made between vector-only transfected IPF cells to the miR-17∼92 transfected IPF cells, *P < 0.004. COL = collagen; CTGF = connective tissue growth factor; VEGF = vascular endothelial growth factor.
Transfection of the miR-17∼92 cluster in IPF fibroblasts reduced vascular endothelial growth factor (VEGF), connective tissue growth factor (CTGF), Col1a1, and Col13a1 expression compared with untransfected, mock-transfected, and vector-transfected cells (Figure 2C). In contrast, gene expression was relatively unchanged in normal fibroblast cell lines transfected with the miR-17∼92 cluster compared with control samples. VEGF and CTGF have been shown to be directly regulated by the miR-17∼92 cluster (19, 21, 22, 24, 25), whereas Col1a is indirectly targeted by miR-17∼92 regulation of TGF-β (23). Because Col13a1 was a predicted target of the cluster, we confirmed that it is directly regulated by the miRNAs contained in the miR-17∼92 cluster (see Figure E4C). As a negative control, HIF1α mRNA expression, highly expressed in lung tissue (36, 37) and not a predicted target of the miR-17∼92 cluster, was unchanged in miR-17∼92–transfected normal and IPF lung fibroblasts (see Figure E4D).
Altered DNA Methylation Patterns of the CpG Island in the miR-17∼92 Cluster Promoter in IPF
Because c-MYC transcriptionally regulates miR-17∼92 cluster expression (19, 20), we evaluated its expression in lung tissue from patients with IPF and found no differences in mRNA or protein expression in lung tissue or fibroblasts from patients with IPF (data not shown), suggesting alternative mechanisms accounted for decreased miR-17∼92 expression in IPF.
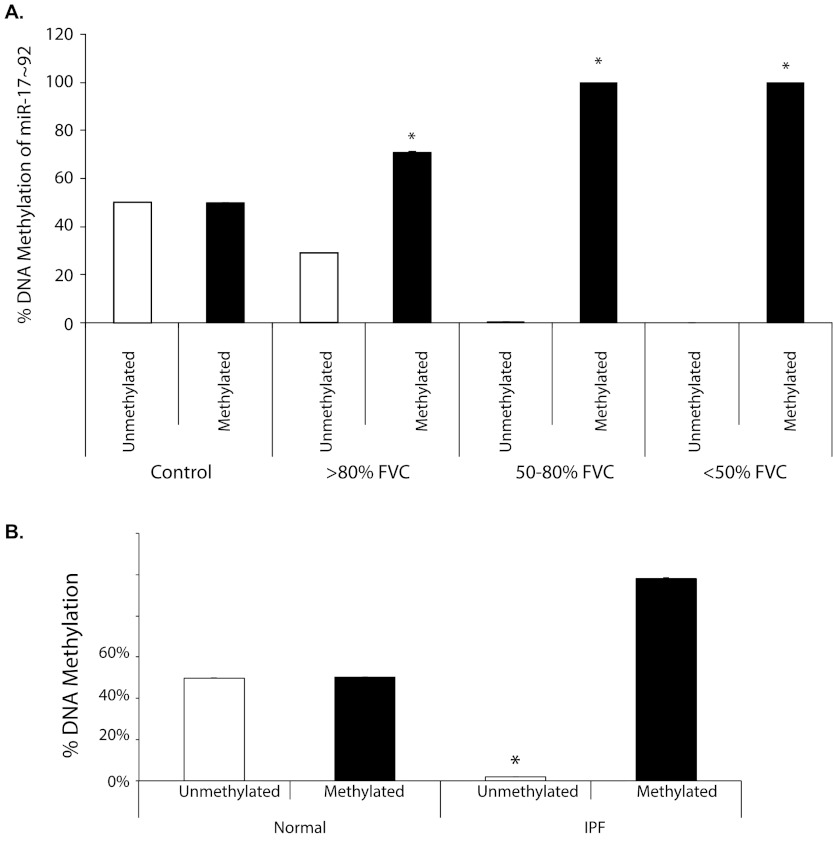
Interestingly, more than 80% of the miR-17∼92 promoter is heavily occupied with a large CpG island. We next hypothesized that this CpG island was aberrantly methylated in lung tissue from patients with IPF, reducing miR-17∼92 cluster expression. In lung tissue from patients with IPF, DNA methylation of the miR-17∼92 promoter was significantly increased compared with control lung tissue (Figure 3A). No unmethylated DNA was detected for the miR-17∼92 promoter in lung tissue from patients with moderate (50–80% FVC) and severe (<50% FVC) IPF, whereas unmethylated DNA in the miR-17∼92 promoter was present in mild disease (>80% FVC). Interestingly, a 20% increase in DNA methylation from baseline seen in mild disease samples seemed sufficient to reduce miR-17∼92 expression (Figure 1A). In contrast, control lung tissue had an equal distribution of unmethylated and methylated DNA in the miR-17∼92 cluster promoter. Similarly, the miR-17∼92 promoter was methylated in lung fibroblasts derived from patients with IPF (Figure 3B). Thus, our data suggested that silencing of the miR-17∼92 cluster occurred through DNA methylation of its promoter.
Figure 3.
Increased DNA methylation of the miR-17∼92 promoter in idiopathic pulmonary fibrosis (IPF). (A) DNA was isolated from control lung tissue (n = 3) and individuals with IPF (n = 3 per severity group). Samples were selected from the cohort used in Figure 1. Data are presented as the average percent of unmethylated or methylated DNA of the miR-17∼92 promoter ± SD. Statistical difference was determined by comparing the % unmethylated with % methylated for each severity group, *P < 0.003. (B) DNA was isolated from normal or IPF fibroblasts. DNA methylation for the miR-17∼92 promoter was determined. Shown is the percent DNA methylation, n = 6 (*P < 0.0001 normal fibroblast compared with IPF fibroblast for unmethylated or methylated DNA).
Inhibition of DNA Methylation in Lung IPF Fibroblasts Leads to Phenotypic and Genetic Changes
We next examined whether 5′-aza-2′-deoxycytidine, a compound that inhibits DNA methylation, restored miR-17∼92 cluster expression in IPF fibroblasts. Treating IPF fibroblasts with 5′-aza-2′-deoxycytidine for 24 hours increased miR-17, -18a, -19b, and -20a expression (Figure 4A). In contrast, there was no change in miR-19a and -92a expression. Interestingly, gene expression was different between miR-17∼92–transfected and 5′-aza-2′-deoxycytidine–treated IPF fibroblasts (Figure 4B). Notably, Col13a1 expression was decreased in miR-17∼92–transfected cells (Figure 2C) but not in 5′-aza-2′-deoxycytidine–treated cells (Figure 4B). Similar to cells transfected with the miR-17∼92 cluster (Figure 2A), IPF fibroblasts treated with 5′-aza-2′-deoxycytidine exhibited a more normal phenotype (see Figure E5A).
Figure 4.
Treatment of idiopathic pulmonary fibrosis (IPF) lung fibroblasts with 5′-aza-2′-deoxycytidine liberates microRNA (miRNA) expression and down-regulates target mRNAs. IPF lung fibroblast cell line was either treated with the vehicle control dimethyl sulfoxide (DMSO) or treated with 0.5 μM 5′-aza-2′-deoxycytidine (5′-Aza) for 24 hours. RNA was isolated and subjected to quantitative real-time polymerase chain reaction analysis for (A) miRNA or (B) fibrotic genes. Data were normalized to (A) RNU48 or (B) CAP-1 expression. Shown is the average relative copy number (RCN) ± SD from two independent experiments. COL = collagen; CTGF = connective tissue growth factor; VEGF = vascular endothelial growth factor.
Increased Expression of DNMT-1 in IPF
To define the mechanisms regulating DNA methylation of the miR-17∼92 cluster in IPF lung tissue, we next examined DNMT-1 expression. DNMT-1 is the most abundant DNMT in mammalian cells (17, 18, 38) and is a predicted target of the miR-17∼92 cluster. Interestingly, DNMT-1 mRNA expression increased as FVC declined in IPF lung tissue and inversely correlated with miR-17∼92 cluster expression (Figure 5A). Similarly, DNMT-1 expression was elevated in IPF lung fibroblast cell lines compared with normal fibroblast cell lines (see Figure E5C). Introduction of the miR-17∼92 cluster in IPF fibroblast cell lines significantly decreased DNMT-1 expression compared with vector control-transfected samples (see Figure E5C). These observations suggest that DNMT-1 contributed to the hypermethylation of the miR-17∼92 promoter region in IPF lung tissue. Notably, treatment of the IPF lung fibroblast cell line with 5′-aza-2′-deoxycytidine also decreased DNMT-1 expression (Figure 5B).
Figure 5.
DNA methyltransferase (DNMT)-1 is altered in idiopathic pulmonary fibrosis (IPF) and a target of the miR-17∼92 cluster. (A) DNMT-1 expression was examined by quantitative real-time polymerase chain reaction from a select cohort of tissue samples shown in Figure 1 (n = 3, per each disease severity group and control tissue samples). Using glyceraldehyde phosphate dehydrogenase as an endogenous control, the average relative copy number (RCN) ± SD was calculated. Significance *P < 0.05 is shown. (B) IPF lung fibroblasts cells were transfected with or without miR-19b or 20a antagomir to knock down (KD) their expression. After treatment with 5′-aza-2′-deoxycytidine (5′ Aza) for 24 hours, RNA was extracted and DNMT-1 expression quantitated by quantitative real-time polymerase chain reaction. DNMT-1 expression was normalized to glyceraldehyde phosphate dehydrogenase in vehicle-treated untransfected cells. Shown is the average fold-change ± SD (n = 3). Significance for 5′ Aza–treated cells compared with 5′Αza/miR-19b KD cells is *P < 0.001. No clinical difference was apparent between 5′ Αza–treated cells and 5′ Αza/miR-20a KD cells. (C) The wild-type (WT) or mutated (mut) 3′ untranslated region for DNMT-1 was cloned into the pGL-3 Firefly luciferase vector and transfected in the presence of the indicated microRNAs (miRNAs) in HEK 293 cells. Irrelevant scrambled miRNA served as a control. Cells were also cotransfected with the Renilla luciferase construct, pRL-TK. Luciferase production was measured first for the Firefly luciferase followed by the Renilla luciferase from the culture supernatant after 24 hours. Firefly luciferase was normalized to the Renilla luciferase. Shown is the average luciferase production from eight independent experiments (± SD). WT DNMT-1 was compared with the mutated DNMT-1 for each specified miRNA, *P < 0.001.
To understand the relationship between DNMT-1 and miR-17∼92 expression, IPF lung fibroblast cell lines were treated with 5′-aza-2′-deoxycytidine in the presence of an antagomir to either miR-19b or -20a to reduce miRNA-specific expression. After treatment with the antagomir to miR-19b, treatment with 5′-aza-2′-deoxycytidine no longer decreased DNMT-1 expression (Figure 5B). In contrast, similar studies using the miR-20a antagomir did not alter DNMT-1 expression in the presence of 5′-aza-2′-deoxycytidine (Figure 5B), indicating that miR-19b primarily regulated DNMT-1 expression.
DNMT-1 Expression Is Directly Regulated by the miR-17∼92 Cluster
We identified seed sequences for miR-17, -19b, -20a, and -92a in the 3′ UTR of DNMT-1. Using a luciferase system, transfection of miR-17, -19b, -20a, or -92a into HEK-293 cells expressing the 3′-UTR wild-type DNMT-1-luc construct reduced cellular luciferase activity, an effect abrogated by mutating the DNMT-1 seed sequence (Figure 5C). As expected, there was no change in DNMT-1 luciferase production in the presence of miR-19a and -18a, which do not have a predicted seed sequence in the 3′ UTR for DNMT-1.
5′-Aza-2′-Deoxycytidine Alters Gene Expression in Murine Model of Pulmonary Fibrosis
Using a murine model of pulmonary fibrosis, we evaluated whether 5′-aza-2′-deoxycytidine treatment altered miR-17∼92 DNA methylation, fibrotic gene expression, and lung fibrosis. Similar to the pathologic fibrotic pattern observed in human IPF, mice given bleomycin twice weekly by intraperitoneal injection for 4 weeks develop subpleural fibrosis of the lung (28). In this model, fibrosis appears in the lungs within 2 weeks of treatment (28). To assess the use of demethylating agents in lung fibrosis, mice were treated for 2 weeks with intraperitoneal bleomycin injections before treatment with 5′-aza-2′-deoxycytidine or vehicle during the final 2 weeks of bleomycin treatment. As shown in Figure 6A, repeated systemic bleomycin treatment reduced miR-17∼92 cluster expression in murine lung tissue, which was abrogated by 5′-aza-2′-deoxycytidine treatment, even when started 2 weeks after bleomycin injections started. Although not significant (P = 0.1348), 5′-aza-2′-deoxycytidine treatment altered lung pathology after 2 weeks of bleomycin (Figures 6B and 6C) and significantly reduced collagen, VEGF, CTGF, and DNMT-1 gene expression (Figures 6D and 6E). Notably, 5′-aza-2′-deoxycytidine alone enhanced the endogenous expression of the miR-17∼92 cluster leading to a reduction in the indicated genes in the lung tissue. As predicted, 5′-aza-2′-deoxycytidine abolished bleomycin-induced DNA methylation of the miR-17∼92 cluster compared with mice treated with bleomycin alone (Figure 6F).
Figure 6.
In vivo treatment of mice with 5′-aza-2′-deoxycytidine attenuates bleomycin-induced fibrotic gene expression. Mice were treated with 0.035 U/kg twice weekly for 4 weeks with bleomycin (n = 4) or phosphate-buffered saline (PBS) alone (n = 4). A set of bleomycin-treated mice were injected with 0.156 mg/kg/wk of 5′-aza-2′-deoxycytidine intraperitoneally for the last 2 weeks of bleomycin treatment (n = 4) and designated as Bleomycin (4 wk) 5′ Aza (2 wk). As a control, mice were treated with PBS alone for 4 weeks or PBS for 2 weeks and then 5′-aza-2′-deoxycytidine for 2 weeks without bleomycin (5′Aza [2 wk]). (A) RNA was extracted from the lung tissue and microRNA (miRNA) expression was examined by quantitative real-time polymerase chain reaction and normalized using snoRNA-202 expression. Shown is the average relative copy number (RCN) ± SD, *P value < 0.001 compared with bleomycin only treated mice. (B) Lung tissue was section and paraffin-embedded then subjected to trichrome staining to detect collagen deposition. The larger image is at ×10 magnification, whereas the inset is a ×4 magnification. Shown are representative images from a mouse from each group. (C) Trichrome sections were blindly assessed by a board-certified pathologist. The average arbitrary score ± SD is shown (n = 4 per group). Significance compared with bleomycin-only treated mice, *P < 0.0075 and **P = 0.1348. (D) RNA was subjected to quantitative real-time polymerase chain reaction for the indicated genes and (E) DNA methyltransferase (DNMT)-1 expression. CAP-1 served as an endogenous control for normalization for fibrotic genes and DNMT-1. Data are expressed as the relative copy number (RCN) ± SD, *P value < 0.001 compared with bleomycin-treated mice. (F) DNA were isolated and subjected to analysis for promoter DNA methylation of the miR-17∼92 cluster. Data presented is the average % unmethylated and % methylated, n = 3 mice per treatment group. *P value < 0.001 compared with bleomycin-only treated mice. COL = collagen; CTGF = connective tissue growth factor; VEGF = vascular endothelial growth factor.
Discussion
The molecular mechanisms underlying IPF are unknown. Lung injury followed by dysfunctional lung repair seems to be a mechanism for lung pathology in IPF (3, 4, 34). Insights from familial IPF also suggest a role for ineffective lung repair and remodeling (12) associated with enhanced DNA methylation (39). Moreover, lung epithelial homeostasis is critical in pulmonary fibrosis, because mutations in surfactant protein C, produced by lung epithelial cells, result in lung fibrosis (13). Given that ineffective lung epithelial cell repair and excessive collagen production are important in IPF (4, 34), we examined mechanisms connecting these processes. Thus, we focused on expression of the miR-17∼92 cluster, which is critically important in lung epithelial cell development (15, 26, 27) and targets critical fibrotic genes (19, 21–25). Here, we report that the miR-17∼92 cluster was decreased in lung tissue from patients with IPF accompanied by enhanced DNA methylation of its promoter. We also found decreased expression of several members from the paralog miR-17∼92 clusters (miR-106a∼363 and miR-106b∼25) (data not shown) suggesting an unlikely compensatory role for these miRNAs. Moreover, a Food and Drug Administration–approved demethylating agent, 5′-aza-2′-deoxycytidine, increased the expression of the miR-17∼92 cluster. Notably, miR-17∼92 expression was reduced in lung tissue from patients with IPF but not in COPD lung tissue.
In IPF, changes in fibrotic gene expression in lung tissue are present, including metalloproteinases, collagen, and growth factors (2, 9–11). We therefore hypothesized that these changes were regulated by miRNAs and may contribute to disease pathogenesis. Notably, TGF-β–stimulated overexpression of miR-21 is important in human and murine models of pulmonary fibrosis, and may amplify fibrogenesis by targeting negative regulators of TGF-β signaling in lung myofibroblasts (40). Targeting miR-21 in mice attenuates the effects of intratracheal bleomycin. Furthermore, several miRNAs are down-regulated in IPF including let-7d and miR-17 and -92a (9, 41, 42).
Highly expressed genes in IPF were subjected to miRNA prediction analysis. Interestingly, miRNAs contained in the miR-17∼92 cluster were predicted not only to target numerous fibrotic genes, but also DNMTs that play important roles in epigenetic gene regulation suggesting that IPF may be a disease of aberrant DNA methylation (43, 44). Thus, we found that the miR-17∼92 cluster specifically targeted DNMT-1. Introducing the miR-17∼92 cluster into IPF lung fibroblasts reduced promoter methylation of the cluster and fibrotic gene expression.
An inverse relationship between the miR-17∼92 cluster and DNMT-1 expression was observed in IPF lung tissue. Importantly, DNMT-1 is recruited to sites of DNA damage where it regulates epigenetic events (18, 38). Notably, α-SMA expression is inversely regulated by DNMT expression in rat ex vivo fibroblasts and TGF-β–induced myofibrobast differentiation (43). Here, we report decreased expression of miR-19b and -20a from human lung tissue from patients with IPF accompanied by enhanced α-SMA expression (Figure 1; see Figure E3). Using either the miR-17∼92 cluster or 5′-aza-2′-deoxycytidine reduced DNMT-1 expression in human cells and reduced actin expression. Notably, we did not examine the relationship with DNMT expression and α-SMA expression in cells from mice with bleomycin-induced pulmonary fibrosis. In light of our findings and that of Hu and colleagues (43), it is possible that subtle differences for α-SMA regulation exist between species.
Proteins that bind methylated DNA can either repress or activate gene expression. These include methyl DNA binding proteins (MBD) 1 and MBD2 and methyl CpG binding protein (MeCP2) (44). Mice lacking MeCP2 have decreased collagen deposition and myofibroblast differentiation in response to intratracheal injection of bleomycin (44). Notably, the miR-17∼92 cluster is predicted to target MBD2 and MeCP2 but not MBD1. Thus, it is conceivable that epigenetic regulation by the miR-17∼92 cluster is critically regulated by targeting multiple proteins.
DNMTs are regulated by miRNAs in human cancer and some miRNAs are regulated by epigenetic events (45). In addition to the regulatory activity of miR-19b in regulating DNMT-1, miR-29b indirectly regulates DNMT-1 expression (45). Cushing and colleagues (46) found that increased TGF-β signaling suppresses miR-29 family members in bleomycin-induced fibrosis. In human IPF lung tissue samples, we and others found changes in miR-29 family members compared with control tissue samples (9, 42). More recently, Xiao and coworkers (47) demonstrated that enforced expression of miR-29 in the lung of mice subjected to intratracheal bleomycin attenuates lung fibrosis. Although miR-148a and -152 also target DNMT-1 (48), we did not detect significant changes in their expression in fibrotic lung tissue (9).
Altering epigenetic changes with the chemotherapeutic drug 5′-aza-2′-deoxycytidine is clinically used in treating humans with myelodysplastic syndrome and acute myelogenous leukemia (49). However, this drug is associated with myelosuppression caused by neutropenia, thrombocytopenia, and anemia. Interestingly, 5′-aza-2′-deoxycytidine treatment of IPF fibroblasts increased the expression of most miRNAs contained in the miR-17∼92 cluster and decreased the expression of fibrotic gene targets. Of note, the fibrotic genes studied here have single or multiple CpG islands. However, the promoters of most of these genes are unmethylated in control and IPF tissue (see Figure E6) suggesting a critical role for miRNAs in the regulation of their expression.
We investigated the preclinical use of 5′-aza-2′-deoxycytidine in murine pulmonary fibrosis induced by intraperitoneal bleomycin. Treatment of bleomycin-treated mice with 5′-aza-2′-deoxycytidine after the initiation of fibrosis reduced fibrotic and DNMT-1 gene expression while restoring miR-17∼92 cluster expression. Although lung pathology was not altered significantly in these mice, our data suggest that 5′-aza-2′-deoxycytidine treatment most likely does not mediate the breakdown of collagen already present but rather prevents the deposition of additional collagen. Bechtel and coworkers (50) found that 5′-aza-2′-deoxycytidine attenuated renal fibrosis by reducing fibrotic genes and decreasing DNMT-1–induced methylation of RASAL1, a regulator of Ras activation. Although TGF-β regulates RASAL1 expression transcriptionally and through DNMT-1, it is important to note that several miRNAs from the miR-17∼92 cluster are predicted to target RASAL1 and TGF-β.
To our knowledge, this represents the first report showing that reexpression of the miR-17∼92 cluster using 5′-aza-2′-deoxycytidine leads to reduced fibrotic gene expression in vitro and in vivo. The current study suggests that epigenetic modifying drugs may have a therapeutic benefit for IPF.
Supplementary Material
Acknowledgments
The authors appreciate the assistance of Ms. Jennifer Price, Battelle Memorial Institute, in isolating RNA from human tissue samples as previously described. They appreciate the animal support provided by Ms. Christie Newland and Ariel Basye from the Center for Critical Care Animal Core, The Ohio State University. They also recognize the biostatisticians Dr. Xiaokui Mo and Mr. Gary Phillips for statistical assistance, Center for Biostatistics, The Ohio State University.
Footnotes
Supported by NIH/NHLBI grants R01 HL102464 and HL067176 (C.B.M.) and the American Thoracic Society (M.G.P.).
Author Contributions: M.G.P. and C.B.M. conceived the ideas, and designed and wrote the paper. M.G.P. and D.D. performed, oversaw, and analyzed studies. K.B. and Y.W. assisted in the polymerase chain reaction studies. V.P.W. assisted in data analysis. G.N. and A.M. assisted in in situ hybridization. C.Z.C.-S. assisted in lung cell isolations. P.Y. assisted in DNA methylation studies. C.L.H. performed pathologic analysis of lung tissue sections. S.P.N.-S. assisted in writing the paper and provided scientific input.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201205-0888OC on January 10, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ley B, Collard HR, King TE., Jr Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2011;183:431–440 [DOI] [PubMed] [Google Scholar]
- 2.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci USA 2000;97:1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magro CM, Waldman WJ, Knight DA, Allen JN, Nadasdy T, Frambach GE, Ross P, Marsh CB. Idiopathic pulmonary fibrosis related to endothelial injury and antiendothelial cell antibodies. Hum Immunol 2006;67:284–297 [DOI] [PubMed] [Google Scholar]
- 4.Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J 2007;30:835–839 [DOI] [PubMed] [Google Scholar]
- 5.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos C, Montano M, Garcia-Alvarez J, Ruiz V, Uhal BD, Selman M, Pardo A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am J Respir Cell Mol Biol 2001;24:591–598 [DOI] [PubMed] [Google Scholar]
- 7.Bringardner BD, Baran CP, Eubank TD, Marsh CB. The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxid Redox Signal 2008;10:287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keane MP, Strieter RM, Lynch JP, III, Belperio JA. Inflammation and angiogenesis in fibrotic lung disease. Semin Respir Crit Care Med 2006;27:589–599 [DOI] [PubMed] [Google Scholar]
- 9.Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhallah D, Price J, Bornman D, Zhang S, et al. Systems biology of interstitial lung diseases: integration of mRNA and microRNA expression changes. BMC Med Genomics 2011;4:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 10.Kaminski N, Rosas IO. Gene expression profiling as a window into idiopathic pulmonary fibrosis pathogenesis: can we identify the right target genes? Proc Am Thorac Soc 2006;3:339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med 2008;5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, III, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med 2007;356:1317–1326 [DOI] [PubMed] [Google Scholar]
- 13.Lawson WE, Grant SW, Ambrosini V, Womble KE, Dawson EP, Lane KB, Markin C, Renzoni E, Lympany P, Thomas AQ, et al. Genetic mutations in surfactant protein C are a rare cause of sporadic cases of IPF. Thorax 2004;59:977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–866 [DOI] [PubMed] [Google Scholar]
- 15.Nana-Sinkam SP, Karsies T, Riscili B, Ezzie M, Piper M. Lung microRNA: from development to disease. Expert Rev Respir Med 2009;3:373–385 [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer 2008;98:1881–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bestor TH, Verdine GL. DNA methyltransferases. Curr Opin Cell Biol 1994;6:380–389 [DOI] [PubMed] [Google Scholar]
- 18.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 1999;27:2291–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, et al. The Myc-miR-17∼92 axis blunts TGF-β signaling and production of multiple TGF-β-dependent antiangiogenic factors. Cancer Res 2010;70:8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005;435:839–843 [DOI] [PubMed] [Google Scholar]
- 21.Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. De-repression of CTGF via the miR-17–92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 2010;29:3411–3422 [DOI] [PubMed] [Google Scholar]
- 22.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 2006;1:e116.1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Shi JY, Zhu GQ, Shi B. Mir-17–92 cluster regulates cell proliferation and collagen synthesis by targeting TGFβ pathway in mouse palatal mesenchymal cells. J Cell Biochem 2012;113:1235–1244 [DOI] [PubMed] [Google Scholar]
- 24.Lungu G, Kuang X, Stoica G, Wong PK. Monosodium luminol upregulates the expression of Bcl-2 and VEGF in retrovirus-infected mice through downregulation of corresponding mirnas. Acta Virol 2010;54:27–32 [DOI] [PubMed] [Google Scholar]
- 25.van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, Cleutjens JP, van Zandvoort MA, Heymans S, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 2011;10:769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17–92 family of miRNA clusters. Cell 2008;132:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microrna miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 2007;310:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baran CP, Fischer SN, Nuovo GJ, Kabbout MN, Hitchcock CL, Bringardner BD, McMaken S, Newland CA, Cantemir-Stone CZ, Phillips GS, et al. The transcription factor ets-2 plays an important role in the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol 2011;45:999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, Thompson JC, Caserta E, Wang H, Chong JL, et al. PTEN in stromal fibroblasts suppresses mammary epithelial tumours. Nature 2009;461:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008;3:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc 2009;4:107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun 2008;373:607–612 [DOI] [PubMed] [Google Scholar]
- 33.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b. Proc Natl Acad Sci USA 2007;104:15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acharya PS, Zukas A, Chandan V, Katzenstein AL, Pure E. Fibroblast activation protein: a serine protease expressed at the remodeling interface in idiopathic pulmonary fibrosis. Hum Pathol 2006;37:352–360 [DOI] [PubMed] [Google Scholar]
- 35.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol 2008;9:446–454 [DOI] [PubMed] [Google Scholar]
- 36.Wiener CM, Booth G, Semenza GL. In vivo expression of mRNAS encoding hypoxia-inducible factor 1. Biochem Biophys Res Commun 1996;225:485–488 [DOI] [PubMed] [Google Scholar]
- 37.Shimoda LA, Semenza GL. HIF and the lung: role of hypoxia-inducible factors in pulmonary development and disease. Am J Respir Crit Care Med 2011;183:152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci USA 2005;102:8905–8909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 2008;39:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. MiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2011;207:1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;182:220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 2011;157:191–199 [DOI] [PubMed] [Google Scholar]
- 43.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol 2010;177:21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am J Pathol 2011;178:1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3a and 3b and indirectly DNMT1. Blood 2009;113:6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. MiR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 2011;45:287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. MiR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 2012;20:1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braconi C, Huang N, Patel T. Microrna-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010;51:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claus R, Almstedt M, Lubbert M. Epigenetic treatment of hematopoietic malignancies: in vivo targets of demethylating agents. Semin Oncol 2005;32:511–520 [DOI] [PubMed] [Google Scholar]
- 50.Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, Salant DJ, Muller CA, Kalluri R, Zeisberg M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 2011;16:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.