Abstract
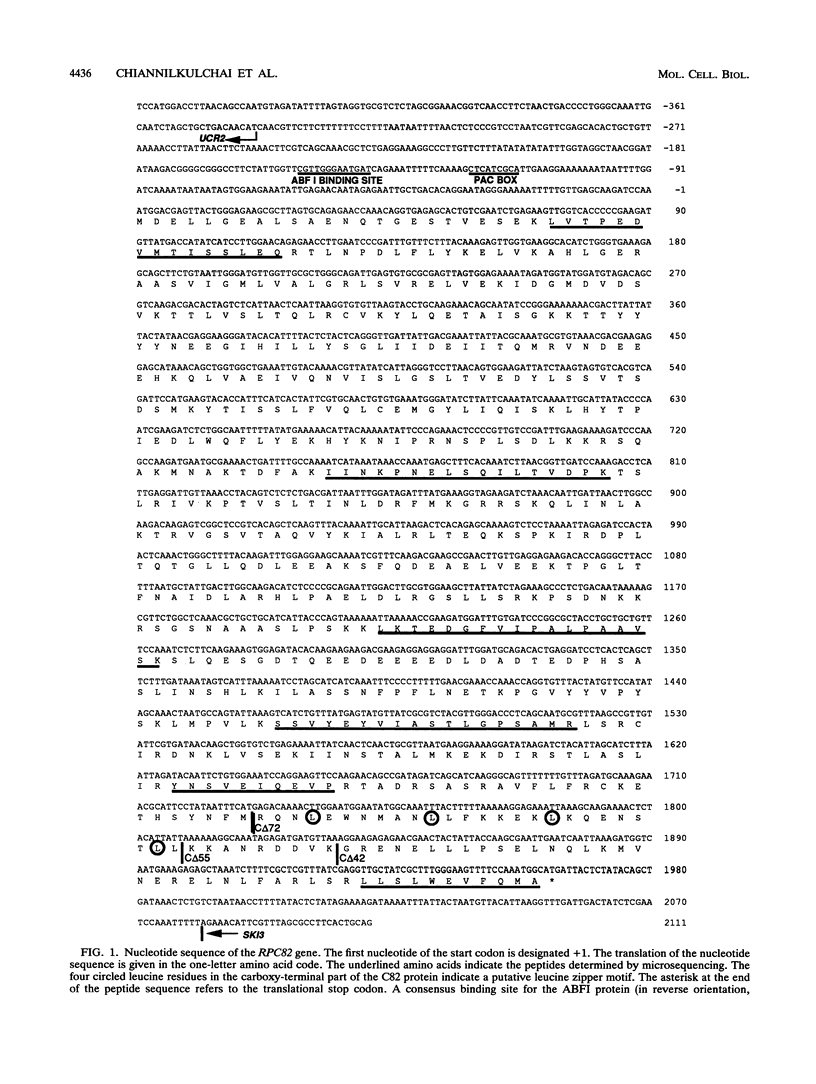
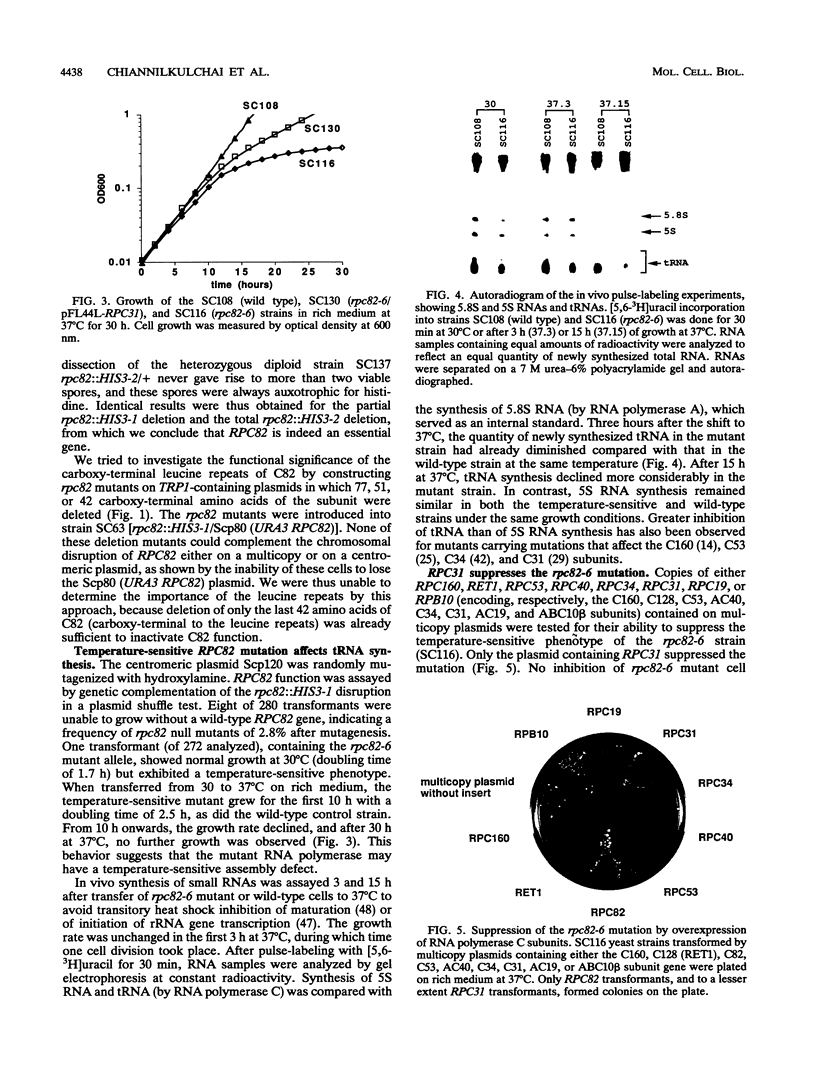
RNA polymerase C (III) promotes the transcription of tRNA and 5S RNA genes. In Saccharomyces cerevisiae, the enzyme is composed of 15 subunits, ranging from 160 to about 10 kDa. Here we report the cloning of the gene encoding the 82-kDa subunit, RPC82. It maps as a single-copy gene on chromosome XVI. The UCR2 gene was found in the opposite orientation only 340 bp upstream of the RPC82 start codon, and the end of the SKI3 coding sequence was found only 117 bp downstream of the RPC82 stop codon. The RPC82 gene encodes a protein with a predicted M(r) of 73,984, having no strong sequence similarity to other known proteins. Disruption of the RPC82 gene was lethal. An rpc82 temperature-sensitive mutant, constructed by in vitro mutagenesis of the gene, showed a deficient rate of tRNA relative to rRNA synthesis. Of eight RNA polymerase C genes tested, only the RPC31 gene on a multicopy plasmid was capable of suppressing the rpc82(Ts) defect, suggesting an interaction between the polymerase C 82-kDa and 31-kDa subunits. A group of RNA polymerase C-specific subunits are proposed to form a substructure of the enzyme.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Archambault J., Schappert K. T., Friesen J. D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol. 1990 Dec;10(12):6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Busch S. J., Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990 Feb;6(2):36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- Della Seta F., Treich I., Buhler J. M., Sentenac A. ABF1 binding sites in yeast RNA polymerase genes. J Biol Chem. 1990 Sep 5;265(25):15168–15175. [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J Biol Chem. 1991 Aug 15;266(23):15300–15307. [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Faye G., Kujawa C., Fukuhara H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S ribosomal RNA in petite mutants of saccharomyces cerevisiae. J Mol Biol. 1974 Sep 5;88(1):185–203. doi: 10.1016/0022-2836(74)90304-0. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Sentenac A. RNA polymerase III (C) and its transcription factors. Trends Biochem Sci. 1991 Nov;16(11):412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Gudenus R., Mariotte S., Moenne A., Ruet A., Memet S., Buhler J. M., Sentenac A., Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E., Saumweber H., Dallendörfer A., Stein H. RNA polymerase III from Drosophila hydei pupae. Purification and partial characterization. Eur J Biochem. 1980 Oct;111(2):395–401. doi: 10.1111/j.1432-1033.1980.tb04953.x. [DOI] [PubMed] [Google Scholar]
- Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985 Dec 5;260(28):15304–15310. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Whelen S., Hall B. D. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J Biol Chem. 1991 Mar 25;266(9):5616–5624. [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991 Sep;11(9):4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Mann C., Micouin J. Y., Chiannilkulchai N., Treich I., Buhler J. M., Sentenac A. RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol Cell Biol. 1992 Oct;12(10):4314–4326. doi: 10.1128/mcb.12.10.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhoc E., Masson J. M., Teissié J. High efficiency transformation of intact yeast cells by electric field pulses. Biotechnology (N Y) 1990 Mar;8(3):223–227. doi: 10.1038/nbt0390-223. [DOI] [PubMed] [Google Scholar]
- Mosrin C., Riva M., Beltrame M., Cassar E., Sentenac A., Thuriaux P. The RPC31 gene of Saccharomyces cerevisiae encodes a subunit of RNA polymerase C (III) with an acidic tail. Mol Cell Biol. 1990 Sep;10(9):4737–4743. doi: 10.1128/mcb.10.9.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosrin C., Thuriaux P. The genetics of RNA polymerases in yeasts. Curr Genet. 1990 May;17(5):367–373. doi: 10.1007/BF00334516. [DOI] [PubMed] [Google Scholar]
- Oudshoorn P., Van Steeg H., Swinkels B. W., Schoppink P., Grivell L. A. Subunit II of yeast QH2:cytochrome-c oxidoreductase. Nucleotide sequence of the gene and features of the protein. Eur J Biochem. 1987 Feb 16;163(1):97–103. doi: 10.1111/j.1432-1033.1987.tb10741.x. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Rhee S. K., Icho T., Wickner R. B. Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast. 1989 May-Jun;5(3):149–158. doi: 10.1002/yea.320050304. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 May 25;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar V. E., Jaehning J. A., Gage L. P., Roeder R. G. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase III from the posterior silk gland of Bombyx mori. J Biol Chem. 1976 Jun 25;251(12):3794–3800. [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R. N., Conesa C., Lefebvre O., Carles C., Ruet A., Quemeneur E., Gagnon J., Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Hager G. L., Weinberg F., Rutter W. J. Molecular structure of yeast RNA polymerase III: demonstration of the tripartite transcriptive system in lower eukaryotes. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1024–1028. doi: 10.1073/pnas.73.4.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veinot-Drebot L. M., Singer R. A., Johnston G. C. Heat shock causes transient inhibition of yeast rRNA gene transcription. J Biol Chem. 1989 Nov 25;264(33):19473–19474. [PubMed] [Google Scholar]
- Veinot-Drebot L. M., Singer R. A., Johnston G. C. rRNA transcription initiation is decreased by inhibitors of the yeast cell cycle control step "start". J Biol Chem. 1989 Nov 25;264(33):19528–19534. [PubMed] [Google Scholar]
- Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol. 1992 Mar;12(3):1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]






