Summary
Although distinct human induced pluripotent stem cell (hiPSC) lines can display considerable epigenetic variation, it has been unclear if such variability impacts their utility for disease modeling. Here, we show that although low passage female hiPSCs retain the inactive X-chromosome of the somatic cell they are derived from, over time in culture they undergo an “erosion” of X-chromosome inactivation (XCI). This erosion of XCI is characterized by loss of XIST expression and foci of H3-K27-trimethylation, as well as transcriptional de-repression of genes on the inactive X that cannot be reversed by either differentiation or further reprogramming. We specifically demonstrate that erosion of XCI has a significant impact on the use of female hiPSCs for modeling Lesch-Nyhan syndrome. However, our finding that most genes subject to XCI are de-repressed by this erosion of XCI suggests that it should be a significant consideration when selecting hiPSC lines for modeling any disease.
Introduction
There is considerable interest in the use of human induced pluripotent stem cells (hiPSC) for the study of X-linked diseases, in particular for the understanding of cellular processes that lead to X-linked mental retardation and autism spectrum disorders (Chiurazzi et al., 2008; Marchetto et al., 2010). However, it remains to be settled how X-chromosome inactivation (XCI) is regulated during reprogramming and during the long-term culture of female hiPSC lines. Thus, it is difficult to interpret phenotypes in female hiPSC lines that are caused by X-linked mutations. For instance, it has been suggested that hiPSC lines derived from girls with Rett syndrome behave like mouse iPS cells in that reactivation of the inactive X-chromosome occurs during reprogramming and that random XCI can then subsequently occur during differentiation (Marchetto et al., 2010). However, other studies have concluded that early passage hiPSC lines contain an inactive X (Xi) chromosome and that this Xi may be the same Xi originally contained within the somatic cell that was reprogrammed (Pomp et al., 2011; Tchieu et al., 2010). If X-linked models of human disease are to be properly interpreted, it will be critical to resolve how the X-chromosome is regulated in female hiPSCs.
Lesch-Nyhan syndrome (LNS) is caused by mutations in the X-linked HPRT gene and is characterized by profound behavioral and neurological symptoms, including mental retardation, self-mutilation, and motor-dysfunction (Jinnah, 2009; Visser et al., 2000). LNS is generally observed in HPRT−/Y males, while HPRT+/HPRT- carrier females are generally non-symptomatic. Mutations in HPRT interfere with the purine salvage pathway (Baumeister and Frye, 1985; McDonald and Kelley, 1971). However, it is unknown why mutations in this pathway most severely affect the nervous system. Unfortunately, HPRT mutant mice do not recapitulate many of the neurological phenotypes observed in patients (Engle et al., 1996; Finger et al., 1988). We reasoned that derivation of hiPSC lines from individuals with HPRT mutations might allow a robust human cellular model of LNS to be established. Furthermore, because mutations in HPRT allow for isolation of somatic cells with a given status of XCI, we hypothesized that LNS hiPSCs might also allow resolution of the ambiguity surrounding XCI during human reprogramming.
Here, we report that hiPSCs can be used to generate an in vitro model for LNS. Additionally, our results confirm that early passage female hiPSC lines contained an inactive X-chromosome (Pomp et al., 2011; Tchieu et al., 2010) and that this inactive chromosome was in every case the same inactive X-chromosome found in the somatic fibroblast that they were derived from. However, as we subjected these hiPSCs to long term culture, we found that female cell lines lost foci of both histone H3 lysine-27 tri-methylation (H3K27me3) and XIST RNA, and that these events coincided with ectopic reactivation of the functional HPRT gene from the inactive X. We further show that this “erosion” of dosage compensation was not reversed by either differentiation or another round of reprogramming, and had phenotypic ramifications when hiPSC were used for modeling LNS. Consequently, female lines that have lost XCI marks no longer exhibited the LNS phenotype when differentiated into neurons. Through multiple lines of evidence, including functional assays of HPRT activity and analysis of X-chromosome-wide levels of DNA methylation and transcription, we show that erosion of dosage compensation results in the gradual de-repression of the vast majority of genes normally subjected to XCI. Thus, if hiPSCs are to be used for disease modeling of X-linked disorders, it will be critical to carefully monitor their state of XCI.
Results
Reprogramming does not change XCI status
To create a stem cell model for LNS, we used retroviral transduction to generate hiPSC lines using fibroblasts isolated from a mutant male patient (HPRT−/Y) (Dimos et al., 2008; Takahashi et al., 2007). We designated these mutant male hiPSC lines LNS-1 and LNS-2. As further controls, we also derived several new male hiPSC lines from BJ fibroblasts and designated these lines WT-M-1 and WT-M-2. We also produced control female hiPSC lines from wi-38 fibroblasts and named these lines WT-F-1 through WT-F-3 (Figures 1A–1C, Figure S1, Figure S2). In addition, for some experiments, we used the male control hiPSC line 20b, which we have recently described (Boulting et al., 2011).
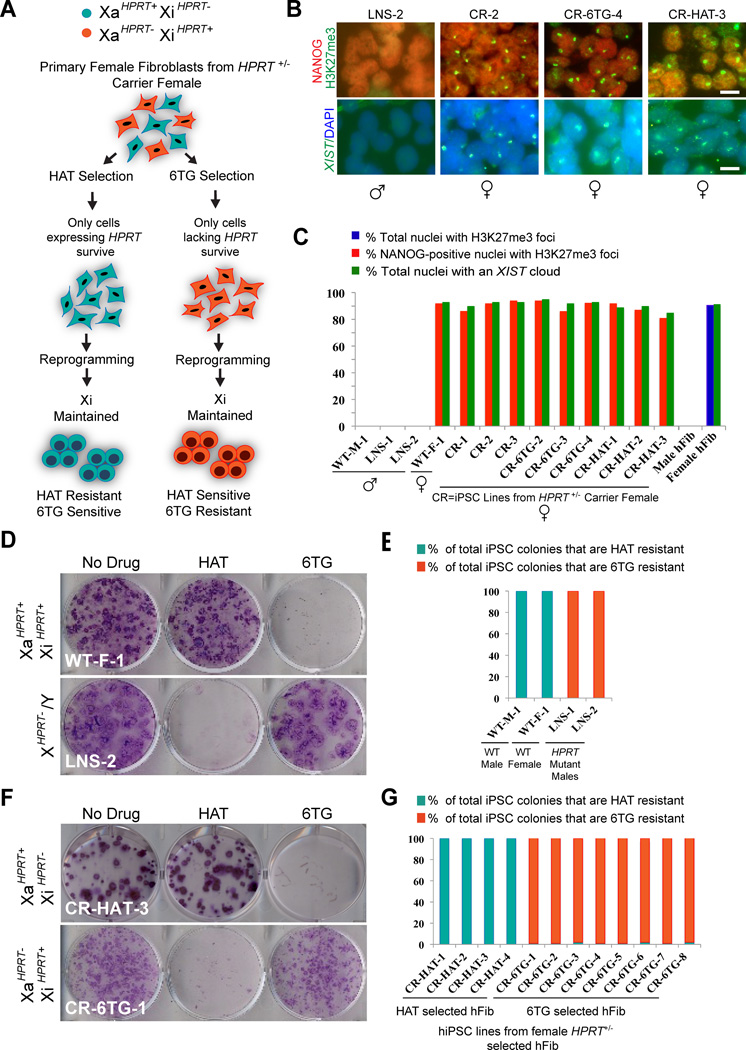
Figure 1. Generation of hiPSC lines from LNS affected males and carrier females.
(A) Strategy for generating fibroblasts with a known inactive X-chromosome for reprogramming to hiPSC and results.
(B) XIST FISH and NANOG/H3K27me3 staining for new hiPSC lines at passage 5–6. (CR=female carrier lines for the HPRT mutation, CR-6TG-1–8=iPS lines derived from 6TG treated fibroblasts, CR-HAT 1–4=iPS lines derived from HAT treated fibroblasts, LNS 1–2=iPS lines derived from HPRT−/Y male fibroblasts). Scale bars=15µm.
(C) Quantification of NANOG-positive cells with H3K27me3 foci and XIST cloud at passage six in newly derived hiPSC lines and in control male and female fibroblasts (hFib).
(D) Crystal violet staining of newly derived HPRT +/+ or HPRT−/Y hiPSC lines at passage 6 after treatment with media containing HAT or 6TG.
(E) Quantification of iPSC colonies that are HAT or 6TG resistant in wildtype and HPRT deficient male iPSC lines.
(F) Crystal violet staining of HPRT +/− hiPSC lines after treatment with media containing HAT or 6TG.
(G) Quantification of hiPSC colonies that are HAT or 6TG resistant in four hiPSC lines from HAT selected fibroblasts (CR-HAT-1–4) and eight hiPSC lines from 6TG selected fibroblasts (CR-6TG-1–8). See also Figures S1 and S2 for additional data.
Because it has been suggested that XCI can be exploited to generate isogenic control and experimental hiPSC lines (Cheung et al., 2010; Tchieu et al., 2010), we also generated hiPSCs from the patient’s phenotypically normal mother (HPRT+/−). HPRT +/− fibroblasts from this woman would be expected to be composed of two cell-types, those that had chosen the functional HPRT allele for inactivation (XaHPRT−XiHPRT+) and those that had inactivated the mutant allele (XaHPRT+XiHPRT−). It is known that cells selecting the functional HPRT allele for inactivation, and therefore lacking HPRT activity, are resistant to the toxic nucleotide analog 6-thio-guanine (6TG). Conversely, cells that express the functional HPRT allele can grow in the presence of hypoxanthine, aminopterin, thymidine (HAT) containing medium. We used this biochemical distinction between cells in the population to select fibroblasts with a known inactive X status, using HAT selection to isolate homogenous populations of XaHPRT+XiHPRT− fibroblasts and 6TG to select for fibroblasts that were XaHPRT−XiHPRT+ (Figure 1A, Figure S1).
To monitor the fate of the inactive X-chromosome during reprogramming of female fibroblasts, we produced hiPSCs by retroviral transduction (Takahashi et al., 2007) using several distinct populations of fibroblasts. We reprogrammed fibroblasts that were subjected to HAT selection, fibroblasts subjected to 6TG selection, and the original non-selected, female carrier fibroblasts (Figure S1, Figure S2). We named the female hiPSC lines derived from the non-selected fibroblasts CR-1 through CR-3 to designate their carrier status. hiPSC lines derived from HAT selected fibroblasts were named CR-HAT-1 through CR-HAT-4, and lines from 6TG selected fibroblasts were named CR-6TG-1 through CR-6TG-8. To determine the X-inactivation status of these new hiPSC lines, we performed XIST RNA FISH as well as co-stained cultures with antibodies specific to NANOG and H3K27me3 (Figures 1B–1C). Male hiPSCs and fibroblasts neither exhibited foci of H3K27me3, nor a “cloud” of XIST RNA (n=3 hiPSC lines, n=2 fibroblasts) (Figure 1A–1B). In contrast, each of the low passage (p5–6) female hiPSC lines was homogenously composed of cells that displayed a cloud of XIST and a focus of H3K27me3, similar in appearance and frequency to those found in female fibroblasts (n=10 hiPSC lines, n=2 fibroblast lines) (Figures 1B–1C).
To examine whether there had been transient reactivation of the Xi during reprogramming, or if instead they had retained the same Xi of the fibroblasts they were generated from, we exposed the newly-generated hiPSC to both HAT and 6TG selection (Figures 1D–1G). As expected, male HPRT−/Y (LNS-1 and LNS-2) hiPSC were HAT sensitive and 6TG resistant, whereas wild type male HPRT+/Y (WT-M1) and female HPRT+/+ (WT-F1) lines were HAT resistant and 6TG sensitive (Figures 1D–1E). When we analyzed low-passage hiPSC lines generated from drug-selected HPRT+/− fibroblasts (CR-HAT-1 through 4 and CR-6TG-1 through 8), we found in all cases that they were uniformly composed of cells that retained the same drug resistance profile as the fibroblasts they were derived from (n=12/12 lines at p5–6) (Figures 1F–1G). In none of the cases did we observe “switching” of a cell line’s drug resistance status, which would have been indicative of a change in XCI status. Thus, our study corroborates recent studies (Pomp et al., 2011; Tchieu et al., 2010), suggesting that hiPSC inherit the inactive X of the donor cell they were produced from.
HPRT deficient hiPSC lines produce abnormal neuronal cultures
To begin investigating whether disease-specific hiPSC lines might be used to model LNS, we performed embryoid body (EB) mediated neuronal differentiation, and then stained cultures with antibodies specific to the neuronal antigens TUJ1, MAP2A, synapsin and tyrosine hydroxylase (Figures 2A–2D, Figure S3A). Strikingly, we found that HPRT−/Y hiPSC lines LNS-1 and LNS-2 (n=2 lines) produced significantly fewer neurons than control wild type hiPSC and human embryonic stem cell (hESC) lines. In addition, the neurons that were generated by these LNS lines displayed significantly shortened neurites relative to the neurons produced from control hiPSCs (Figures 2A–2D). This LNS “neuronal phenotype” was not significantly affected by co-culture with neural supportive glial cells (Figures S3B–S3C).
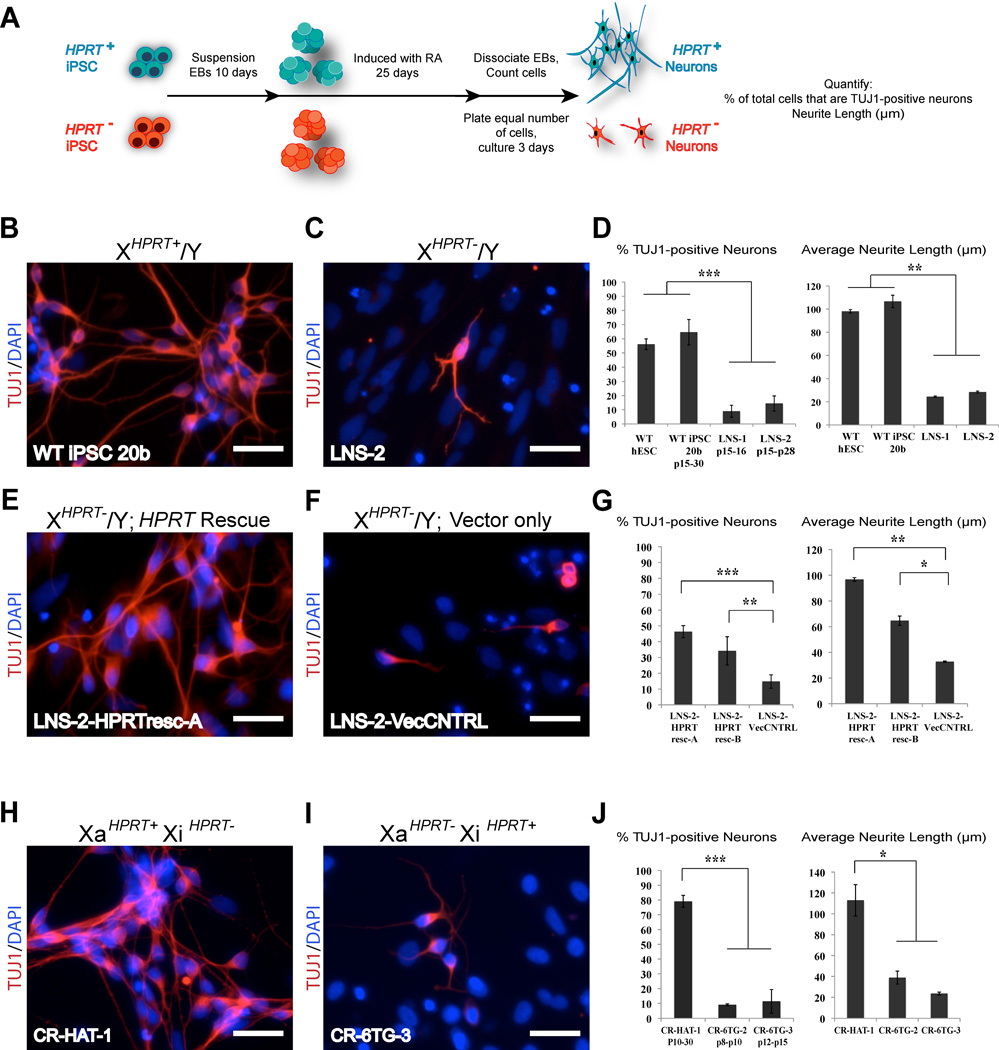
Figure 2. Neuronal differentiation of HPRT deficient hiPS cells can be used to model LNS.
(A) Neuronal differentiation of HPRT positive and negative hiPSC and our results (RA, retinoic acid).
(B) Neurons generated from the HPRT positive, wildtype male hiPSC line 20b, stained for TUJ1. Scale bars = 50µm.
(C) Neurons generated from the HPRT mutant male hiPSC line LNS-2 stained for TUJ1. Scale bars = 50µm.
(D) Quantification of TUJ1 positive neurons and average neurite length in neuronal cultures generated from HPRT+ lines hiPSC 20b and hESC HUES 51, along side diseased HPRT- lines LNS-1 and LNS-2. Error bars represent mean +/− SD.
(E) Neurons derived from the LNS-2-HPRTresc iPSC line that was transduced with lenti virus carrying the intact HPRT cDNA, stained for TUJ1. Scale bars = 50µm.
(F) Neurons derived from the LNS-2-VecCNTRL hiPSC line that was transduced with an empty lenti virus, stained for TUJ1. Scale bars = 50µm.
(G) Quantification of TUJ1 positive neurons and average neurite length in neuronal cultures generated from two independently derived LNS-2 HPRT rescue lines (LNS2-HPRTresc-A and B) and from the vector control hiPSC line (LNS-2-VecCNTRL). Error bars represent mean +/− SD.
(H) Neurons generated from female carrier, HPRT positive, phenotypically wildtype hiPSC CR-HAT-1 at passage 20, stained for TUJ1. Scale bars = 50µm.
(I) Neurons derived from of the isogenic female carrier, HPRT negative, “mutant” iPS CR-6TG-3 at passage 13, stained for TUJ1. Scale bars = 50um.
(J) Quantification of TUJ1 positive neurons and average neurite length in neuronal cultures derived from isogenic HPRT positive and negative female iPSCs. Error bars represent mean +/− SD. Statistical significance was analyzed using 2-tailed Student Test; *P<0.05, **P<0.01, ***P<0.001. See also Figures S3 and S4 for additional data.
Recently, we have shown that distinct hiPSC lines, even lines derived from the same individual, can display variable propensities for differentiation down the neuronal lineage (Boulting et al., 2011). We therefore reasoned that a definitive means of determining whether the differences we observed in neuronal cultures were the result of varying levels of HPRT activity, was to restore HPRT activity in mutant lines and see if this rescued neuronal number and neurite length to control levels. To this end, we transduced the HPRT deficient LNS-2 male hiPSC line with either a control lenti-viral vector or an identical vector harboring the HPRT cDNA. To determine whether transduction with the HPRT lenti-virus could restore HPRT activity to the cells, we exposed the HPRT transduced (LNS-2-HPRTresc) and control transduced LNS-2 cell line (LNS-2-VecCNTRL) to HAT selection (Figure S4A). Like the parental LNS-2 line, the line subjected to control transduction remained HAT sensitive, while the line transduced with the HPRT vector produced HAT resistant colonies. To determine whether rescue of HPRT activity was sufficient to rescue the neuronal phenotype, we differentiated two independently generated rescue sub-lines (LNS-2-HPRTresc-A, and LNS-2-HPRTresc-B) side by side with the vector only control line (LNS-2-VecCNTRL). As expected, LNS-2-VecCNTRL hiPSC line produced neuronal cultures with few neurons that possessed short neurites, while each of the two rescued LNS-2-HPRTresc hiPSC lines produced neuronal cultures that contained significantly more neurons with significantly longer neurites. In fact, the transgenic rescue restored these properties to those found using wild-type control hiPSCs (Figures 2E–2G, Figure S4B).
As another means of demonstrating the phenotypes we observed were due to loss of HPRT function, we tested the disease modeling utility of our isogenic female carrier lines. For these experiments, we assayed neuronal cultures derived from both HAT resistant “wildtype” (XaHPRT+XiHPRT−) lines CR-HAT-1, and 6TG resistant “mutant” (XaHPRT−XiHPRT+) lines CR-6TG-2 and CR-6TG-3. As one might predict by their pattern of XCI, the low passage XaHPRT+XiHPRT− CR-HAT-1 line generated normal neuronal cultures that were comparable to WT control lines. In contrast, the low passage XaHPRT−XiHPRT+ cell lines CR-6TG-2 and CR-6TG-3 behaved like HPRT−/Y mutant male lines, producing significantly fewer neurons, with significantly shorter neurites (Figures 2H–2J). As these cell lines were isogenic, other than expression of X-linked genes, these results further support the notion that differences in the neuronal cultures we observed were not the derivative of autosomal genetic variation amongst the iPSC lines.
Overall, our findings suggest that hiPSCs can provide a model for studying the neuronal basis of LNS that overcomes limitations of models that have been previously described (Cristini et al., 2010; Guibinga et al., 2009).
Dosage compensation gradually erodes in female hiPSC lines
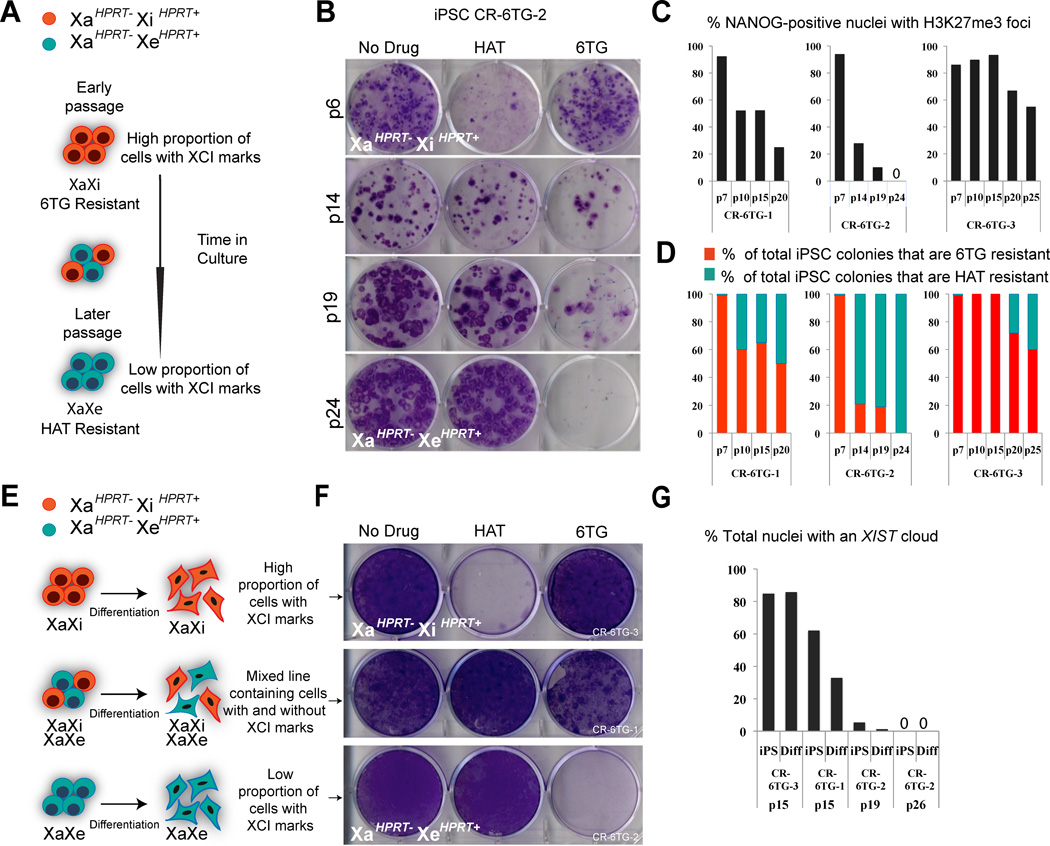
Although we observed what seemed to be normal dosage-compensation in low-passage hiPSC, as we expanded these cells lines, we found that an ever increasing portion of cells within each line gradually lost H3K27me3 foci and XIST clouds (Figures 3A, 3C, 3G). It has been suggested that loss of these marks of XCI in somatic cells and in human pluripotent stem cells does not directly result in loss of dosage compensation (Brown and Willard, 1994; Csankovszki et al., 2001; Silva et al., 2008; Tchieu et al., 2010). However, when we used the HAT and 6TG selections to assay for the functional state of HPRT activity over time in culture, we observed that originally homogeneous 6TG resistant cells lines (XaHPRT−XiHPRT+) such as CR-6TG-2, gradually became “mixed” cell lines composed of both HAT resistant and 6TG resistant sub-populations. These mixed cell-lines eventually converted into cell cultures that were completely HAT resistant. This gradual transition in HPRT activity was observed in 3/3 independent hiPSC lines and was strongly correlated with the loss of H3K27me3 foci and loss of XIST clouds. (Figures 3B–3D, 3G). Loss of these marks of XCI over time was not due to selective pressure to reactivate HPRT expression, as we observed that HAT resistant isogenic (XaHPRT+ XiHPRT−) lines such as CR-HAT-1 and CR-HAT-2 also lost foci of H3K27me3 over time in culture (Figure S5A).
Figure 3. Female hiPSC lines lose marks of XCI with time in culture and ectopically express HPRT from the Xi.
(A) Diagram depicting our observation that dosage compensation erodes on the X-chromosome over time in culture.
(B) Crystal violet staining of hiPSC line CR-6TG-2 at early, mid, and late passages, after treatment with either HAT or 6TG.
(C) Quantification of H3K27me3 foci in NANOG-positive cells for hiPSC lines show decrease in XCI marks over time in culture.
(D) Quantification of 6TG and HAT resistant colonies for hiPSC lines over time in culture.
(E) Diagram depicting our observation that hiPSC lines do not change their XCI state during differentiation.
(F) Crystal violet staining of differentiated cells generated from female iPSC lines that were originally HPRT negative (XaHPRT−/XiHPRT+), with different proportions of XIST.
(G) Quantification of XIST clouds in hiPSC lines and their differentiated derivatives. See Figure S5 for additional data.
We reasoned that there were two potential models that could explain our finding. Either as we cultured these hiPSC lines, they gradually reactivated their inactive X, becoming further reprogrammed and more like mouse ES cells, as has been suggested (Chin et al., 2009). Or instead, that as we passaged these lines, loss of XCI marks was occurring inappropriately, as has been observed in certain hESC lines (Dvash et al., 2010; Enver et al., 2005; Shen et al., 2008; Silva et al., 2008).
To begin distinguishing between these two models, we first examined whether low-passage (XIST+) and high-passage (XIST-) cell lines were equivalently reprogrammed. Important indicators of the extent of reprogramming include the examination of whether retroviral transgenes encoding pluripotency factors have been silenced, and whether transcription of their endogenous genetic counterparts has been activated. Quantitative RT-PCR analysis indicated that even in low-passage hiPSC lines, expression of retroviral OCT4, SOX2 and cMYC had been extinguished, while expression of the endogenous loci had been induced to levels similar to those found in both late passage hiPSC lines, established hiPSC lines, and hESC lines (Figure S5B). Although we did detect modest levels of viral KLF4 expression in all of our hiPSC cell lines, transcriptional levels were rather invariant between high and low-passages of the same cell line suggesting that they were independent of the extent of reprogramming. Thus, even our low-passage lines were sufficiently reprogrammed to have silenced the transgenes that encode for core reprogramming factors, and to have activated expression of endogenous loci involved in regulating pluripotency. These findings suggest that ongoing reprogramming, as measured by this assay, was not likely to be the cause of the shift in X-linked gene expression we observed.
It has been suggested that under certain culture conditions, hiPSCs can take on properties more similar to mouse ES cells (Hanna et al., 2010), and that this may change their state of XCI as well as eliminate their dependence on Tgf-β and FGF signaling for self-renewal (Hanna et al., 2010). To determine whether our hiPSC were taking on more “naive” properties, we asked whether their self-renewal was still dependent on Tgf-β and FGF signaling. To this end, we carried out colony forming assays with each line at both low and high passages, in the presence and absence of chemical inhibitors of Tgf-β and FGF signaling. We found that regardless of whether we inhibited Tgf-β or FGF signaling, colony formation was similarly compromised at both low and high passage, suggesting that our cell lines were not taking on more “naive” characteristics as they lost XCI marks (Figure S5C).
As an additional means of testing if our cells were taking on a more “naive” mouse-like character, we asked if, as in mouse pluripotent stem cells, XCI would take place upon differentiation of lines that lost marks of the inactive X. We carried out differentiation experiments both in vitro and in vivo within teratomas. Initially, we took individual passages of hiPSC lines that were originally uniformly XaHPRT−XiHPRT+, but had acquired declining levels of XCI marks, split each batch of cells in half, and then carefully quantified XCI status by XIST FISH in the first half of the cells. We then took the second half of the batch of each passage of cells, differentiated them within EBs, then again quantified their XCI status by XIST FISH as well as assessed their state of HPRT activity (Figure 3E–3G). We found that low passage hiPSC that were 6TG resistant and possessed an inactive X (XaHPRT−XiHPRT+), produced 6TG resistant, HAT sensitive differentiated cells, with an inactive X, indicating they continued to maintain the same inactive X throughout differentiation. However, when we differentiated later passages of the same lines that had become HAT resistant, lost XIST expression, and had de-repressed HPRT expression from the Xi, the resulting differentiated progeny were also devoid of XIST and remained HAT resistant and 6TG sensitive (Figure 3E–3G).
We next proceeded to determine whether in vivo differentiation in the context of a long-term teratoma was able to reset XCI in hiPSC lines that had lost XIST clouds. To this end, we injected a passage of the CR-6TG-2 cell line that had become devoid of clouds of XIST RNA, as well as passages of the CR-6TG-3 and CR-HAT-1 lines that still retained XIST expression, into nude mice. Following teratoma formation, we examined differentiated cells from each teratoma for marks of XCI. As predicted by our in vitro differentiation experiments, hiPSCs that contained XIST clouds produced teratomas containing cells that retained XIST clouds. However, hiPSC lines that had lost XIST clouds, produced teratomas containing cells that remained devoid of XIST clouds (Figure S5D). Thus, these results show that once foci of H3K27me3 and XIST clouds had been lost, and HPRT expression was de-repressed in a hiPSC line, it could not be restored by differentiation, either in vitro or in vivo.
In aggregate, our results suggest that instead of becoming more like mouse ES cells, our hiPSCs were undergoing a sort of “erosion” of dosage compensation. Although this process seemed indicative of an epigenetic rather than a genetic phenomenon, we did ask whether mutations in protein coding portions of the genome might have accumulated during either reprogramming or the passaging process that could have contributed to the effect. For this purpose, we took passages of the hiPSC lines 29A and 29e (Boulting et al., 2011) that had recently been subjected to whole exome sequencing (Gore et al., 2011) and assessed whether they still contained marks of XCI. We found that both lines were largely devoid of cells that still contained XIST clouds and H3K27me3 foci, and these lines did not harbor any common mutations that could be contributing to the erosion of XCI we observed (Figures S5E–S5F).
From these experiments, we can conclude that XCI in female hiPSC lines does not behave similarly to that in mouse ES or iPSCs. Instead, it seems that although low passage XaHPRT−XiHPRT+ hiPSCs differentiate into cells that are also XaHPRT−XiHPRT+, over time in culture, the cells gradually underwent a process of transcriptional de-repression on their inactive X. We have termed this process “erosion” of dosage compensation (Xe = Xeroded). As hiPSCs were cultured, they eventually became more and more homogeneously XaHPRT−XeHPRT+. These XaHPRT−XeHPRT+ cells did not regain dosage compensation upon differentiation and instead became differentiated cells that were also XaHPRT−XeHPRT+ (Figures 3A, 3E).
Erosion of dosage compensation in hiPSC impacts LNS disease modeling
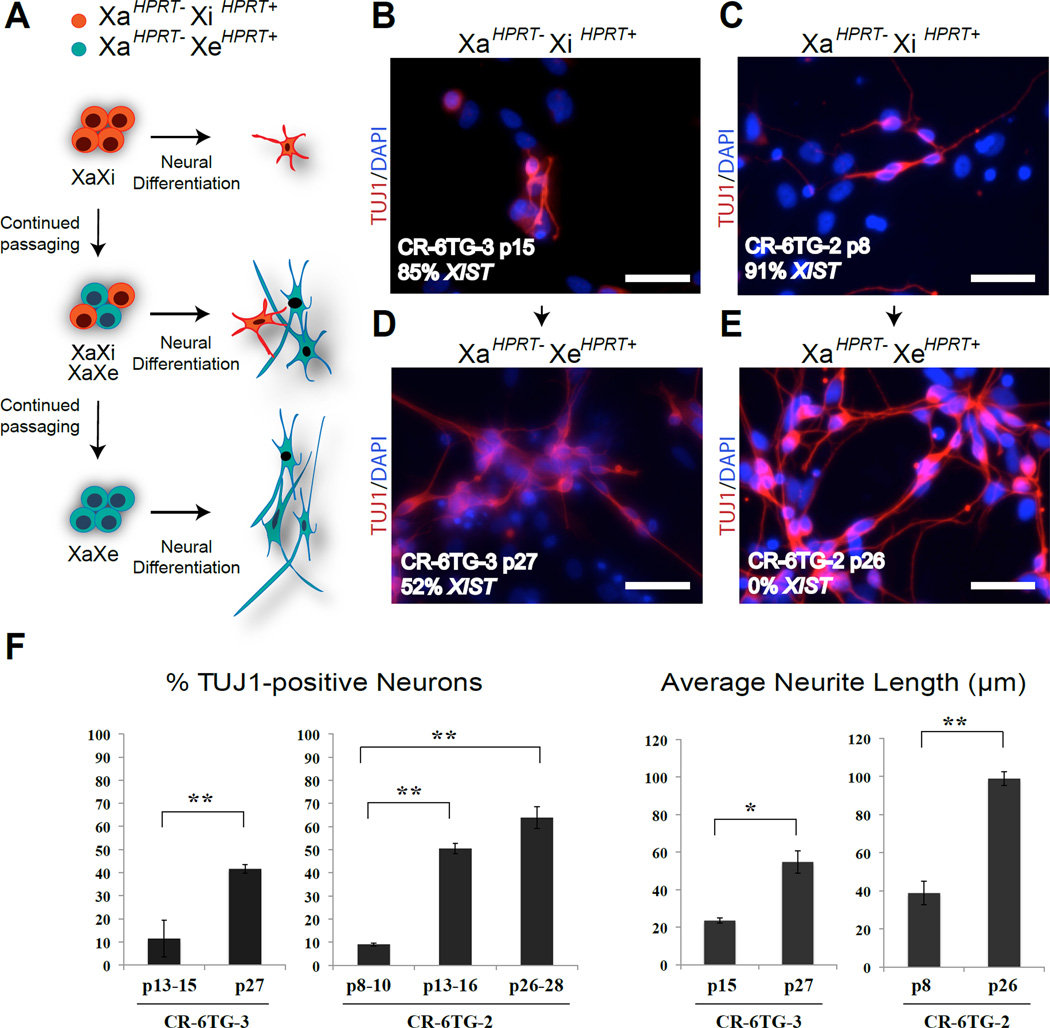
We next asked whether the erosion of dosage compensation we had observed could affect the phenotype of our LNS disease model. We performed neuronal differentiation using escalating passages of two “mutant” female carrier hiPSC lines, that had each initially been uniformly composed of cells with the inactive X carrying the normal HPRT allele (XaHPRT−XiHPRT+) (Figures 4A–4C). At low passage, both of these cell lines produced neuronal cultures with a phenotype similar to those exhibited by mutant male neurons (Figure 4B–4C, and 4F). However, when we differentiated later passage of the same lines that had begun to undergo erosion of dosage compensation, the resulting neuronal cultures contained a mixture of neurons that displayed the mutant phenotype and a mixture of neurons that appeared normal (Figures 4D and 4F and Figure S6). Examination of XCI by staining for H3K27me3 in these neuronal cultures demonstrated that neurons with a mutant phenotype tended to contain marks of an inactive X, while neurons that displayed a more wild type appearance, had lost their focus of H3K27me3, suggesting erosion of dosage compensation had occurred (Figure S6). When yet higher passage cultures of hiPSC that had lost all marks of XCI were differentiated, the resulting neuronal cultures were indistinguishable from those produced from wild type (HPRT+) hiPSC (Figure 2, Figures 4E–4F).
Figure 4. Erosion of dosage compensation affects LNS neuronal disease phenotypes.
(A) Diagram depicting our observation that erosion of dosage compensation rescues the neuronal phenotype caused by the X-linked HPRT mutation.
(B) Neurons derived from hiPSC line CR-6TG-3 at low passage stained for TUJ1. Scale bars=50µm
(C) Neurons derived from of the hiPSC line CR-6TG-2 at low passage stained for TUJ1. Scale bars = 50µm.
(D) Neurons derived from higher passage hiPSC CR-6TG-3 that have undergone erosion of XCI in a portion of their cells, stained for TUJ1. Scale bars = 50µm.
(E) Neurons derived from higher passage hiPSC CR-6TG-2 that have undergone erosion of dosage compensation in all their cells, stained for TUJ1. Scale bar=50µm.
(F) Quantification of TUJ1 positive neurons and neurite length in neuronal cultures derived from lower and higher passages of hiPSC lines CR-6TG-2 and CR-6TG-3. Error bars represent mean +/− SD. Statistical significance was analyzed using 2 tailed Student Test; *P < 0.05, **P<0.01, ***P<0.001. See also Figures S6 and S7 for additional data.
We reasoned that the shifting phenotype we observed could have been due to some independent or unexplained function of increased passage that was unrelated to dosage compensation. To rule out this possibility, we began with an intermediate passage hiPSC line that was a mixture of 6TG resistant (XaHPRT−XiHPRT+) and HAT resistant (XaHPRT−XeHPRT+) cells. We then used 6TG selection to purify the XaHPRT−XiHPRT+ sub-population and HAT selection to purify the XaHPRT−XeHPRT+ sub-population (Figure S7A–S7B). We confirmed that the 6TG resistant population was uniformly composed of cells with an inactive X, while the HAT resistant population was composed of cells that lacked H3K27me3 foci and XIST clouds (Figure S7C). When we performed neuronal differentiation of these two distinct sub-populations, the 6TG resistant XaHPRT−XiHPRT+ cells exhibited the mutant phenotype. In contrast, the neuronal cultures produced from the HAT resistant XaHPRT−XeHPRT+ sub-population were again identical to normal “wild type” controls (Figures S7D–S7F). Therefore, we conclude that the erosions of dosage compensation we observed at the HPRT locus was not reversed by neuronal differentiation and as a result, eliminated the LNS related neuronal phenotype originally observed upon differentiation of this cell line.
Erosion of dosage compensation is a major source of variation among human pluripotent stem cell lines
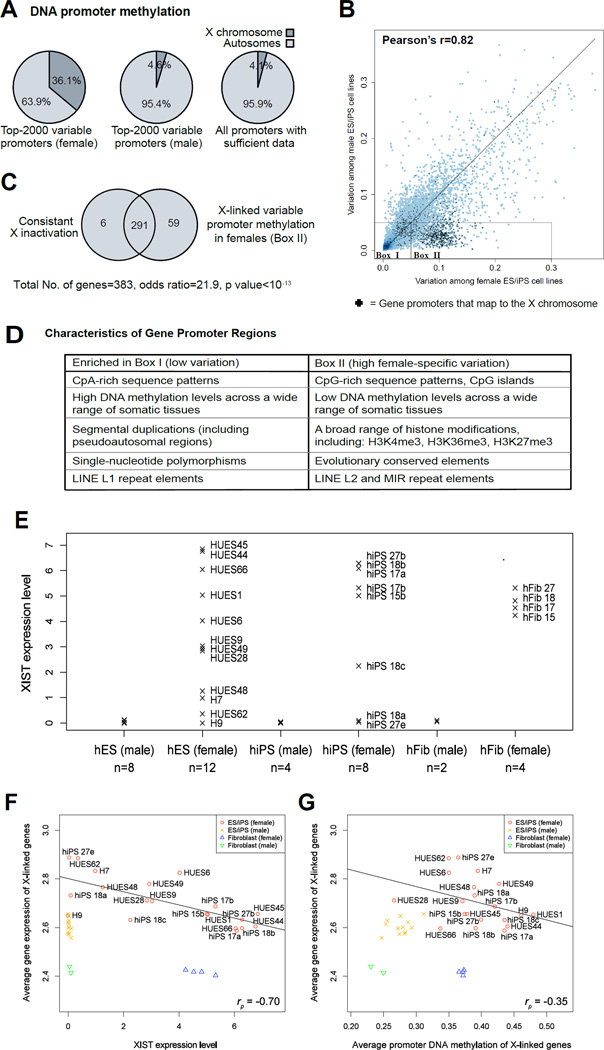
To determine whether the de-repression of gene silencing we observed for HPRT was reflective of broader erosion in dosage compensation, we turned to a large dataset of genome-wide DNA methylation and transcriptional profiles that we reported recently (Bock et al., 2011). This dataset catalogs genome-wide DNA methylation patterns and gene expression levels for 20 hESC lines and 12 hiPSC lines (Bock et al., 2011; Boulting et al., 2011). Although we had previously noted that nearly 50% of the loci with highest variation in DNA methylation were located on the X-chromosome (Bock et al., 2011), we had not explored the functional ramifications of this finding.
Interestingly, when we focused our analysis specifically on female pluripotent cell lines, 36.1% of gene promoters with the most variable methylation were found on the X-chromosome. In contrast, only 4.6% of the most variably methylated promoters were found on the X-chromosome in male cell lines (Figure 5A). This sex-specific difference was also clearly detectable when we plotted the promoter-associated levels of DNA methylation variation for male lines against female cell lines (Figure 5B).
Figure 5. Loss of X-inactivation is widespread among human ES and iPS cell lines.
(A) Percentage of the most DNA-methylation-variable gene promoters that are located on the X-chromosome, separately shown for female ES/iPS cell lines (left) and for male ES/iPS cell lines (center). For comparison, the background distribution of all gene promoter regions with sufficient DNA methylation data is shown (right).
(B) Correlation between DNA methylation variation among female ES/iPS cell lines and DNA methylation variation among male ES/iPS cell lines. Variation was measured as the standard deviation of promoter methylation levels and plotted for all genes with sufficient DNA methylation data. Dark blue regions indicate high point densities, and black crosses denote genes that map to the X-chromosome. Boxes I and II highlight genes with low vs. high variation among female ES/iPS cell lines.
(C) Overlap between genes that are subject to consistent X-inactivation (left) and genes that exhibit high levels of promoter-associated DNA methylation variation in female ES/iPS cell lines (right). Significance was established using Fisher’s exact test. X-inactivation data was obtained from a comprehensive mapping of the human X-chromosome (Carrel and Willard, 2005).
(D) Promoter characteristics of X-linked genes with low vs. high levels of DNA methylation variation among female ES/iPS cell lines, as indicated by boxes I and II in panel E. The table summarizes the results of an EpiGRAPH analysis (Bock et al., 2011) testing ~1,000 genomic attributes for significant enrichment or depletion between the two sets of gene promoter regions.
(E) XIST expression levels in male and female hES, hiPS, and human fibroblast (hFib) cell lines, based on microarray data (Bock et al., 2011) and relative to a scale from 0 (no expression) to 10 (maximal expression) across all genes.
(F) Correlation between cell-line specific XIST expression levels and mean gene expression levels across all X-linked genes, based on microarray data (Bock et al., 2011). A linear regression line was calculated across all female ES/iPS cell lines (red), and the corresponding Pearson correlation coefficient is shown at the bottom right of the diagram.
(G) Correlation between mean DNA methylation levels across all X-linked gene promoters and mean gene expression levels averaged across all X-linked genes. DNA methylation and gene expression data for 20 ES cell lines, 12 iPS cell lines and 6 fibroblast cell lines were obtained from a recent study reporting a reference map of human pluripotent cell lines (Bock et al., 2011). See also Table S1 for additional data.
We also found that the vast majority of gene promoters that exhibited substantially higher levels of DNA methylation variation in female compared to male cell lines (Box II) were not only located on the X-chromosome (Figure 5B), but were subject to X-inactivation (Figure 5C). Furthermore, these promoters exhibited characteristics consistent with genomic regions known to be effectively repressed by DNA methylation (Figure 5D). In contrast, those X-linked gene promoters that exhibited similar levels of variation in male and female cell lines (Box I in Figure 5B) were more likely to evade X-inactivation and were characterized by distinct gene promoter properties (Figure 5D). A full-list of genes with promoters that are subject to erosion of dosage compensation (Box II) and those that seem more recalcitrant to the effect (Box I) have been listed in supplemental table 1 (Table S1). Overall, 91% of the genes known to be subject to X inactivation with sufficient information for us to examine, displayed this variable promoter methylation (Figure 5C).
Consistent with our findings in LNS hiPSC, we observed that female pluripotent cell lines varied in their XIST expression levels (Figure 5E). The cell-line specific differences in X-inactivation status appeared to have widespread functional consequences: Female lines with low XIST expression were characterized by a chromosome-wide increase in X-linked transcription relative to male lines (Pearson's r=-0.70) (Figure 5F). In addition, we observed a significant negative correlation between the average promoter DNA methylation levels and the average gene expression levels for all X-linked genes in female hESC and hiPSC lines (Pearson's r=-0.35) (Figure 5G). This latter finding suggests that erosion of dosage compensation involves transcriptional de-repression and loss of the associated promoter DNA-methylation.
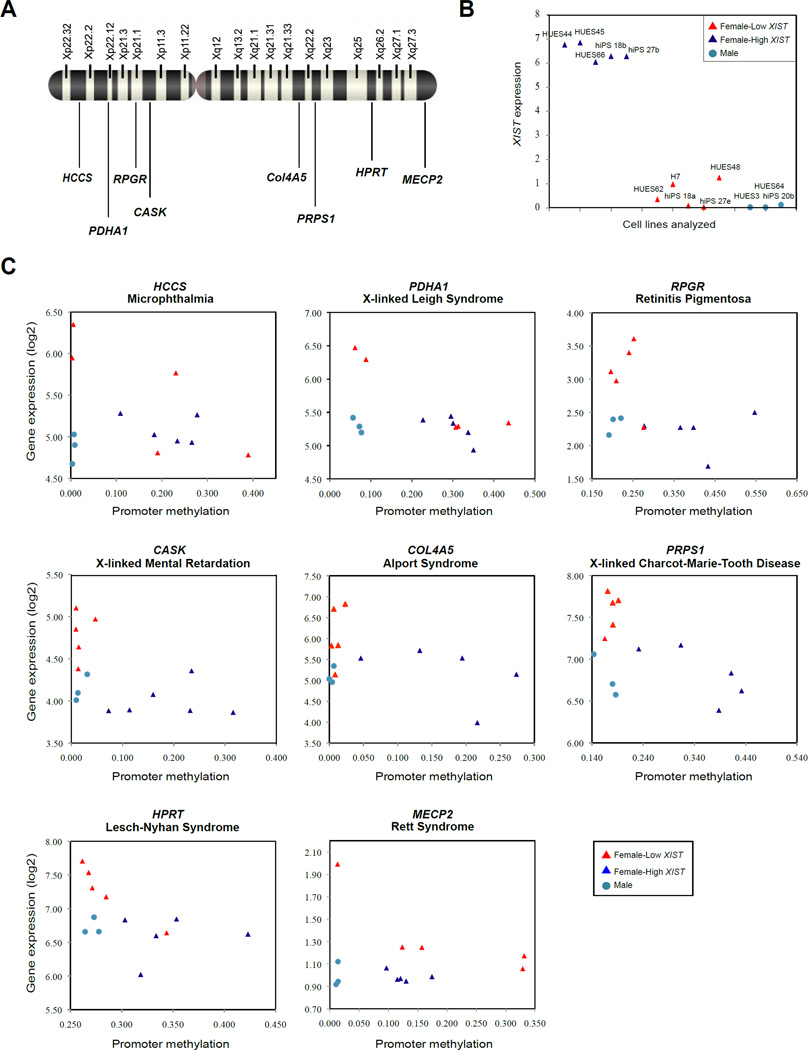
We next decided to take advantage of our genomic analysis to determine whether expression levels of other X-linked loci commonly mutated in disease were similarly affected by this phenomenon. If so, it would suggest that the erosion of dosage compensation reported here and its negative impact on modeling LNS would extrapolate to those conditions as well. To this end, we compared five human pluripotent stem cell lines with high XIST levels to five stem cell lines with little or no XIST expression. We plotted the levels of transcription of 8 such genes found in distinct regions of the X-chromosome (HCCS, PDHA1, RPGR, CAST, COL4A5, PRPS1, HPRT, MECP2) in a given cell-line against that cell line’s average relative gene promoter methylation (Figures 6A–6C). It was immediately apparent that many female cell lines that had lost XIST expression, had demethylated their former Xi and had de-repressed expression of these disease-specific genes (Figure 6C). This suggests that the use of hiPSC for disease modeling would also be impacted for at least these additional genes.
Figure 6. Erosion of dosage compensation affects many disease-linked genes across the X-chromosome.
(A) Diagram of the human X-chromosome and location of eight X-linked disease specific genes examined in this section.
(B) Five female hES and hiPS cell lines with high XIST expression and five female cell lines with no XIST expression (Bock et al., 2011) chosen for comparison of erosion of X-linked disease genes in (A).
(C) Correlation between mean promoter DNA methylation levels and gene expression for the genes in (A) and hESC/hiPSC in (B).
In summary, our bioinformatic analyses show that the majority of female pluripotent cells exhibit signs of erosion in dosage compensation. Furthermore, loss of XIST expression in the female pluripotent cells did not simply modify the expression of only a small number of genes, but instead appeared to lead to a broad de-repression of X-linked genes. This increase in X-linked expression levels is also correlated with widespread loss of repressive DNA methylation from the (formerly inactive) X-chromosome. Thus, our data demonstrate that erosion of dosage compensation in hiPSC is not only relevant to the study of LNS, but has a broad range of implication for the study of many X-linked diseases.
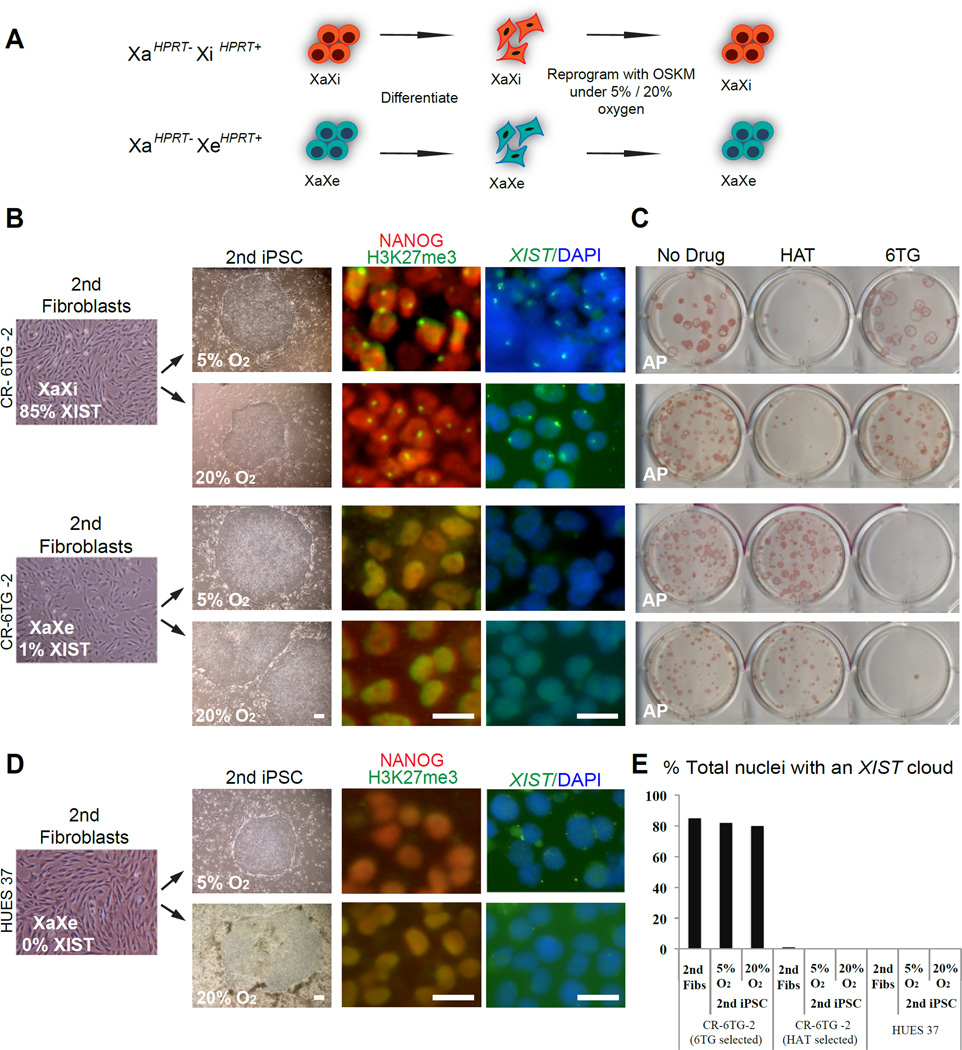
Once dosage compensation has eroded, further reprogramming cannot restore it
We have thus far demonstrated that during prolonged culture female pluripotent cells undergo gradual erosion in dosage compensation that cannot be restored by differentiation. We next considered whether further reprogramming might restore normal dosage compensation. We took an XaHPRT−XiHPRT+ hiPSC line that was 6TG resistant, as well as HAT resistant derivatives of this line that had undergone XCI erosion (XaHPRT−XeHPRT+), differentiated both lines into embryonic fibroblasts, transduced them with reprogramming retroviruses and selected secondary hiPSC lines. We then analyzed the very first passages of the resulting secondary hiPSC lines for both HPRT gene activity and for the presence of marks of the inactive X, H3K27me3 foci and XIST clouds (Figure 7A–7B). Regardless of whether it was at low (physiological) oxygen or high (atmospheric) oxygen conditions, when we performed a second round of reprogramming on cells that had an inactive X, the secondary hiPSCs again maintained the inactive X that had been silent in both their primary fibroblast and hiPSCs predecessors. In contrast, when we performed an additional round of reprogramming on cells that had undergone erosion of dosage compensation, the secondary derivatives of these cells also displayed eroded dosage compensation, did not restore marks of XCI, and continued to ectopically express HPRT from what was originally the Xi (Figures 7A–7C, 7E)
Figure 7. Additional round of reprogramming does not restore XCI.
(A) Diagram depicting experimental approach and results obtained from reprogramming of secondary fibroblast (2nd fibroblasts) with normal or aberrant XCI marks under both physiological (5%) and ambient (20%) oxygen conditions.
(B) XIST high and XIST low sub-lines of the secondary iPSC line CR-6TG-2 differentiated into secondary fibroblasts, and staining for H3K27me3/NANOG and XIST FISH in newly reprogrammed secondary iPSC cells. Scale bars=20 µm.
(C) Alkaline Phosphatase (AP) staining of 5% and 20% oxygen hiPSC colonies following treatment with HAT or 6TG.
(D) Human ES line HUES 37 differentiated into secondary fibroblasts and staining for H3K27me3/NANOG and XIST FISH in iPSC derived from HUES 37 secondary fibroblasts. Scale bars=20 µm.
(E) Quantification of XIST cloud in secondary fibroblasts (2nd Fibs) and secondary iPSC (2nd iPSC) under 5% or 20% Oxygen.
Furthermore, we found that the failure of reprogramming to restore the inactive X-chromosome was not unique to derivatives of hiPSC but also held true for reprogramming of fibroblasts derived from hESCs. When we took fibroblasts derived from high passage HUES 37 cells that had eroded XCI, reprogramming could neither restore XIST clouds or foci of H3K27me3 (Figures 7D–7E).
Discussion
Here, we have demonstrated that HPRT null male hiPSC lines can be used to produce a new robust model system for the study of Lesch-Nyhan Syndrome. The development of this cellular based human LNS model is in and of itself significant, as previous efforts to model this condition in the mouse have been unsuccessful (Engle et al., 1996; Finger et al., 1988). Our experiments clearly demonstrate that the loss of HPRT activity directly results in a significant loss of neurons and decrease in neurite length. Moreover, we show that these neuronal defects are cell autonomous and can be rescued by exogenous expression of the functional HPRT. It remains unclear why the malfunction in HPRT affects most severely the neuronal tissue, and weather the purine salvage pathway plays a significant role in these cells or if HPRT-deficient neurons are merely more sensitive to the accumulation in uric acid or other environmental stressors. Our system provides a platform where these questions can now be further explored.
We have further shown that XaHPRT− XiHPRT+ female hiPSC lines with proper dosage compensation also produce the LNS neuronal phenotype seen using male lines. However, over time in culture, we observed a gradual decrease in the frequency of female hiPSCs that contained XIST and H3K27me3 foci, which was highly correlated with ectopic HPRT expression from the inactive X. Although others have noted variation between cell lines in the proportion of human pluripotent cells with these XIST and H3K27me3 marks (Lengner et al., 2010; Shen et al., 2008; Silva et al., 2008; Tchieu et al., 2010), the functional ramification of losing XIST expression has not been previously explored. The lack of motivation for investigating this phenomenon might be explained by studies in both the mouse and in human somatic cells, which demonstrated that even when XIST expression was eliminated from the inactive X, repressive DNA methylation and transcriptional repression were retained (Brown and Willard, 1994; Csankovszki et al., 2001). However, we found that in human pluripotent stem cell lines this does not seem to be the case. Instead, our findings demonstrated that loss of XIST and the heterochromatin modifications of the inactive X in female stem cells were associated with chromosome-wide erosion in dosage compensation.
Our studies have shown that erosion in dosage compensation is not a benign effect and instead has substantial ramifications for the utility of human pluripotent stem cell lines. These results clearly demonstrate that this loss of dosage compensation can occlude the LNS phenotypes and may interfere with the use of hiPSC for the study of other X-linked diseases. Furthermore, our bioinformatic analyses suggest that this phenomenon likely affects most of the genes subject to XCI (91%) we examined, and that this seems to be a universal effect impacting many pluripotent stem cell lines.
Extrapolating from our experience with modeling the X-linked disease Lesch- Nyhan Syndrome and based on our genomic analysis, we predict that a similar phenomenon would occur when using female pluripotent stem cells to model any X-linked disease caused by mutations in genes subject to XCI. These findings provide a potential explanation for the variations in the Rett Syndrome phenotypes, the XCI state, and MECP2 expression observed by various groups when they used XMECP2− /XMECP2+ hiPSC for disease modeling (Cheung et al., 2010; Kim et al., 2011; Marchetto et al., 2010).
Because erosion of dosage compensation was such a widespread phenomenon amongst female pluripotent cell lines, and because it seemed to have such substantial functional ramifications, we have begun to contemplate steps that might be taken to restore proper XCI. As differentiation of cell lines could not restore XCI, we considered the possibility that an additional round of reprogramming might. Unfortunately, this was not the case, as further reprogramming did not seem to affect the proportion of cells that did or did not contain an XIST cloud, or a focus of repressive H3K27me3. Lines with a clearly inactive X retained it, while signs of XCI and gene silencing were not restored to lines that displayed eroded dosage compensation. Thus, once XCI is lost, it cannot be reestablished using standard procedures such as in vitro differentiation or a second round of iPS reprogramming.
The findings reported here raise the intriguing question of why erosion of dosage compensation is occurring in female pluripotent cells, while it does not occur for instance in somatic fibroblasts. It could be that under the rapid growth conditions, which human pluripotent stem cells are adapted to, the expression of XIST or perhaps some other mechanistic regulator of XCI, is dynamically unstable. It is also possible that there are genes expressed from the X-chromosome that provide pro-survival or pro-growth signals for pluripotent stem cells that might select for cells that have lost XCI. The observation that certain regions of the X-chromosome are among those most commonly duplicated in stem cell lines with abnormal karyotypes is consistent with the notion that increased X-chromosome gene dosage might have a selective advantage (Baker et al., 2007). If this is true, there may be a selective pressure for cells to undergo erosion of XCI.
The results reported here are consistent with Nazor and colleagues studies of the genome-wide profiling of transcription and DNA methylation levels in 205 pluripotent cell lines and 130 somatic controls (Nazor et al., 2012). Similar to our findings, they observed that the X-chromosome harbors the vast majority of epigenetic variation between female pluripotent cell lines and that this variation results in the de-repression of specific X-linked disease genes in as many as 60 percent of pluripotent cultures.
Together our studies underscore the importance of monitoring epigenetic variation during the use of pluripotent stem cells in disease modeling efforts. Although the erosion of dosage-compensation we report here would most obviously impact X-linked disease models, it is clear that the levels of hundreds of genes that could have modifying effects on other disease-phenotypes are also fluctuating significantly. In the short-term, it will therefore be critical to be aware of the state of XCI in any given female cell line during its use in disease modeling efforts. In the longer-term, it would be desirable to identify culture conditions that can protect female cell lines from this erosion of XCI.
Experimental Procedures
Human iPSC derivation and culture
All human ESC work was previously approved by Harvard's Embryonic Stem Cell Research Oversight Committee (ESCRO). All animal research was performed under the oversight of the Institutional Animal Care and Use Committee (IACUC) at Harvard University. Human fibroblasts were commercially obtained from Coriell Cell Repository and ATCC and grown under standard conditions. To produce hiPSC colonies, human fibroblasts were transduced with viruses containing OCT4, SOX2,KLF4, and cMYC as previously described (Boulting et al., 2011; Dimos et al., 2008). hiPSC colonies were manually picked based on morphology within 3–4 weeks. All hiPSC lines were continuously cultured under standard conditions on irradiated mouse embryonic fibroblasts, and manually passaged every 6–8 days throughout the work in this study. Differentiation of human pluripotent stem cells was carried out by EB suspension in media containing serum. After 8–10 days, cells were plated onto gelatin-coated plates and grown for another 8 –10 days before analysis. To test for dependency on FGF and TGFβ, hiPSC were cultured in media containing the inhibitor SB431542 (Sigma) or media without FGF containing the inhibitor PD173074 (Sigma). For the generation of rescue lines, The HPRT negative hiPSC, LNS-2, was plated on matrigel (FISHER) in mTESR media (stem cell technologies) and transduced with virus containing the intact HPRT gene or the empty viral vector. Cells with HPRT activity were selected with mTESR media containing HAT, then used for neural differentiation. All cultures were maintained at 37°C, 5% CO2, 20% O2, unless noted otherwise.
6TG and HAT selections
To produce pure population of HPRT - or HPRT + fibroblasts from the HPRT +/− carrier cells (Corierll: GM00026), cells were incubated with media containing either 6TG (30–60 µM 2-amino-6-mercaptopurine; Sigma) or HAT (1×10–4M hypoxanthine, 4×10–7M aminopterin, 1.6×10–5M thymidine; Invitrogen) respectively, for a period of six days. Each selected sub-line was then expanded and then transduced with viruses for the production of hiPSC. To assay for HPRT activity in established hiPSC lines, equal number of manually dissected colony clumps were placed in three wells of a six well plate. 2–3 days after passaging, colonies were marked and scored to ensure equal survival of colonies used for drug selection. Selection was carried out in standard hESC media containing either 6TG or HAT for a period of five to six days, after which cells were fixed and stained with Crystal Violet. Colonies were manually scored for each drug selection, and proportion of colonies was compared to that of cells not treated with either drug.
Differentiation of human pluripotent cells into neurons
hESC/iPSC were grown to 80% confluence and then manually gridded and dissociated. Equal number of clumps were resuspended in hESC media without bFGF or plasmanate, in low attachment 6-well plates (Corning) in order to form embryoid bodies (EB). After ten days, EBs were induced toward neuronal identity for another 25 days using retinoic acid (1 uM, Sigma) in N2B27 media containing DMEM:F-12, 1% N2 Supplement (Invitrogen), 1% B27 supplement (Invitrogen), 0.2 mM ascorbic acid (Sigma-Aldrich), 0.16% D-Glucose (Sigma-Aldrich), 1% nonesesential amino acids (Invitrogen), 2 mM glutamax-I (Invitrogen), 50 units/ml penicillin and 50 g/ml streptomycin (Invitrogen). After a total of 35 days of differentiation, the EBs were dissociated with 20 units of papain and 1000 units of Deoxyribonuclease I (Worthington Biochemical Corporation). The resulting single cells were then washed with D-MEM/F12 and then resuspended in N2B27 media supplemented with neurotrophic factors (GDNF, BDNF, CNTF; 10 ng/ml, R&D Systems). The cells were counted and equal number of cells were then plated on Poly-D-Lysine/Laminin 8 well CultureSlides (BD Biosciences) coated with matrigel (FISHER). All comparative differentiation experiments were carried out with all experimental lines and controls in parallel, and all stained neuronal cultures were imaged and scored blindly. TUJ1 positive neurons and neurite lengths were analyzed using ImageJ software.
Immunocytochemistry
Pluripotent, differentiated, or neuronal cultures were fixed in 4% paraformaldehyde. Fixed cultures were permeabilized with either 0.5% Tween or 0.2% Triton-X in PBS, blocked in 10% serum, and then incubated in blocking solution containing primary antibodies. Cells were incubated in secondary antibodies, then washed and mounted in Vectashield with Dapi (Vector Laboratories) and imaged. Alkaline phosphatase activity was detected in live cultures using the alkaline phosphatase substrate kit (Vector, SK-5100).
RNA FISH and qPCR
RNA FISH was carried out as previously described (Erwin and Lee, 2010). Briefly, cells were were trypsinized, spun unto slides (cytospin), washed in PBS, placed in cytoskeletal buffer containing 0.5% Triton X, and then fixed in 4% paraformaldehyde. DNA probes were generated to the first 5kb of human XIST exon 1, labeled with fluorescein-12-dUTP using the Prime-It Fluor Labeling kit (Stratagene). Hybridizations were carried out in buffer containing human Cot1 DNA (Invitrogen) at 37C overnight. Following washes, slides were mounted with Vectashield (Vector Labs) containing DAPI and imaged. For immunostaining, slides were blocked in 10% FBS and then stained as described above. 500–1000 nuclei were scored for each line for XIST and H3K27me3 assays. For RNA analysis, total RNA was isolated from cultures using Trizol (Invitrogen) and synthesized into cDNA with iScript (Bio-Rad). qRT-PCR was then performed using SYBR green (Bio-Rad) and the iCycler system (Bio-Rad). Quantitative levels for all genes were normalized to endogenous GAPDH expression. Primer sequences can be provided upon request.
Teratoma Assay
hiPSC were trypsinized and injected into the kidney of SCID mice (SHO). Tissue masses were formed within 8–10 weeks. Masses were extracted, fixed, sectioned and stained. Cells representing all three germ layers were identified under the microscope.
Supplementary Material
Acknowledgments
The authors wish to thank all K.E. lab members for their technical support and helpful discussions, as well as J.F. Loring and M.H. Stewart for critical reading of this manuscript. This work was supported by HHMI, the Stowers Medical Institute, NYSTEM-NYSCF, NICHD grant #HD045732-03, and NIGMS P01 grant GM099117 to K.E. This work was also supported by NIH grants U01ES017155 and P01GM099117 to A.M. C.B. was supported by a Feodor Lynen Fellowship (Alexander von Humboldt Foundation) and a Charles A. King Trust Postdoctoral Fellowship (Charles A. King Trust, N.A., Bank of America, Co-Trustee). A.S.B. is supported by the Boehringer Ingelheim Fonds, E.K. is a NYSCF Postdoctoral Fellow, A.M. is supported by the Pew Charitable Trusts, and K.E. is an HHMI early career scientist and fellow of the MacArthur Foundation.
References
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Baumeister AA, Frye GD. The biochemical basis of the behavioral disorder in the Lesch-Nyhan syndrome. Neurosci Biobehav Rev. 1985;9:169–178. doi: 10.1016/0149-7634(85)90043-0. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of Human ES and iPS Cell Variation Enable High-Throughput Characterization of Pluripotent Cell Lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011 doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2010;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur J Hum Genet. 2008;16:422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- Cristini S, Navone S, Canzi L, Acerbi F, Ciusani E, Hladnik U, De Gemis P, Alessandri G, Colombo A, Parati E, et al. Human Neural Stem Cells: a model system for the study of Lesch-Nyhan Disease neurological aspects. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq072. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS ONE. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle SJ, Womer DE, Davies PM, Boivin G, Sahota A, Simmonds HA, Stambrook PJ, Tischfield JA. HPRT-APRT-deficient mice are not a model for lesch-nyhan syndrome. Hum Mol Genet. 1996;5:1607–1610. doi: 10.1093/hmg/5.10.1607. [DOI] [PubMed] [Google Scholar]
- Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Dawud RA, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- Erwin JA, Lee JT. Characterization of X-chromosome inactivation status in human pluripotent stem cells. Current protocols in stem cell biology Chapter 1 . Unit 1B.6. 2010 doi: 10.1002/9780470151808.sc01b06s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger S, Heavens RP, Sirinathsinghji DJ, Kuehn MR, Dunnett SB. Behavioral and neurochemical evaluation of a transgenic mouse model of Lesch-Nyhan syndrome. J Neurol Sci. 1988;86:203–213. doi: 10.1016/0022-510x(88)90099-8. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibinga GH, Hsu S, Friedmann T. Deficiency of the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) dysregulates neurogenesis. Mol Ther. 2009;18:54–62. doi: 10.1038/mt.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng A, Saha K, Kim J, Lengner C, Soldner F, Cassady J, Muffat J, Carey B, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnah HA. Lesch-Nyhan disease: from mechanism to model and back again. Dis Model Mech. 2009;2:116–121. doi: 10.1242/dmm.002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of Pre-X Inactivation Human Embryonic Stem Cells under Physiological Oxygen Concentrations. Cell. 2010 doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JA, Kelley WN. Lesch-Nyhan syndrome: altered kinetic properties of mutant enzyme. Science. 1971;171:689–691. doi: 10.1126/science.171.3972.689. [DOI] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Müller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10(this issue):620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X Chromosome Skewing during Culture and Reprogramming of Human Somatic Cells Can Be Alleviated by Exogenous Telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JE, Bar PR, Jinnah HA. Lesch-Nyhan disease and the basal ganglia. Brain Res Brain Res Rev. 2000;32:449–475. doi: 10.1016/s0165-0173(99)00094-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.